Abstract
Oocyte maturation and early embryonic development require the cytoplasmic polyadenylation and concomitant translational activation of stored maternal mRNAs. ePAB [embryonic poly(A)-binding protein, also known as ePABP and PABPc1-like] is a multifunctional post-transcriptional regulator that binds to poly(A) tails. In the present study we find that ePAB is a dynamically modified phosphoprotein in Xenopus laevis oocytes and show by mutation that phosphorylation at a four residue cluster is required for oocyte maturation. We further demonstrate that these phosphorylations are critical for cytoplasmic polyadenylation, but not for ePAB’s inherent ability to promote translation. Our results provide the first insight into the role of post-translational modifications in regulating PABP protein activity in vivo.
Keywords: cytoplasmic polyadenylation, mRNA translation, oocyte maturation, poly(A)-binding protein (PABP), phosphorylation, post-translational modification
INTRODUCTION
Oocyte maturation, fertilization and early embryogenesis occur in the absence of transcription and are thus reliant on changes in the translation, localization or stability of stored maternal mRNAs to achieve changes in patterns of protein synthesis [1]. These complex cascades of post-transcriptional control are mediated by the action of regulatory RNA-binding proteins.
Maternal mRNAs are frequently stored with short poly(A) tails, and an increase in poly(A) tail length is required for translational activation of many key mRNAs. Activation by cytoplasmic polyadenylation (reviewed in [2]) occurs in a strict temporal order and is critical for invertebrates through to mammals. Although best studied during oocyte maturation, it also occurs in early embryos and in other cell types, such as neurons [3].
Cytoplasmic PABPs [poly(A)-binding proteins] are a family of multifunctional RNA-binding proteins that bind poly(A) tails and act as central regulators of mRNA translation and stability [4]. PABP binding allows mRNAs to adopt a closed-loop conformation via interactions between PABP and initiation factors bound to the 5′ cap. This activates mRNA translation and regulates mRNA deadenylation and stability.
Vertebrates express a specific PABP, ePAB (embryonic PABP) [5–7], which predominates in oocytes and early embryos. ePAB interacts with 5′-end-bound eIFs (eukaryotic initiation factors) including eIF4G, a component of the cap-binding complex eIF4F [8,9]. Thus ePAB enhances translation [9] and also protects mRNAs from deadenylation [6]. In addition, ePAB interacts with factors involved in cytoplasmic polyadenylation {e.g. CPEB (cytoplasmic polyadenylation element-binding protein) 1/Symplekin/CPSF (cleavage and polyadenylation specificity factor)-containing complexes [10] and Pum (Pumilio) [11,12]} and promotes cytoplasmic polyadenylation [10]. However, it is unclear how its participation in different complexes is regulated or whether PTMs (post-translational modifications) of ePAB or other PABPs regulate their different activities in vivo.
Xenopus laevis oocyte maturation offers an ideal opportunity to identify and probe the functional consequences of PABP PTMs in vivo, due to its experimental tractability and the importance of well characterized signalling cascades. In the present study, we find that X. laevis ePAB is a phosphoprotein whose phosphorylation is up-regulated during oocyte maturation. Hyperphosphorylated ePAB binds to translating mRNAs, as well as protein complexes associated with mRNA translation and polyadenylation. Crucially, we identify several phosphorylation sites within ePAB and show that blocking their phosphorylation disrupts oocyte maturation. Furthermore we find that these phosphorylations are dispensable for ePAB-dependent translational activation, but are required for the cytoplasmic polyadenylation of key maternal mRNAs.
EXPERIMENTAL
Antibodies and constructs
Anti-CPEB1 antibody [13] and Myc-xPAIP2 plasmid {xPAIP2 is X. laevis PAIP2 [poly(A)-interacting protein 2]} [10] were provided by Joel Richter (Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, MA, U.S.A.). Anti-PAIP2 [14] and anti-Dazl (Daz-like) [15] antibodies were provided by Nahum Sonenberg (Department of Biochemistry, McGill University, Montreal, QC, Canada) and Masakane Yamashita (Department of Biological Sciences, Hokkaido University, Sapporo, Japan) respectively. An anti-ePAB antibody, a His6-ePAB expression vector [6] and an anti-rRNA antibody (Y10b) [16] were provided by Joan Steitz (Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, U.S.A.). Anti-Symplekin (BD Biosciences), anti-Pum2 (Bethyl Laboratories) and anti-Myc antibodies (Sigma) were purchased. For ePAB expression, the ePAB ORF (open reading frame) (amplified from the His6-ePAB expression vector [6]) was cloned in-frame with a FLAG-tag into pcDNA3.1 using engineered EcoRI and XbaI restriction sites (pcDNA3.1-ePAB). pcDNA3.1-4×Ala-ePAB was prepared using site-directed mutagenesis following the manufacturer’s instructions (Stratagene). pLG-MS2 {luc (luciferase)-MS2 [17]}, pMSPN, pMS2-ePAB [9] and pCSFV-lacZ (β-galactosidase [18]) were as described previously. pMS2-4×Ala-ePAB was generated by ligating a KpnI-NsiI fragment encompassing the mutagenized codons from pcDNA3.1-4×Ala-ePAB into pMS2-ePAB.
