Abstract
Although hydroxychloroquine is used for treatment of numerous autoimmune disorders the mechanism is unclear. We here demonstrate that hydroxychloroquine preferentially induces apoptosis of CD45RO+ memory and effector T cells by inhibiting the survival pathway of autophagy.
Keywords: Hydroxychloroquine, autoimmunity, autophagy, apoptosis, T cells
To the Editor:
Autoimmune diseases, such as multiple sclerosis, systemic lupus erythematosus (SLE), inflammatory bowel disease, and rheumatoid arthritis (RA), are the result of inappropriate immunes responses against “self”. Hyperactivated, and likely autoreactive, effector T cells are common in autoimmunity, indicating that T cell homeostasis is disturbed. Impaired apoptosis of self-reactive, effector T cells has been proposed as a driving mechanism of autoimmunity.1 Consequently, a better understanding of the mechanisms regulating T cell homeostasis and identifying possible ways to manipulate it pharmacologically for therapeutic purpose remain an unbridged knowledge gap.
T cell homeostasis is, in part, regulated by the balance between apoptosis and autophagy.2 During autophagy, double membrane vesicles with cytoplasmic material (autophagosomes) are formed, which will fuse with lysosomes for degradation of their content. Autophagy has been described as a powerful mechanism of cell survival, postponing the onset of apoptosis upon cellular stress.3 Autophagy has furthermore been implicated in the development of immunity, and may therefore be an important therapeutic target for autoimmune diseases.4 Although the role of autophagy in the innate immune system is gaining clarity, its role in adaptive immunity remains poorly understood. Autophagy has been reported to occur in CD4+ T cells, although human data are scarce. Mice in which autophagy genes are specifically knocked-out in CD4+ T cells display reduced CD4+ T cell numbers, demonstrating that autophagy is important for T cell survival.5
Here we investigated if hydroxychloroquine (HCQ) can modulate autophagy in human CD4+ T cells and consequently cellular homeostasis, thereby exploring a novel therapeutic mechanism for this commonly used drug. HCQ is a potent autophagy inhibitor, which affects lysosomal acidification, and thereby inhibits endogenous protein degradation, resulting in an increase in the number of autophagic compartments.6
HCQ is also used for treatment of autoimmune diseases including RA, Sjörgen’s syndrome, and SLE.7 Although HCQ has been demonstrated to inhibit pro-inflammatory cytokine production by macrophages and antigen presentation, potential effects on T cells remain elusive.7
To assess if HCQ can modulate autophagy in T cells, human peripheral blood mononuclear cells (PBMC) were cultured in the presence of increasing concentrations of HCQ. The concentrations utilized in our experiments (0.7–20 µM) are comparable those in the blood of HCQ-treated patients (0–23 µM).8 Autophagosomes were stained with a specific Cyto-ID autophagy detection kit9 (see Methods) and the presence of autophagosomes in CD4+ T cells was determined by flow cytometry. HCQ treatment increased the number of autophagosomes in CD4+ T cells in a dosedependent fashion, indicating HCQ treatment indeed inhibits autophagy, resulting in the accumulation of autophagosomes (Fig. 1A, B). To confirm and extend these data, sorted CD4+ T cells were treated with HCQ, stained for autophagosomes, and analyzed by confocal microscopy. The number of autophagosomes per cell was increased upon HCQ treatment (Fig. 1C, D). Furthermore, Western blot analysis showed that the autophagosome markers LC3-II and p62 increased upon HCQ treatment in sorted CD4+ T cells (Fig. 1E). Taken together, these data demonstrate that HCQ treatment inhibits completion of the autophagy process in primary human CD4+ T cells, resulting in the accumulation of autophagosomes.
Figure 1. HCQ impairs autophagy in CD4+ T cells.
(A) Autophagy in human CD4+ cells, cultured in the presence of HCQ (hydroxychloroquine), n=8. (B) Histograms of (A). (C) Autophagosome visualization in CD4+ cells, cultured with 20 µM HCQ. (D) Quantification of (C), n=3. (E) p62, LC3, and β-actin expression in CD4+ cells, n=4. (F) Autophagy in human CD4+CD45RA+CD45RO- and CD4+CD45RA-CD45RO+ cells, cultured with HCQ, n=7.*p<0.05
Since CD4+CD45RO+ memory and effector T cells are among the drivers in the pathogenesis of autoimmune diseases, we hypothesized that the rate of the autophagy process (also known as the autophagic-flux) in these cells could differ from naive CD4+CD45RA+CD45RO- cells. Human PBMC were treated with HCQ, and autophagosomes in CD45RA+ and CD45RO+ T cells were analyzed. While HCQ-treatment led to a rapid accumulation of autophagosomes in CD4+CD45RO+ T cells, there was only a mild increase in CD4+CD45RA+ cells (Fig. 1F). These data demonstrate that the autophagic-flux is lower in CD45RA+ naive cells compared to CD45RO+ memory and effector T cells.
