Abstract
Conantokins are venom peptides from marine cone snails that are NMDA receptor antagonists. Here, we report the characterization of a 24 AA conantokin from Conus brettinghami (1), conantokin-Br (con-Br), the first conantokin that does not have the conserved glutamate residue at position 2. Molecular modeling studies suggest that con-Br has a helical structure between residues 2–13. In contrast to other characterized conantokins, con-Br has a high potency for NMDA receptors with NR2D subunits. To identify determinants for NR2D potency, we synthesized chimeras of con-Br and conantokin-R (con-R), the latter has a ~30-fold lower potency for the NR2D subtype. The characterization of two reciprocal chimeras (con-Br/R and con-R/Br), comprising the first 9–10 N-terminal AAs of each conantokin followed by the corresponding C-terminal AAs of the other conantokin demonstrates that determinants for NR2D selectivity are at the N-terminal region. Additional analogs comprising 1–3 amino acid substitutions from each peptide into the homologous region of the other led to the identification of a key determinant; a Tyr residue in position 5 increases potency for NR2D, while Val at this locus causes a decrease. The systematic definition of key determinants in the conantokin peptides for NMDA receptor subtype selectivity is an essential component in the development of conantokin peptides that are highly selective for each specific NMDA receptor subtype.
The glutamate receptor superfamily comprises a large number of different genes, each encoding a subunit that assembles to form the functional receptor complex, believed to be composed of four primary subunits (2). Glutamate receptors are traditionally divided into three classes, AMPA receptors, NMDA receptors and kainate receptors, based initially on pharmacological criteria (3–5). Each class has separate, but overlapping physiological roles and corresponding links to various pathologies in the nervous system. Dysfunction among all three classes has been implicated in epilepsy (6), Parkinson’s disease and Alzheimer’s disease, (7), as well as excitotoxic neuronal death (8). Additionally, NMDA receptors are thought to be involved in chronic pain (9) and mechanisms of drug and alcohol addiction (10, 11).
NMDA receptors have many properties that distinguish them from other members of the glutamate receptor superfamily (i.e., voltage-dependent Mg2+ inhibition, permeability to Ca2+, and the requirement of the co-agonists glycine or D-serine), and multiple NMDA receptor subunit genes contribute to their functional diversity (12). The NR1 subunit, which binds glycine, has eight splice variants, and is ubiquitously expressed in the central nervous system (5). Four separate genes (NR2A-NR2D) encode different isoforms of the NR2 subunit, which binds glutamate; expression of different NR2 genes varies according to both regional and temporal expression patterns in the brain (12). In addition, two NR3 genes have been identified (NR3A-NR3B), which also vary in their regional and temporal expression patterns (13). The assembly of a functional tetrameric complex is thought to require the presence of two obligatory NR1 subunits and a combination of modulatory NR2 or NR3 subunits; studies examining receptor subunit assembly indicate that NR1 subunits form heterodimers with NR2 or NR3 subunits at the initial stage in assembly (14).
Although much remains to be elucidated, pharmacological evidence suggests that different NMDA receptor subunit combinations may play different roles in the physiology and corresponding pathology of the central nervous system. For instance, NMDA receptor antagonists that selectively inhibit receptors containing the NR2B subunit have been implicated in the treatment of epilepsy and chronic pain, whereas more broadly selective NMDA receptor antagonists generally result in toxicity or intolerable side effects in the same assays (15). Conversely, more broadly targeted NMDA receptor antagonists were effective at preventing axonal de-myelination in models of ischemia, whereas NR2B-specific antagonists had no effect on myelin degradation, suggesting the antagonism of non-NR2B subtypes mediates this protective effect (16).
Interestingly, evidence suggests the effects of NR2B-selective antagonists show correspondence to temporal and regional expression patterns in the brain. For instance, NR2B expression is enriched in hippocampal and temporal regions in adult rats (17), corresponding to areas of the brain affected in many types of epilepsy, and suggesting a possible mechanism of action. It remains to be determined whether antagonists targeted to other NR2 subunits, such as NR2D, which is enriched in midbrain regions (12), affect the physiology and pathophysiology associated with these brain regions.
In order to understand the function of each individual receptor subtype in the intact nervous system, the standard approach that has been employed is gene knockout technology. As is the case for all ion channel complexes with multiple subunits, glutamate receptor gene knockouts need to be complemented with pharmacological agents that are as highly subtype selective as possible. It is particularly desirable to find ligands which bind at subunit interface sites that permit the ligand to interact with determinants on two different subunits. There is a fundamental difficulty with using knockout technology exclusively to assess ion channel function for those families in which a wide variety of different receptor subunits can be assembled as heteromeric complexes in vivo. Ablating the function of a gene that encodes a single type of subunit can result in a very complex phenotype, in which multiple functional complexes with different subunit compositions and physiological roles are affected. The phenotype of NMDA receptor NR1 subunit gene knockout mice, for instance, is not straightforward – concomitant expression of the NR2B subunit is markedly reduced, and neonates do not survive long after birth (18).