Animals and oocyte collection and manipulation
X. laevis were housed in accordance with guidelines from the Institutional Animal Care and Use Committee (U.S.) or the Home Office (U.K.). X. laevis ovaries were surgically removed and treated with type IV collagenase (2 mg/ml for 3 h at 25 °C) to isolate oocytes. However, for maturation experiments, including xPAIP2 and ePAB overexpression, oocytes were manually defolliculated prior to injection. Oocytes were maintained in vitro in OR2+ medium (5 mM Hepes/KOH, pH 7.8, 82.5 mM NaCl, 2.5 mM KCl, 1 mM Na2HPO4, 1 mM MgCl2 and 1 mM CaCl2) at 18 °C. Oocyte maturation was induced using 10 μg/ml final concentration of progesterone for ~5–7 h and scored by the appearance of a white spot [GVBD (germinal vesicle breakdown)] on the animal pole.
Exogenous protein expression and tethered-function analysis
For protein expression, mRNAs encoding the indicated proteins were in vitro transcribed with T7 RNA polymerase. After transcription, RNAs were treated with DNase I, extracted with phenol/chloroform/3-methyl-1-butanol (25:24:1 by vol.), and RNA concentrations were determined using a spectrophotometer. A 46 nl volume of 2 mg/ml Myc–xPAIP2 RNA (high) or 0.5 mg/ml RNA encoding Myc–xPAIP2, FLAG–ePAB or FLAG–4×Ala-ePAB, as appropriate, were injected into the oocyte cytoplasm followed by a 6 h incubation. Tethered function assays (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/445/bj4450093add.htm) were performed as described previously [17] using the plasmids detailed above. Maturation was scored by the appearance of a white spot on the animal pole.
Sucrose gradient analysis
Sucrose gradient analysis was performed essentially as described [19]. Oocytes were lysed in polysome gradient buffer [250 mM KCl, 2 mM MgCl2, 20 mM Hepes, pH 7.4, 0.5 % NP-40 (Nonidet P40), 2.5 mM DTT (dithiothreitol), 150 μg/ml cycloheximide with 5 mM EDTA where indicated] after whole oocytes were pre-incubated with 10 μg/ml cycloheximide as described previously [20]. Samples were separated over a linear 10–40 % sucrose gradient prepared in polysome gradient buffer by centrifugation at 40 000 rev./min in an SW41 rotor at 4 °C for 2 h. Fractions of 100 μl were manually collected and analysed for RNA concentration in a spectrophotometer.
Polyadenylation assays
Ligation-dependent RT (reverse transcription)–PCR was performed essentially as described [20]. The P1 oligonucleotide (5′-P-GGTCACCTTGATCTGAAGC-3′) [21] was blocked at the 3′ end using terminal nucleotide transferase (Invitrogen) and cordycepin (Sigma) as described in the manufacturer’s instructions. Gene-specific primers were labelled using T4 PNK (polynucleotide kinase) (USB) and γ-[32P]ATP and used in the PCR reaction. PCR reactions were analysed by non-denaturing PAGE (10% gels) after 30 cycles.
SDS/PAGE, 2D (two-dimensional)-PAGE and Western blotting
Oocytes were lysed in RIPA buffer (50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 25 mM sodium 2-glycerophosphate, 10 mM sodium tetrapyrophosphate, 1 mM sodium orthovanadate, 2 mM DTT, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS and 10 nM Calyculin A) plus Complete protease inhibitors (Roche) and were Freon-extracted [22]. For 2D-PAGE analysis, cleared lysates were precipitated and resolubilized in DeStreak 2D rehydration buffer (GE Healthcare) containing 0.5% pH 6–11 ampholytes (GE Healthcare) before isoelectric focusing on 7 cm pH 6–11 IPG strips (immobilized pH gradient strips; Invitrogen) followed by SDS/PAGE on 4–12% acrylamide ZOOM gels (Invitrogen) according to the manufacturer’s instructions. Western blotting was performed as described previously [6]. For 1D (one-dimensional) analyses, lysates were run on 8% Tris/glycine gels until a pre-stained 50 kDa marker reached the bottom of the gel. Alkaline phosphatase treatment for 1D SDS/PAGE experiments was performed for 30 min at 30 °C; where indicated, 2-glycerophosphate and potassium fluoride were added at 2 mM (phosphatase inhibitors). For 2D SDS/PAGE, oocytes were lysed as for IP (immunoprecipitation) except that phosphatase-treatment lysis buffer (25 mM Hepes, pH 7.8, 300 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100 and 0.1 mM DTT) containing EDTA-free Complete protease inhibitors was used. Either active or heat-inactivated (95 °C for 5 min) shrimp alkaline phosphatase (0.1 units per μg of protein) was incubated with lysate for 2 h at 37°C. Lysates were then prepared for 2D analysis as above.