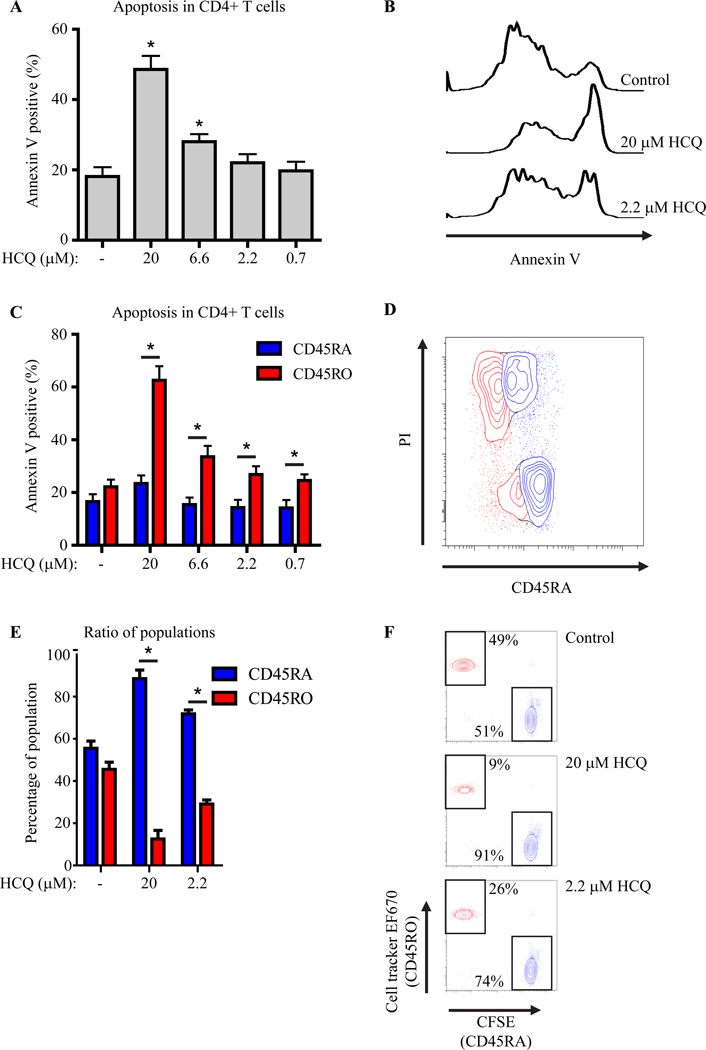
Autophagy can function as a survival mechanism and prevent apoptosis.3 Furthermore, reduced apoptosis of T cells has been proposed as a key mechanism of autoimmunity.1 Since we observed that HCQ can abrogate autophagy in CD4+ T cells, we determined if HCQ treatment would result in more apoptosis of these cells as a consequence of impaired autophagy. Human PBMC were treated with HCQ and apoptosis of CD4+ T cells was determined by annexin V staining. HCQ treatment dose-dependently increased apoptosis in CD4+ T cells (Fig 2A, B). Since the autophagic flux was differentially regulated in CD45RA+ and CD45RO+ populations, the percentage of annexin V positive cells was analyzed in these two subsets. Although HCQ only mildly induced apoptosis in CD45RA+ cells, apoptosis in CD45RO+ cells was significantly greater (Fig. 2C). These results were confirmed using a propidium iodide (PI) staining (Fig. 2D). To verify that this difference in apoptosis induction between CD4+CD45RA+ and CD4+CD45RO+ T cells would also result in different survival patterns of T cell subsets after HCQ treatment, CD4+CD45RA+ and CD4+CD45RO+ T cells were sorted, both stained with a different cell tracker dye for identification, and cultured at a one-to-one ratio in the presence HCQ for four days. As expected, the CD45RA/CD45RO ratios were significantly altered after HCQ treatment, favoring CD45RA+ T cell survival (Fig. 2E, F). Importantly, the cellular CFSE or cell tracker content was not diluted indicating that the observed differences were not the result of altered proliferation. Collectively, these data demonstrate that CD4+CD45RO+ cells normally undergo more autophagy than CD4+CD45RA+ T cells, and that inhibition of autophagy in these cells by HCQ results in apoptosis.
