Abstract
Viral infection may induce the cell-surface expression of PVR (CD155) that, upon recognition by its cognate activating DNAM-1 receptor present on cytotoxic lymphocytes, may promote antiviral immune responses. Here we show that expression of the human immunodeficiency virus type 1 (HIV-1) Vpr protein in Jurkat T cells increases cell-surface and total PVR levels. Analysis of mutated Vpr variants indicated that Vpr uses the same protein surfaces, and hence probably the same mechanisms, to upregulate PVR and arrest the cell cycle in the G2 phase. Moreover, we found that PVR upregulation by Vpr relied on the ability of the protein to activate the ATR kinase that triggers the DNA damage response pathway and G2 arrest. Finally, we showed that Vpr contributes to PVR up-modulation in HIV-infected CD4+ T lymphocytes and inhibits the PVR downregulating activity of the viral Nef protein.
Human cells may respond to viral infection, tumour transformation or other stress by expressing cell-surface ligands for activating receptors of cytotoxic NK and T cells, thus eliciting recognition and elimination by the immune system. Work from our own and other laboratories has shown that upon infection with the human immunodeficiency virus type 1 (HIV-1), CD4+ T cells express ligands for the NKG2D activating receptor, particularly ULBP2, becoming susceptible to lysis mediated by NK cells that are all NKG2D+ (Cerboni et al., 2007b; Ward et al., 2009). Furthermore, we recently reported that primary CD4+ T lymphocytes respond to HIV-1 infection by upregulating cell-surface PVR (poliovirus receptor, CD155 or Necl-5) (Matusali et al., 2012), a nectin-like protein that functions as ligand for the DNAM-1 (DNAX accessory molecule-1 or CD226) activating receptor of NK and CD8+ T cells (Takai et al., 2008). However, some HIV-1 encoded proteins, including Nef, Vpu and Vif, have evolved the capacity to inhibit the cell-surface expression of NKG2D ligands and/or PVR, hence reducing NKG2D- and DNAM-1-mediated NK-cell lysis of infected cells (Cerboni et al., 2007b; Matusali et al., 2012; Norman et al., 2011). Therefore, understanding the mechanisms that regulate the expression of activating ligands in HIV-infected cells may provide novel strategies to strengthen antiviral immune responses.
Increasing lines of evidence indicate that the expression of NKG2D and DNAM-1 ligands is induced by the DNA damage response (DDR) pathway, specifically by the activation of the DNA damage sensor kinases ATM (ataxia telangiectasia-mutated) and ATR (ATM and Rad3-related) (Ardolino et al., 2011; Cerboni et al., 2007a; Gasser et al., 2005; Soriani et al., 2009). During infection with HIV-1, the viral Vpr protein induces a G2 cell-cycle arrest through a multistep process that involves Vpr association to the cellular DDB1–CUL4A E3 ubiquitin ligase complex via DCAF-1 (initially named VprBP) and induction of the DDR pathway via ATR activation (Andersen et al., 2008). It was recently shown that, by inducing the DDR pathway via ATR, Vpr also upregulates several NKG2D ligands (MICA/B and ULBP1-3 proteins) (Richard et al., 2010; Ward et al., 2009). On the basis of these findings, we sought to investigate whether Vpr plays a role in PVR up-modulation.