Oocyte IF (immunofluorescence)
Ovaries were manually dissected from the surrounding tissue. Dehydration and fixation were performed in methanol (glutaraldehyde fixation resulted in gross morphological disruptions and IF artefacts). Samples were submitted to the Yale Pathology Labs for sectioning. IF was performed on fixed, paraffin-embedded and thin-sectioned (5 μm) samples. Paraffin was removed in xylene with serial washes in ethanol. Slides were rehydrated using TBSN (20 mM Tris/HCl, pH 7.6, 150 mM NaCl and 0.5% NP-40). Primary antibodies were used at 1:100 dilution and incubations were performed overnight at 4 °C. Washes were completed in TBSN prior to secondary antibody incubation (goat anti-rabbit- or anti-mouse-conjugated AlexaFluor 660) for 2 h at 4 °C. All subsequent washes were in PBS. IF was detected on an Axioplan II microscope (Carl Zeiss), and images were processed and pseudocoloured using ImageJ software.
IP and affinity selection
For IP, oocytes were homogenized in lysis buffer (150 mM NaCl, 25 mM Hepes/KOH, pH 7.8, 1 mM MgCl2 10% glycerol, 2 mM EDTA, 0.5% Triton-X100, 2 mM sodium orthovanadate, 2 mM 2-glycerophosphate, 0.5 mM PMSF and 1 mM DTT) with 20 μg/ml RNase I, which cleaves between all ribonucleotides, and clarified by centrifugation for 5 min at 13 000 g. IPs were performed in lysis buffer and antibodies were immobilized on Protein A–Sepharose, except for the anti-Myc antibody (Protein G–Sepharose) (GE Healthcare). Washes were performed with lysis buffer. In all cases, to ensure equal loading, pellet samples were loaded in their entirety.
Oligo(dT) selection was performed as above using oligo(dT) cellulose (GE Healthcare), but the lysate was not pre-treated with RNase, unless indicated. m7GTP-Sepharose selection was performed as described previously [23]. In all cases, to ensure equal loading, pellet samples were loaded in their entirety.
RESULTS
ePAB phosphorylation is dynamic
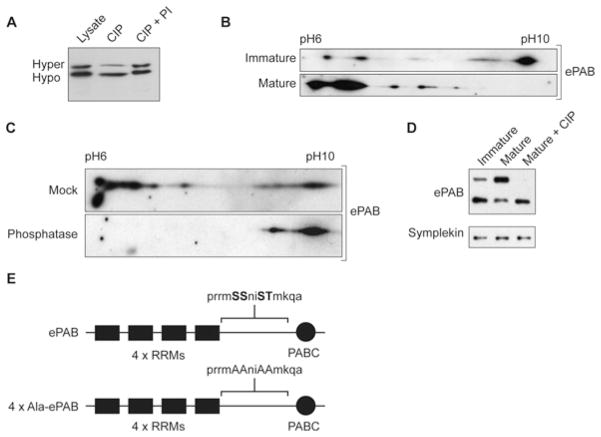
Key mediators of gene expression are regulated by phosphorylation in X. laevis oocytes (i.e. CPEB1 [24]) prompting us to examine the modification status of ePAB. Immunoblotting of oocyte lysates revealed that ePAB was detectable as two forms of differing electrophoretic mobility (Figure 1A), with MS confirming their identity (K. Friend, unpublished work). Phosphatase treatment of lysates (Figure 1A) reduced the relative abundance of the upper band, consistent with phosphorylation. This effect was blocked by phosphatase inhibitors.
Figure 1. ePAB is dynamically phosphorylated during oocyte maturation.
(A) Immature oocyte lysates were untreated (Lysate), or treated with calf alkaline phosphatase (CIP) or CIP in the presence of inhibitors (CIP + PI) and immunoblotted for ePAB. Hyperphosphorylated (Hyper) and hypophosphorylated (Hypo) ePAB are indicated. SDS/PAGE was run under modified conditions (8 % gel, extended electrophoresis time) compared with studies that did not report multiple ePAB electrophoretic forms [6,8,9]. (B) Immunoblots of 2D SDS/PAGE of ePAB from immature and mature oocyte lysates. The pI range is indicated. (C) Mature oocyte lysates were treated with shrimp alkaline phosphatase (Phosphatase) or heat-inactivated phosphatase (Mock) prior to analysis as in (B). (D) Immature or mature oocyte lysates were immunoblotted for ePAB to resolve hyperphosphorylated (Hyper) and hypophosphorylated (Hypo) forms, after CIP treatment where indicated. (E) Schematic showing the phosphorylations identified at Ser460, Ser461, Ser464 and Thr465 (upper-case bold letters) in the proline-rich region between the RRMs (RNA recognition motifs) and the PABC (PABP C-terminal domain) domain of ePAB, which were mutated to alanine residues (upper-case letters) in the present study.
2D-gel electrophoresis (Figure 1B) showed that a proportion of ePAB was detected close to its predicted pI value of pH 9.37, whereas the remainder had a reduced pI, indicating multiple PTMs, with a stoichiometry consistent with that observed in Figure 1(A). Intriguingly, upon oocyte maturation the bulk of ePAB exhibited a massively reduced pI (lower panel) consistent with enhanced post-translational modification. In order to determine the contribution of phosphorylation to this pI change, extracts from matured oocytes were phosphatase-treated, causing the majority of ePAB to resolve close to its unmodified pI in 2D-gel electrophoresis (Figure 1C, lower panel) and decreasing the abundance of the upper band in 1D SDS/PAGE (Figure 1D). Taken together, these results indicate that ePAB is present as hypo- and hyper-phosphorylated forms in immature stage VI oocytes. Although multiple phosphorylation events are predicted by the large pI distribution of the multiple ePAB species resolved at both stages, in mature oocytes hyperphosphorylated forms predominate. These findings demonstrate that ePAB phosphorylation is dynamically regulated during oocyte maturation, when the adenylation status and translation of many mRNAs are dramatically altered.