Figure 2. HCQ treatment preferentially induces apoptosis in CD4+CD45RA-CD45RO+ T cells.
(A) Apoptosis (by Annexin V staining) in human CD4+ cells, cultured in the presence of HCQ, n=8. (B) Histograms of (A). (C) Apoptosis in human CD4+CD45RA+CD45RO- and CD4+CD45RA-CD45RO+ cells, n=7. (D) FACS-plot of HCQ treated CD4+CD45RA+CD45RO- (blue) and CD4+CD45RA-CD45RO+ (red) cells stained with propidium iodide (PI). (E+F) Sorted CD4+CD45RA+ (CFSE) or CD4+CD45RO+ (EF670) cells were mixed at a 1:1 ratio and cultured in the presence of HCQ, (n=4).
Here we demonstrate that in human T cells there is a basal level of autophagy and that the inhibition of autophagy by hydroxychloroquine induces apoptosis. Autophagic flux and the rate of apoptosis induces by autophagy inhibition with hydroxychloroquine are markedly higher in CD45RO+ T cells as compared to naive CD45RA+ T cells. These results improve our understanding of the mechanism of action of HCQ, a drug used in several autoimmune diseases. Impaired apoptosis of T cells has previously been reported to be involved in the development of autoimmunity.1 Since virtually all activated self-reactive T cells in patients with autoimmune disease are CD45RA-negative and CD45RO-positive, HCQ treatment will preferentially induce apoptosis in these cells while only mildly affecting the naive T cell repertoire. These data provide a novel mechanism by which autoimmunity can be modulated using HCQ and validate autophagy as a promising therapeutic target.
Methods
Autophagy detection for FACS
Sorted CD4+ T cells or total PBMC were cultured in the presence of hydroxychloroquine (HCQ) for 18 hour. Subsequently cells were stained with Cyto-ID autophagy detection kit (Enzo life sciences) according to the manufacturer’s protocol. In short, cells were washed twice and stained with the autophagy specific dye diluted in supplemented culture medium (1:500) at 37°C for 30 minutes. Cells were washed three times and analyzed directly by flow cytometry.
Statistical analysis
Hypotheses were tested with two-tail t-tests in single pairwise comparisons or multiple orthogonal comparisons. ANOVA post-hoc tests were used for correction of multiple non-orthogonal comparisons: Dunnett’s when multiple conditions were tested against a single control condition; Sidak’s when a subset of conditions was selected for pairwise comparisons; and Tukey’s when all possible combinations were tested. One-sample two-tail t-tests were used for comparisons against a fixed value (100%). Dependent samples were analyzed with paired t-tests or repeated measures ANOVA. p-values, calculated with Prism (Graphpad), are coded by asterisks: <0.05 (*). Statistical difference in Figure 1D was calculated based upon it’s poisson distribution:
E1 = λt1 = (x1 + x2) × t1/(t1 + t2)
V1 = (x1 + x2) × t1 × t2/(t1 + t2)2
TS = (X1 – E1)2/V1
Acknowledgments
JVL and BJP are supported by the Dutch Arthritis Foundation.
RS and MR are partially supported by the Bartman Foundation.
ML is supported by NIH Grant AG007996
SA is partially supported by the Bartman Foundation and NIH
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pope RM. Apoptosis as a therapeutic tool in rheumatoid arthritis. Nat Rev Immunol. 2002;2:527–535. doi: 10.1038/nri846. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Zvi I, Kivity S, Langevitz P, Shoenfeld Y. Hydroxychloroquine: from malaria to autoimmunity. Clin Rev Allergy Immunol. 2012;42:145–153. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munster T, Gibbs JP, Shen D, Baethge BA, Botstein GR, Caldwell J, et al. Hydroxychloroquine concentration relationships in patients with rheumatoid arthritis. Arthritis Rheum. 2002;46:1460–1469. doi: 10.1002/art.10307. [DOI] [PubMed] [Google Scholar]
- 9.Lee SJ, Lee GM. Monitoring of autophagy in Chinese hamster ovary cells using flow cytometry. Methods. 2012;56:375–382. doi: 10.1016/j.ymeth.2011.11.006. [DOI] [PubMed] [Google Scholar]