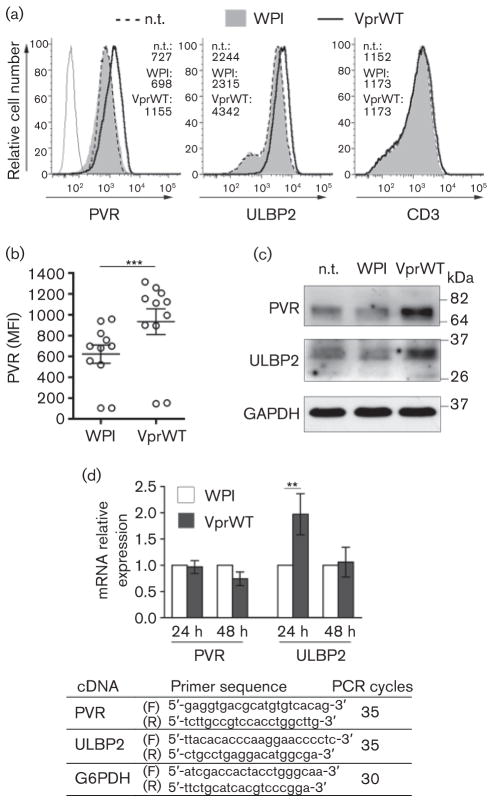
To this end, the Jurkat T-cell line was transduced as described previously (Richard et al., 2010) with the WPI lentivirus either empty or encoding the Vpr protein of the HIV-1 NL4-3 strain (VprWT). The self-inactivating HIV-1-derived WPI lentiviral vector allows co-expression from a human EF1α promoter of a cDNA and the GFP marker from a downstream internal ribosome entry site (IRES). At 48 h after transduction, when the vast majority of cells were vital (at least 80 %) and gated live cells were all GFP-positive (data not shown), we analysed the cell-surface PVR expression by flow cytometry as described elsewhere (Matusali et al., 2012). Results showed that the geometric mean fluorescence intensity (MFI) for PVR was increased similarly to ULBP2 on Vpr-expressing cells when compared to control cells (WPI and not transduced (n.t.) cells), with a mean 50 % up-modulation in 11 independent experiments (PVR MFI mean±SEM: 621±87 vs 933±124; P=0.001 by Wilcoxon signed rank test), while another membrane protein, CD3, was not affected (Fig. 1a, b). As for ULBP2, the effect of Vpr on cell-surface PVR correlated with an overall protein increase (by 45±25 % in three independent experiments) observed by immunoblotting total cell extracts with antibodies against PVR, ULBP2 or, as internal control, against the constitutively expressed glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Fig. 1c). We also measured the amounts of mRNA encoding PVR and ULBP2 [normalized by glucose 6-phosphate dehydrogenase (G6PDH) mRNA] by standard semiquantitative reverse transcriptase (RT)-PCR. Briefly, aliquots (2 μg) of total RNA were used to generate cDNA using random hexamers and reagents from BIOLINE, and increasing amounts of cDNA were amplified by PCR using reagents from GE Healthcare. For each target, forward and reverse primers were designed across exons to avoid amplification of genomic DNA, as validated in pilot assays, and the number of cycles was set to exponential phase of the PCR to allow semiquantitative comparisons among samples (Fig. 1d). The ULBP2 mRNA but not PVR mRNA was significantly more abundant in Vpr-expressing cells compared to control cells, indicating that Vpr does not upregulate PVR at the transcriptional level as opposed to what has been reported for ULBP2 and other NKG2D ligands (Richard et al., 2010; Ward et al., 2009) (Fig. 1d). Thus, it is likely that Vpr induces PVR expression acting at a post-transcriptional level such as, for instance, miRNA-mediated regulation of mRNA translation, an additional mechanism that was shown to control NKG2D ligand expression (Eissmann et al., 2010; Stern-Ginossar et al., 2008).
Fig. 1.
HIV-1 Vpr upregulates PVR acting at a post-transcriptional level. (a–c) Jurkat T cells were transduced with 0.3 m.o.i. of WPI or VprWT lentivirus or not transduced (n.t.) and analysed 48 h later. (a) Histograms show the fluorescence distribution of cells labelled with mAbs against PVR (SKII.4) or ULBP2 (MAB165903; R&D System) and Alexa647-conjugated goat anti-mouse IgG (GAM) (Invitrogen) or with anti-CD3 APC-conjugated mAb (EuroBioSciences) analysed by flow cytometry with a FACSCanto (BD Bioscience). The mean fluorescence intensity (MFI) values are indicated. Dashed line, filled grey histogram, and solid line represent n.t. cells, WPI- and VprWT-transduced cells, respectively. Upon staining with isotype control mAb the three cell samples were identical, so only WPI-transduced cells stained with control IgG are shown (grey line). (b) Scatter dot plot shows the MFI of PVR expression in Jurkat cells transduced with WPI or VprWT lentivirus in 11 independent experiments; mean±SEM are also indicated. ***, P≤0.001 by Wilcoxon signed rank test. (c) Equal amounts of total cell lysates (30 μg) were separated by 10 % SDS-PAGE in non-reducing conditions, then analysed by standard immunoblotting using anti-PVR (5D1), anti-ULBP2 (AF1298; R&D Systems) and anti-GAPDH (MAB374, Millipore) antibodies. Molecular mass standards are indicated (kDa). One out of three independent experiments is shown. (d) RT-PCR was used to assess PVR and ULBP2 mRNA expression normalized to G6PDH mRNA at 24 h and 48 h after transduction of Jurkat cells with WPI or VprWT. Relative mRNA expression in VprWT-transduced cells (filled bar) compared with WPI-transduced cells (open bar) is shown. Error bars are ±SEM. Results shown are representative of three independent experiments. **, P≤0.01 by Wilcoxon signed rank test. Forward (F) and reverse (R) primer sequences and number of PCR cycles used for each cDNA are shown.