Hyperphosphorylated ePAB is in multiple complexes
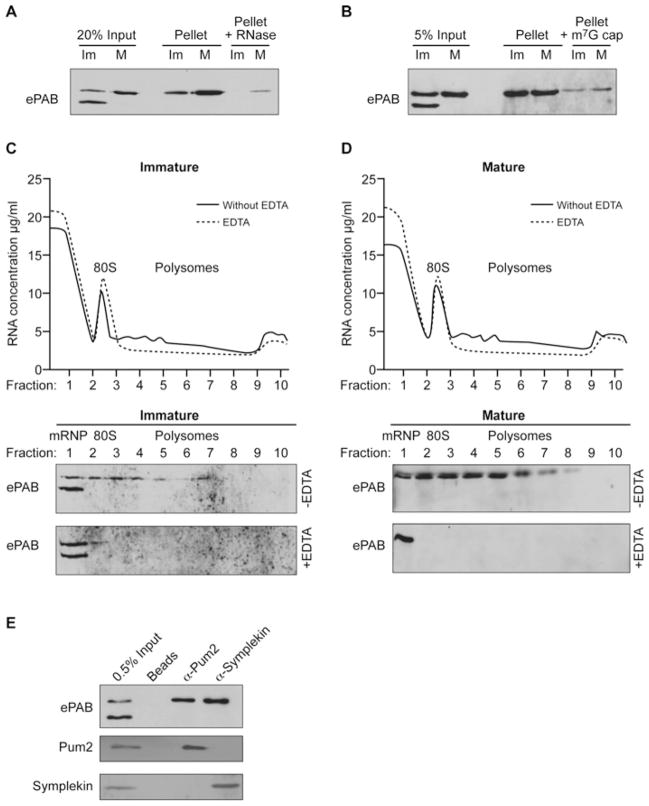
The regulated phosphorylation of ePAB suggested a potential role in controlling one or more of its activities, leading us to examine the localization of hyper- and hypo-phosphorylated forms of ePAB and their association with functional complexes. Immunohistochemistry with anti-ePAB antibodies revealed that ePAB is present at higher levels in the animal pole of immature oocytes (Figure 2A). However Western blotting of hemisected oocytes showed that neither form was specifically enriched within either hemisphere (Figure 2B), indicating that ePAB phosphorylation status does not determine its subcellular localization. In contrast, oligo-dT selection of polyadenylated mRNAs from immature or mature oocytes only co-purified hyperphosphorylated ePAB (Figure 3A), suggesting that hyper-phosphorylation may confer, or be a consequence of, poly(A)-binding. Similarly, hyper- but not hypo-phosphorylated ePAB was found to associate with cap-binding complexes (eIF4F) isolated by m7GTP-chromatography (Figure 3B), and actively translating polyribosomes in an EDTA-sensitive manner (Figures 3C and 3D). Intriguingly, proteins associated with cytoplasmic polyadenylation complexes, namely Pum2 and Symplekin, also appeared to exclusively interact with hyperphosphorylated ePAB (Figure 3E). Thus ePAB phosphorylation plays a role in its associations with RNA–protein complexes that are involved in both translation and cytoplasmic polyadenylation.
Figure 2. Subcellular localization of ePAB is not dependent on its phosphorylation.
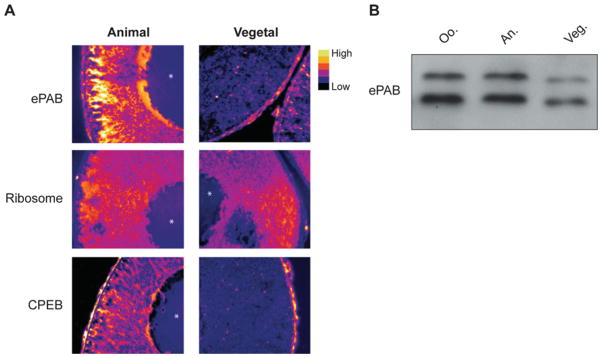
(A) The subcellular localization of ePAB, CPEB and ribosomes was visualized in immature (stage VI) oocytes by immunohistochemistry. Images were pseudocoloured according to signal intensity and the position of germinal vesicles is indicated (*). Ribosomes were detected using an anti-rRNA antibody [16], and CPEB distribution has been described previously [30]. (B) Immature oocytes were dissected into animal (An.) and vegetal (Veg.) halves, and extracts were prepared from each half as well as from whole oocytes (Oo.) and were immunoblotted for ePAB.
Figure 3. Hyperphosphorylated ePAB is associated with mRNA translation and cytoplasmic polyadenylation complexes.