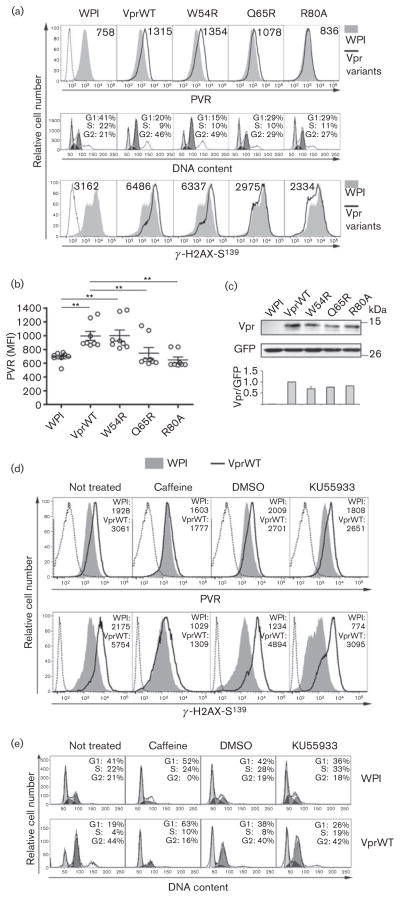
Next, cell-surface PVR levels were monitored in cells expressing Vpr mutants that lost the capacity to interact with UNG2 DNA repair enzyme (VprW54R), with DCAF-1 and E3 ligase (VprQ65R), or affected a putative activation domain without inhibiting DCAF-1 binding and E3 ligase recruitment (VprR80A) (Belzile et al., 2007; Mansky et al., 2000) (Fig. 2a, b). To confirm the previously characterized phenotype of Vpr variants, Jurkat cells transduced with WPI or lentivirus expressing wt or mutated Vpr proteins were fixed with 70 % ethanol at 4 °C for 1 h, then either stained with propidium iodide and analysed for the cell-cycle profile or permeabilized with 0.2 % Triton X-100 and reacted with monoclonal antibody (mAb) against the phosphorylated histone 2A variant X (γ-H2AX), an ATR/ATM substrate, followed by incubation with secondary antibody, and finally analysed for intracellular γ-H2AX levels (Fig. 2a). Compared to VprWT, the VprQ65R and VprR80A mutants that were strongly attenuated in their ability to promote G2 arrest and γ-H2AX accumulation in agreement with previous reports (Belzile et al., 2007), were also defective for PVR up-modulation, while VprW54R that maintained the function in the cell-cycle block and H2AX phosphorylation, was as active as VprWT at upregulating PVR (Fig. 2a, b). The functional defects of VprQ65R and VprR80A could not be ascribed to protein instability since the steady-state level of these mutated proteins was comparable to that of VprWT and VprW54R, as shown by Western blotting (Fig. 2c). These results indicate that Vpr uses the same protein surfaces, and hence probably the same molecular interactions, to upregulate PVR and activate the DNA damage/stress checkpoint. Apparently, for both Vpr activities, the ability to recruit DDB1-CUL4A E3 ligase (lost by VprQ65R but maintained by VprR80A protein) is required but not sufficient, while association with UNG2 is dispensable.
Fig. 2.
Upregulation of PVR by Vpr requires the interaction with E3 ligase and induction of ATR-mediated DDR and G2 arrest. (a) Top panels: Jurkat cells transduced with WPI (filled grey histogram), VprWT or Vpr-mutated (VprW54R, VprQ65R and VprR80A) lentiviruses (solid lines) were stained and analysed for PVR expression as in Fig. 1(a). Middle and bottom panels: transduced cells were fixed, and either incubated with propidium iodide to analyse by flow cytometry the percentages of cells in G1, S, and G2 phases of the cell cycle (middle), or permeabilized with 0.2 % Triton X-100 and stained with anti-phospho S139-H2AX mAb (JBW301, Upstate Biotechnology) and Alexa647-GAM before flow cytometric analysis (bottom). WPI-transduced cells stained with isotype control IgG (representative of all isotype control cell samples) are shown by a dotted line (top and bottom panels). (b) Scatter dot plot shows PVR MFI on cells transduced with the indicated lentiviruses determined as in (a), top panels, in eight independent experiments. Mean±SEM are also indicated. **, P≤0.01 by Wilcoxon signed rank test. (c) Total lysates of Jurkat cells transduced as in (a and b) were separated by 14 % SDS-PAGE and immuno-blotted using guinea pig anti-Vpr serum, and anti-GFP monoclonal antibody (Clontech). Molecular mass standards are indicated (kDa). The signals obtained in three independent experiments were quantified and expressed as Vpr/GFP ratios (VprWT/GFP ratio was set to 1; bottom panel shows mean±SD). (d, e) Jurkat cells were transduced with WPI or VprWT lentiviruses and cultivated for 48 h with and without the addition of 5 mM caffeine, 10 μM KU55933 or DMSO in the last 18 h. Cells were stained and analysed for the expression of cell-surface PVR and intracellular γ-H2AX (d) and for the cell-cycle profile (e) as described in (a). Results shown are representative of at least three independent experiments.
Jurkat cells transduced with WPI or VprWT lentivirus were then cultivated in the presence or absence of 5 mM caffeine (Sigma-Aldrich), an inhibitor of ATR and ATM, or with 10 μM KU55933 (Selleckchem), a specific ATM inhibitor, or equivalent amounts of DMSO (KU55933 solvent) and analysed by flow cytometry for cell-surface PVR and intracellular γ-H2AX levels (Fig. 2d), as well as for the cell-cycle profile (Fig. 2e). Such treatments were performed for the last 18 h of culture and did not affect cell growth and vitality, as determined in pilot assays (data not shown). Of note is that treatment with caffeine impaired the capacity of Vpr to upregulate PVR, while addition of KU55933 had no effect (Fig. 2d, top panels). In line with H2AX being a common target of ATR and ATM, both drugs reduced basal γ-H2AX levels if compared with untreated/DMSO controls (Fig. 2d, bottom panels). However, reduction of γ-H2AX levels by caffeine was significantly higher in cells expressing Vpr compared to control WPI-expressing cells (80 % vs 50 % inhibition), while KU55933 decreased γ-H2AX levels to the same extent (by 30–40 %) in cells with and without Vpr expression. Percentage of inhibition was calculated using the formula 100−[(MFI on drug-treated cells/MFI on control cells) × 100]. As expected, caffeine but not KU55933 completely abrogated the G2 arrest activity of Vpr (Fig. 2e). Overall, these data indicate that Vpr upregulates PVR in a manner that relies on the ability of the protein to activate the ATR DNA damage sensor kinase, not ATM, and trigger a G2 cell-cycle arrest. Furthermore, our results support the existence of a link between PVR upregulation and the G2 phase of the cell cycle that was described in previous studies (Ardolino et al., 2011; Soriani et al., 2009).
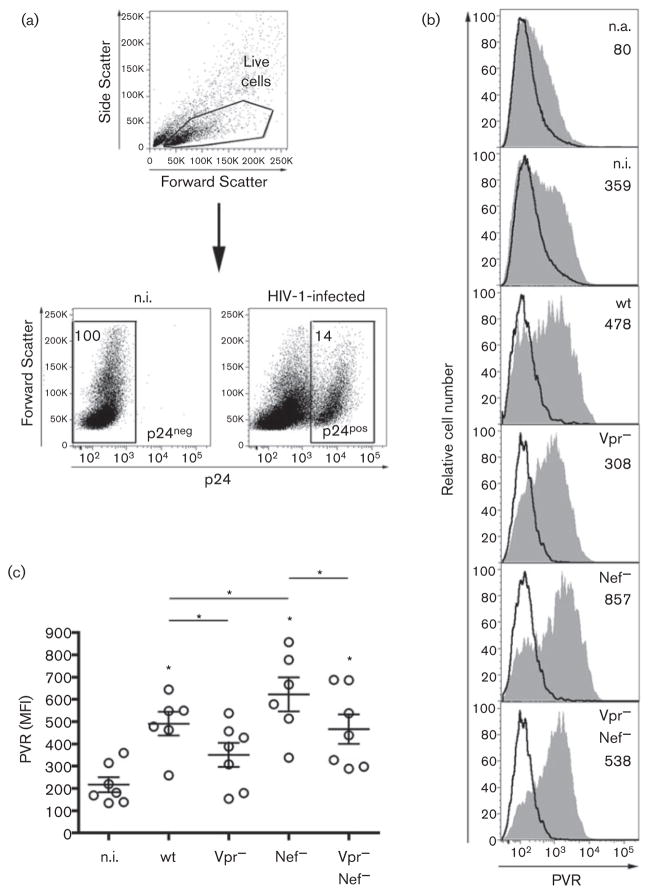