(A) Poly(A) RNA was selected from immature (Im) or in vitro matured (M) oocytes using oligo(dT) cellulose (pellet) and was Western blotted for co-isolated ePAB. RNA-dependence was established by RNase I pre-treatment (Pellet + RNase). (B) Western blot of ePAB isolated by m7GTP-Sepharose chromatography (Pellet) from lysates (Input) of immature (Im) or mature (M) oocytes. Excess m7G cap analogue (~100-fold) reduces ePAB-association with the resin (Pellet + m7G cap). (C) Lysates derived from immature oocytes were separated over a 10–40 % sucrose gradient, and the positions of the 80S peak, polyribosomes (Polysomes) and messenger ribonucleoproteins (mRNPs) are indicated (upper panel). EDTA disrupts polyribosomes and was omitted (without EDTA) or added (EDTA) to lysates. The resulting fractions were Western blotted for ePAB (lower panels). (D) As for (C) except that matured oocytes were used. (E) Lysate (Input) from immature oocytes was incubated with anti-Pum2 or anti-Symplekin antibodies or beads alone (Beads) as indicated, and co-precipitating proteins were detected by immunoblotting.
Mapped phosphorylations are needed for oocyte maturation
Since ePAB phosphorylation was concomitant with its association with functional complexes, MS of purified ePAB was used to identify phosphorylated residues (Supplementary Figure S2A at http://www.BiochemJ.org/bj/445/bj4450093add.htm), identifying two sites: Ser461 and Ser464 (Figure 1E and Supplementary Figure S2B). Interestingly, two additional putative sites were identified in a high-throughput phosphoproteomic screen [25]. Since all four sites lie within a six amino acid stretch, their potential physiological relevance was examined by mutating them to alanine to block their phosphorylation (4×Ala-ePAB, Figure 1E).
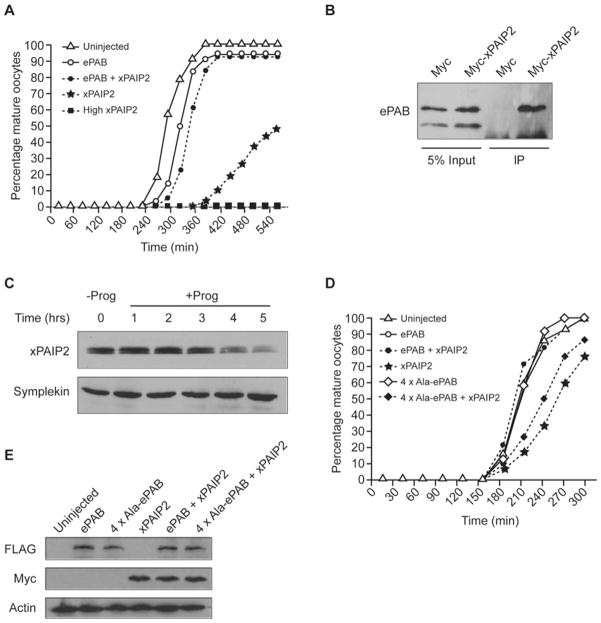
Intriguingly, perturbing PABP function by overexpressing the inhibitory protein xPAIP2 blocks oocyte maturation [10]. As ePAB is the predominant PABP in oocytes it may underlie this effect, providing a potential in vivo assay for its function. To investigate this, Myc–xPAIP2 mRNA was injected to block maturation (Figure 4A) as described previously [10], although lower amounts significantly delayed, rather than blocked, maturation. Importantly, maturation could be rescued by co-expression of FLAG–ePAB, showing that ePAB function is sequestered by xPAIP2 and is required for oocyte maturation. Since this provides an in vivo assay to test whether ePAB phosphorylations are required for oocyte maturation, we examined the interaction of xPAIP2 with endogenous hypo- and hyper-phosphorylated ePAB. Only hyperphosphorylated ePAB co-precipitated with Myc–xPAIP2 (Figure 4B) and, intriguingly, during maturation xPAIP2 levels were reduced (Figure 4C). This reduction may serve to increase the pool of hyperphosphorylated ePAB available for interaction with complexes required for translation and/or cytoplasmic polyadenylation.
Figure 4. ePAB phosphorylation is required for oocyte maturation.
(A) Immature oocytes were left uninjected or injected with the indicated mRNAs. Myc–xPAIP2, alone or in combination with ePAB, was injected at 0.5 mg/ml except where specifically stated (high, 2 mg/ml). Maturation was induced by progesterone (t = 0) and scored by GVBD. The GVBD50 value for uninjected oocytes was 280 min, and only uninjected is depicted at times before which no oocytes had undergone GVBD. (B) Myc–xPAIP2, but not Myc-tag alone (Myc), immunoprecipitates hyperphosphorylated ePAB from immature oocytes. Co-precipitating ePAB was detected by immunoblotting. Myc–xPAIP2 co-migrated with antibody light chain and was not visualized. (C) Immature oocytes were untreated (−Prog) or progesterone-treated (+ Prog) for the indicated times prior to immunoblotting lysates for xPAIP2 (which runs as a doublet) and Symplekin (loading control). (D) As in (A), using 0.5 mg/ml Myc–xPAIP2, with GVBD50 of uninjected oocytes occurring at 210 min. ePAB is largely obscured by uninjected and 4×Ala-ePAB data points. (E) Lysates from (D) (t = 0) were immunoblotted using anti-FLAG or anti-Myc antibodies to detect exogenous protein expression. Endogenous actin was used as a loading control.