We have previously shown that the HIV-1 Nef protein downregulates cell-surface PVR through mechanisms not yet identified, hence reducing DNAM-1-mediated NK-cell killing of infected T cells (Matusali et al., 2012). Therefore, to evaluate Vpr activity on PVR in the context of HIV-1 replication, the level of cell-surface PVR was tested on primary CD4+ T lymphocytes infected with HIV-1 (NL4-3 strain) either wt or defective for the expression of Vpr (Vpr−), Nef (Nef−) or both proteins (Vpr−Nef−). Stocks of viruses were prepared as previously described (Matusali et al., 2012) using proviral plasmid pBR-NL4-3 and its derivatives with the vpr gene deleted (Δvpr), with a stop codon mutation in the nef gene (nef*), or with both defects (Δvpr nef*). Purification of CD4+ T cells, spin infection with HIV-1 followed by T-cell activation with Staphylococcus aureus enterotoxin B, irradiated allogeneic PBMCs and interleukin-2 (IL-2), and flow cytometric analysis of cells at day 5 post-infection (p.i.), were performed as described elsewhere (Cerboni et al., 2007b; Matusali et al., 2012). Fig. 3 shows that cells infected with Vpr− virus displayed reduced cell-surface PVR levels compared to wt-infected cells (P=0.03), indicating that Vpr increases PVR expression on CD4+ T cells during HIV-1 infection. In line with this observation and with the capacity of Nef to downregulate PVR, the expression of PVR was up-modulated on cells infected with Nef− (P=0.03) but not with Vpr−Nef− virus if compared to wt-infected cells. These results indicate that, analogously to NKG2D ligands, PVR is exposed to a dual regulation of opposite sign during the course of HIV-1 infection: a negative regulation due to the Nef protein and a positive regulation exerted by Vpr. Upregulation of PVR and NKG2D ligands may alert the immune system and thus represents for HIV-1 a deleterious consequence of Vpr-mediated activation of DDR and G2 arrest, a viral activity that is highly conserved and assumed to be important for LTR transcription (i.e. virus production) (Goh et al., 1998). As a countermeasure, the Nef protein, among its multiple abilities to impair antiviral immune responses (reviewed by Kirchhoff, 2010), has evolved the capacity to reduce the cell-surface expression of PVR and NKG2D ligands and, as a result, to decrease the susceptibility of HIV-1-infected cells to NK-cell-mediated lysis (Cerboni et al., 2007b; Matusali et al., 2012). Since T cells infected with Vpr−Nef− virus showed higher PVR levels compared to not infected (n.i.) cells (P=0.02), an additional, Vpr-independent mechanism apparently contributes to enhance PVR expression during HIV-1 infection. We speculate that the activation of DDR components triggered by HIV-1 DNA integration into the genome of primary T cells (Lau et al., 2005) may possibly take part in PVR upregulation. Attempts to enhance expression of PVR and other activating ligands with DDR-based treatments and to impede ligand down-modulation by Nef, represent novel attractive approaches to improve recognition and elimination of HIV-1-infected cells by the immune system.
Fig. 3.
Vpr induces up-modulation of PVR during HIV-1 infection. (a–c) Freshly isolated CD4+T cells were infected with HIV-1 either wt or defective for the expression of Vpr (Vpr−), Nef (Nef−) or both proteins (Vpr−Nef−) or not infected (n.i.), activated and, after 5 days, stained to measure surface expression of PVR and intracellular p24 Gag capsid antigen accumulation as described previously (Cerboni et al., 2007b; Matusali et al., 2012). (a) Gating strategy for n.i. and HIV-infected T cells: live cells were first identified by physical parameters and then analysed for intracellular p24 expression to distinguish infected (p24-positive, p24pos) from uninfected cells (p24-negative, p24neg). The percentage of p24neg and p24pos is indicated. (b) PVR expression (filled grey histograms; MFI values are indicated) was evaluated on p24neg cells neither infected nor activated (n.a.), p24neg n.i. cells, and on p24pos cells infected with the indicated viruses. Staining of cells with isotype control IgG is shown by a solid line. (c) Scatter dot plot depicts PVR expression (MFI) on T cells derived from seven donors in seven independent experiments like the one in panel (b). Mean±SEM are also indicated. Statistical differences between n.i. and infected samples (* on top of the cluster) or between infected samples are indicated. *, P≤0.05 by Wilcoxon signed rank test.