Subsequently, the ability of FLAG–4×Ala-ePAB to rescue delayed oocyte maturation was compared with FLAG–ePAB (Figure 4D). Critically, this mutant could not restore normal maturation, despite equivalent expression of both FLAG-tagged proteins (Figure 4E). Neither protein alone affected the kinetics of oocyte maturation indicating that FLAG–4×Ala-ePAB does not act as a dominant negative (Figure 4D). Thus ePAB phosphorylation at one or more of Ser460, Ser461, Ser464 and Thr465 sites is critical for oocyte maturation.
Phospho-site mutations do not alter translation
To investigate which molecular functions of ePAB require phosphorylation at these residues, the inherent ability of 4×Ala-ePAB to mediate translation was examined by tethering (Supplementary Figure S1), an assay previously applied to characterize ePAB function in X. laevis oocytes [9]. Translation of luc–MS2 reporter was stimulated to equivalent extents by MS2–ePAB and MS2–4×Ala-ePAB (Figure 5), indicating that the mapped phosphorylations are not required for the basal role of ePAB in translation. Since ePAB-mediated translational stimulation increases during oocyte maturation [9], the activity of MS2–4×Ala-ePAB was also examined in mature oocytes. However, MS2–ePAB and MS2–4×Ala-ePAB exhibited equivalent increases in activity, suggesting that these phospho-residues do not underlie the observed increase in ePAB-mediated translational stimulation.
Figure 5. ePAB phosphorylation is not required to promote translation.
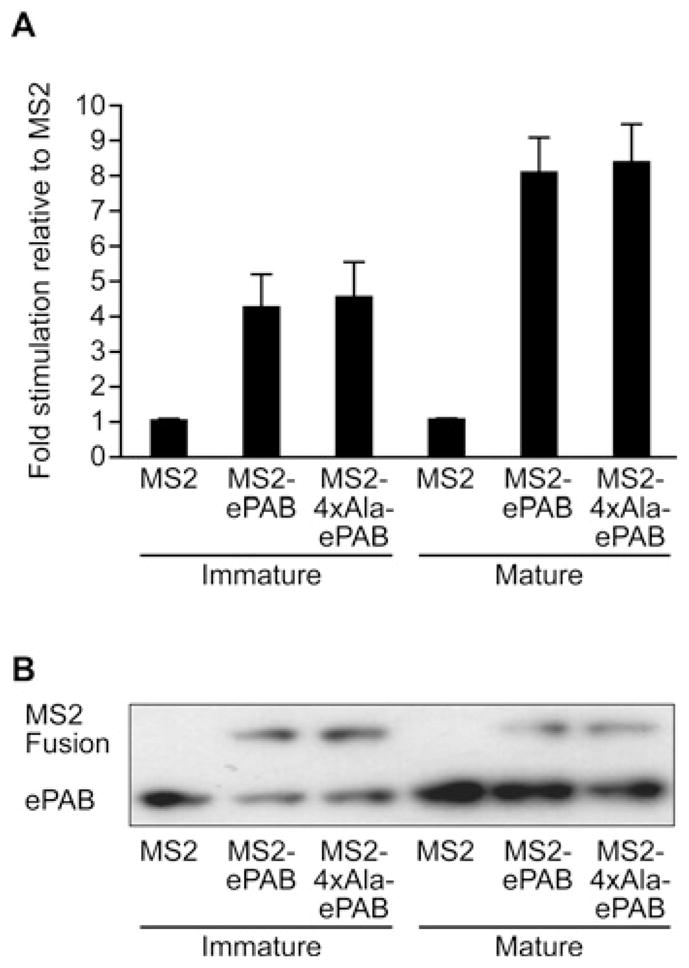
(A) Immature oocytes expressing MS2, MS2–ePAB or MS2–4×Ala-ePAB were co-injected with a non-adenylated PABP-dependent luc reporter mRNA containing MS2-binding sites and PABP-independent control mRNA (depicted in Supplementary Figure S1B at http://www.BiochemJ.org/bj/445/bj4450093add.htm). Half of the oocytes were matured, and control-adjusted fold-stimulation is depicted. (B) Lysates from (A) were immunoblotted for expression of MS2–ePAB fusions, which do not resolve as a doublet owing to the increase in mass (14 kDa) conferred by the MS2 protein fusion.
Phospho-mutants delay cytoplasmic polyadenylation
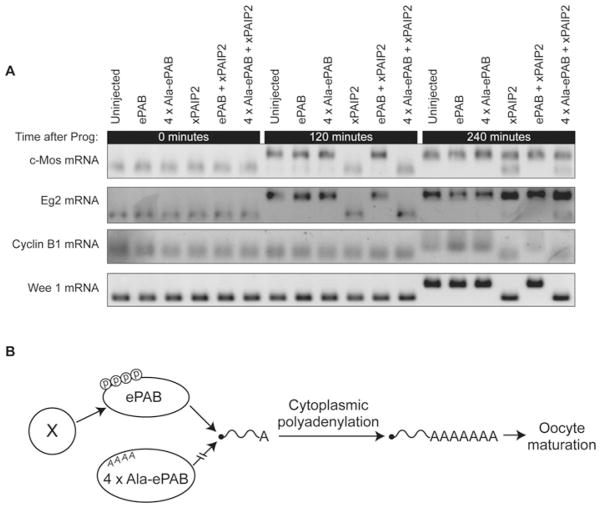
Since the inability of 4×Ala-ePAB to rescue oocyte maturation (Figure 4) is not due to abrogated ability to stimulate translation (Figure 5), we investigated whether these phosphorylations are required for cytoplasmic polyadenylation. Oocytes were injected with mRNAs encoding Myc–xPAIP2 and either FLAG–ePAB or FLAG–4×Ala-ePAB (as in Figure 4), and the effect on cytoplasmic polyadenylation of maternal mRNAs during maturation was examined by ligation-dependent RT–PCR [20] (Figure 6A). Myc–xPAIP2 expression significantly inhibited polyadenylation of all of the examined endogenous mRNAs [c-Mos, Wee1, cyclin B1 and Eg2 (Aurora kinase) mRNAs], an effect that was rescued by simultaneous expression of FLAG–ePAB but, critically, not FLAG–4×Ala-ePAB. Thus the contribution of ePAB phosphorylation to oocyte maturation can be explained by its critical role in promoting the cytoplasmic polyadenylation of key meiotic mRNAs.
Figure 6. ePAB phosphorylation is required for cytoplasmic polyadenylation.
(A) Oocytes were treated as described in Figure 2(D), and ligation-dependent RT–PCR was used to analyse poly(A)-tail lengths at the indicated times after progesterone (Prog) addition. (B) Model for the functional requirement of specific ePAB phosphorylations during oocyte maturation. ePAB is phosphorylated (circled P) at Ser460, Ser461, Ser464 and Thr465 by one or more kinases (X). Blocking these phosphorylations by alanine substitution (A) prevents ePAB promoting (broken arrow) the cytoplasmic polyadenylation of key mRNAs (e.g. c-Mos) whose poly(A)-dependent translational activation and protein products are required for maturation.
DISCUSSION
PABPs are multifunctional post-transcriptional regulators although it has yet to be determined how their different functions, requiring numerous protein–protein interactions, are co-ordinated. In the present study, we report that PABP activity can be regulated by phosphorylation and show that ePAB phosphorylation is a critical component of the molecular circuitry required for the resumption of meiosis and oocyte maturation.
A critical early event in oocyte maturation is the cytoplasmic polyadenylation of c-Mos mRNA, the translation of which initiates a signalling cascade leading to the adenylation and translational activation of other key mRNAs (e.g. cyclin B1). Our results significantly extend previous observations that ePAB can associate with cytoplasmic polyadenylation protein complexes (e.g. CPEB1, CPSF, Symplekin and Gld2), and that PABP depletion interferes with adenylation of reporter mRNAs in vitro [10]. We also provide evidence that phosphorylation(s) at Ser460, Ser461, Ser464 and/or Thr465 of ePAB are required for cytoplasmic polyadenylation of mRNAs including c-Mos (see Figure 6B for a model).
Interestingly, although our results support a direct role for phosphorylation at these specific residues in cytoplasmic polyadenylation, these modifications were not required for the participation of ePAB in translation complexes (Figure 5) and are unlikely to underlie the reported increase in translation activation following oocyte maturation [9]. Rather, this may be due to the reduction in xPAIP2 levels during maturation (Figure 4). However, our results do show that endogenous hypophosphorylated ePAB is not associated with polyribosomes or cap complexes (Figure 2), suggesting that phosphorylation at other residues may affect the interaction of ePAB with the translation machinery. In keeping with this, 2D-PAGE supports the existence of multiple ePAB phosphorylations in addition to the four sites tested in the present study (Figure 1), and phosphorylation of plant PABP1 at unknown sites apparently influences its interactions with translation initiation factors in vitro [26]. However, cap-binding complexes in plants and metazoans differ, and it remains unclear whether plant PABP1 phosphorylation affects translation rates.
ePAB associates with the cytoplasmic polyadenylation machinery and is transferred to the newly synthesized poly(A) tails, protecting them from deadenylase action and enhancing adenylation rates [6,10]; however, the understanding of this process remains in its infancy. Our observation that ePAB phosphorylation is critical for oocyte maturation (Figure 4) through effects on cytoplasmic polyadenylation (Figure 6) raises important new questions. First, are all or only some of these phosphorylations required for cytoplasmic polyadenylation, and which kinase(s) regulates this process? Secondly, how is the transfer of ePAB from polyadenylation complexes to poly(A) tails co-ordinated, and are the identified phosphorylations required for this step? Thirdly, do these phosphorylations directly affect interactions mediated through the proline-rich region, a domain whose function and interactions are poorly understood, or do they license modification at distinct sites?
In summary, we describe the first functional role for specific post-translational modifications of PABPs, finding them to be critical for oocyte meiotic maturation. The importance of our findings is underscored by observations that plant PABP1 is also a phosphoprotein [27], that mammalian PABP1 (PABPC1) [28] is extensively modified during mitosis, and that yeast [29] and sea urchin PABPs [29] are also post-translationally modified. Thus post-translational modifications appear likely to regulate the action of multiple PABP family members in response to different biological stimuli, drawing attention to a need for further study of the roles of post-translational modifications in co-ordinating PABP functions.
Supplementary Material
Acknowledgments
We thank Joan A. Steitz for critical reading of the paper prior to submission and J. Richter, N. Sonenberg and M. Yamashita for materials.