Acknowledgments
For kindly providing reagents, we thank: M. Colonna, Washington University, St. Louis, MO, USA (anti-PVR SKII.4 antibody); F. Kirchhoff, University of Ulm, Germany (pBR-NL4-3 and its derivatives Δvpr, nef*, and Δvpr nef*); A. Nomoto, University of Tokyo, Japan (5D1 mAb); D. Trono, Ecole Polytechnique Fédérale de Lausanne, Switzerland (WPI, psPAX2); S. Gessani, Istituto Superiore di Sanità, Rome, Italy (guinea pig anti-Vpr serum). We also thank G. W. G. Wilkinson and P. Tomasec, Cardiff University, UK, for help with anti-PVR immunoblotting and Marc-Antoine de La Vega, IRCM, Montréal, Canada, for generating the VprW54R-encoding construct. The work was supported by grants of the Italian Ministry of Health, ‘Programma Nazionale di Ricerca sull’AIDS’ in collaboration with ISS, Ricerca Finalizzata, and Ricerca Corrente co-funded by the Italian 5 × 1000 contribution to M. D.
References
- Andersen JL, Le Rouzic E, Planelles V. HIV-1 Vpr: mechanisms of G2 arrest and apoptosis. Exp Mol Pathol. 2008;85:2–10. doi: 10.1016/j.yexmp.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardolino M, Zingoni A, Cerboni C, Cecere F, Soriani A, Iannitto ML, Santoni A. DNAM-1 ligand expression on Ag-stimulated T lymphocytes is mediated by ROS-dependent activation of DNA-damage response: relevance for NK–T cell interaction. Blood. 2011;117:4778–4786. doi: 10.1182/blood-2010-08-300954. [DOI] [PubMed] [Google Scholar]
- Belzile JP, Duisit G, Rougeau N, Mercier J, Finzi A, Cohen EA. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 2007;3:e85. doi: 10.1371/journal.ppat.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK- cell lysis. Blood. 2007a;110:606–615. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- Cerboni C, Neri F, Casartelli N, Zingoni A, Cosman D, Rossi P, Santoni A, Doria M. Human immunodeficiency virus 1 Nef protein downmodulates the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity. J Gen Virol. 2007b;88:242–250. doi: 10.1099/vir.0.82125-0. [DOI] [PubMed] [Google Scholar]
- Eissmann P, Evans JH, Mehrabi M, Rose EL, Nedvetzki S, Davis DM. Multiple mechanisms downstream of TLR-4 stimulation allow expression of NKG2D ligands to facilitate macrophage/NK cell crosstalk. J Immunol. 2010;184:6901–6909. doi: 10.4049/jimmunol.0903985. [DOI] [PubMed] [Google Scholar]
- Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Lau A, Swinbank KM, Ahmed PS, Taylor DL, Jackson SP, Smith GC, O’Connor MJ. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat Cell Biol. 2005;7:493–500. doi: 10.1038/ncb1250. [DOI] [PubMed] [Google Scholar]
- Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. The interaction of Vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusali G, Potestà M, Santoni A, Cerboni C, Doria M. The human immunodeficiency virus type 1 Nef and Vpu proteins down-regulate the natural killer cell-activating ligand PVR. J Virol. 2012;86:4496–4504. doi: 10.1128/JVI.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JM, Mashiba M, McNamara LA, Onafuwa-Nuga A, Chiari-Fort E, Shen W, Collins KL. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat Immunol. 2011;12:975–983. doi: 10.1038/ni.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J, Sindhu S, Pham TN, Belzile JP, Cohen EA. HIV-1 Vpr up-regulates expression of ligands for the activating NKG2D receptor and promotes NK cell-mediated killing. Blood. 2010;115:1354–1363. doi: 10.1182/blood-2009-08-237370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, Cippitelli M, Fionda C, Petrucci MT, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113:3503–3511. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- Ward J, Davis Z, DeHart J, Zimmerman E, Bosque A, Brunetta E, Mavilio D, Planelles V, Barker E. HIV-1 Vpr triggers natural killer cell-mediated lysis of infected cells through activation of the ATR-mediated DNA damage response. PLoS Pathog. 2009;5:e1000613. doi: 10.1371/journal.ppat.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]