FUNDING
This work was funded by the National Institutes of Health [grant numbers K08HD046581-01 and R01HD059909 (to E.S.)], and by Medical Research Council (MRC) Unit funding [grant number U1276.00.002.00011.01] and an MRC Senior Non-Clinical Fellowship (to N.K.G.).
Abbreviations used
- CPEB
cytoplasmic polyadenylation element-binding protein
- CPSF
cleavage and polyadenylation specificity factor
- 1D
one-dimensional
- 2D
two-dimensional
- DTT
dithiothreitol
- eIF
eukaryotic initiation factor
- GVBD
germinal vesicle breakdown
- IF
immunofluorescence
- IP
immunoprecipitation
- luc
luciferase
- NP-40
Nonidet P40
- PABP
poly(A)-binding protein
- ePAB
embryonic PABP
- PAIP
poly(A)-interacting protein
- PTM
post-translational modification
- Pum
Pumilio
- RT
reverse transcription
- xPAIP2
Xenopus laevis PAIP2
Footnotes
AUTHOR CONTRIBUTION
Kyle Friend, Matthew Brook, Betul Bezirci and Michael Sheets generated data. Matthew Brook and Nicola Gray were mainly responsible for writing the paper. Nicola Gray and Emre Seli directed the project.
References
- 1.Vasudevan S, Seli E, Steitz JA. Metazoan oocyte and early embryo development program: a progression through translation regulatory cascades. Genes Dev. 2006;20:138–146. doi: 10.1101/gad.1398906. [DOI] [PubMed] [Google Scholar]
- 2.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang YS, Richter JD. Analysis of mRNA translation in cultured hippocampal neurons. Methods Enzymol. 2007;431:143–162. doi: 10.1016/S0076-6879(07)31008-2. [DOI] [PubMed] [Google Scholar]
- 4.Brook M, Smith JWS, Gray NK. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction. 2009;137:595–617. doi: 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- 5.Guzeloglu-Kayisli O, Pauli S, Demir H, Lalioti MD, Sakkas D, Seli E. Identification and characterization of human embryonic poly(A) binding protein (EPAB) Mol Hum Reprod. 2008;14:581–588. doi: 10.1093/molehr/gan047. [DOI] [PubMed] [Google Scholar]
- 6.Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seli E, Lalioti MD, Flaherty SM, Sakkas D, Terzi N, Steitz JA. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci USA. 2005;102:367–372. doi: 10.1073/pnas.0408378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosson B, Couturier A, Le Guellec R, Moreau J, Chabelskaya S, Zhouravleva G, Philippe M. Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biol Cell. 2002;94:217–231. doi: 10.1016/s0248-4900(02)01195-4. [DOI] [PubMed] [Google Scholar]
- 9.Wilkie GS, Gautier P, Lawson D, Gray NK. Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol Cell Biol. 2005;25:2060–2071. doi: 10.1128/MCB.25.5.2060-2071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 12.Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 14.Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 15.Mita K, Yamashita M. Expression of Xenopus Daz-like protein during gametogenesis and embryogenesis. Mech Dev. 2000;94:251–255. doi: 10.1016/s0925-4773(00)00295-1. [DOI] [PubMed] [Google Scholar]
- 16.Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith RW, Anderson RC, Smith JW, Brook M, Richardson WA, Gray NK. DAZAP1, an RNA-binding protein required for development and spermatogenesis, can regulate mRNA translation. RNA. 2011;17:1282–1295. doi: 10.1261/rna.2717711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillian-Daniel DL, Gray NK, Astrom J, Barkoff A, Wickens M. Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol Cell Biol. 1998;18:6152–6163. doi: 10.1128/mcb.18.10.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlesworth A, Cox LL, MacNicol AM. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J Biol Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rassa JC, Wilson GM, Brewer GA, Parks GD. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology. 2000;274:438–449. doi: 10.1006/viro.2000.0494. [DOI] [PubMed] [Google Scholar]
- 22.Gurdon JB, Wickens MP. The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- 23.Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- 24.Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- 25.McGivern JV, Swaney DL, Coon JJ, Sheets MD. Toward defining the phosphoproteome of Xenopus laevis embryos. Dev Dyn. 2009;238:1433–1443. doi: 10.1002/dvdy.21941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le H, Browning KS, Gallie DR. The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J Biol Chem. 2000;275:17452–17462. doi: 10.1074/jbc.M001186200. [DOI] [PubMed] [Google Scholar]
- 27.Gallie DR, Le H, Caldwell C, Tanguay RL, Hoang NX, Browning KS. The phosphorylation state of translation initiation factors is regulated developmentally and following heat shock in wheat. J Biol Chem. 1997;272:1046–1053. doi: 10.1074/jbc.272.2.1046. [DOI] [PubMed] [Google Scholar]
- 28.Brook M, McCracken L, Reddington JP, Lu ZL, Morrice NA, Gray NK. The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem J. 2012;441:803–812. doi: 10.1042/BJ20111474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drawbridge J, Grainger JL, Winkler MM. Identification and characterization of the poly(A)-binding proteins from the sea urchin: a quantitative analysis. Mol Cell Biol. 1990;10:3994–4006. doi: 10.1128/mcb.10.8.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.