One often-effective approach to obtaining subtype selective pharmacological agents to complement gene knockout technology is to start with the pharmacologically-active components of animal venoms. Unfortunately, there have not been a large number of venom components characterized to date that target glutamate receptors. The polyamine toxins found in spider venoms are one such class (19). A promising general source that needs to be explored further are the venoms of cone snails; these have proven to be very rich in families of peptides targeted to ion channels. There are two Conus peptide families that target glutamate receptors identified so far, the conantokins, that act as NMDA receptor antagonists (15, 20–22), and the recently identified con-ikot-ikots, which are targeted to AMPA receptors (C. Walker et. al., submitted for publication). All members of the conantokin family target NMDA receptors, but with differing subtype selectivity; conversely, the con-ikot-ikots appear to exclusively target AMPA receptors, although many properties of this peptide family remain to be investigated (23–25).
For both these families, the number of Conus species that have been identified with venom components targeted to glutamate receptors has been surprisingly small. Among the conantokins, peptides from only six species have been characterized (20, 24–28), and within the con-ikot-ikot family, a peptide from a single Conus species has been characterized.
We recently initiated an expanded effort to systematically examine new Conus species (29) for venom components that selectively target glutamate receptor subtypes. Previous work has used synthetic peptide analogues to identify some of the determinants of NR2A and NR2B subunit selectivity in conantokins (30) (reviewed in 31). To expand upon this approach we have begun to search for sequence variations among native conantokins that govern target specificity.
In this report, we describe a new peptide, conantokin-Br, from Conus brettinghami (1) that has a number of properties that are novel, including higher relative potency for the NR2D subunit than has been previously reported for conantokin peptides. We also characterized chimeras between conantokin-Br and a conantokin with different subunit selectivity, conantokin-R. This provided an opportunity to assess the relative importance of primary sequence variations with respect to activity on different NMDA receptor subtypes. This study has led to a number of structure/function insights for developing ligands with divergent selectivity for different NMDA receptor subtypes based on the conantokin family of conopeptides.
Materials and Methods
Isolation of the conantokin-Br clone
An oligo (DHOG 450 5' GCCCGTGCCTAGGATTA 3') was designed to the signal sequence of known conantokins, conantokin-R and conantokin-G. This oligo was used to probe a cDNA library constructed from the venom duct of C. brettinghami using Southern hybridization as previously described (32). The identified clone was sequenced using Sequenase version 2.0.
Peptide synthesis and cleavage
Native peptides and analogs were synthesized using Fmoc-protected amino acids in an ABI model 430A automated peptide synthesizer, courtesy of Dr. Bob Schackmann and Scott Endicott of the Peptide/DNA Core Facility at the University of Utah. Each peptide was cleaved from 25–50 mg resin using 1 mL of Reagent K (trifluoroacetic acid/H2O/1,2-ethanedithiol/phenol/thioanisole, 82.5:5:2.5:5:5 by volume). Each peptide was agitated for a minimum of 1.5 h at room temperature, with additional 0.5h for each arginine residue in the individual amino acid sequence. The cleavage mixture was filtered and precipitated with cold methyl-tert-butyl ether (MTBE). The crude peptides were then collected by centrifugation at 5000 × g for 8 min, and washed two times with cold MTBE. The washed peptide pellet was dissolved in 60% acetonitrile in 0.1% trifluoroacetic acid and purified using a Vydac C18 semi-preparative HPLC column (10 × 250 mm, 5 um particle size). Each peptide was eluted from the column using a linear gradient of 10% buffer B (90% ACN, 0.1% TFA) to 50% buffer B in 40 min, at 4ml/min.
Oxidative peptide folding
Conantokins containing two C-terminal cysteines (con-R and con-R variants) were oxidized using either Clear-Ox resin (Peptides International, Inc.), as described previously (33), or 1:1 oxidized/reduced glutathione. To prepare Clear-Ox resin for the oxidation reaction, 20 mg of the resin was activated in dichloromethane for 30 min and washed two times each with 1 ml dimethyl formamide, 1 ml methanol, and 1 ml acetonitrile H2O (1:1) solution. For the Clear-Ox reaction, 200 nmoles of peptide per reaction were oxidized for 2 h in a solution containing 20 mg of the activated resin and 100 mM Tris HCl. The reaction was then quenched by adding formic acid (8% final concentration), filtered under vacuum, and washed with 0.01% TFA in H2O to remove the peptide from the resin. For the glutathione reaction, 200 nmoles of peptide per reaction were oxidized using a mixture of the following reagents: 0.01% TFA in H2O, 1M tris HCl/100mM EDTA, 10 mM oxidized/10 mM reduced glutathione, nanopure H2O (1:1:1:7; final peptide concentration: 20 µM). Glutathione reactions were carried out for 45–60 min and quenched with 8% formic acid. Folded peptides were purified on a Vydac C18 semi-preparative HPLC column (10 × 250 mm, 5um particle size) and eluted using a gradient of 10% buffer B to 50% buffer B in 40 min, at 4ml/min.
Heterologous expression of NMDA receptors in Xenopus oocytes
The rat NMDA receptor clones used were NR2A, NR2B, NR2C, NR2D, and NR1-3b; GenBank numbers AF001423, U11419, U08259, U08260, and U08266, respectively. In vivo, the NR1-3 and NR1-b splice variants are expressed in the hippocampus and cerebral cortex of adult rats (34); the NR2 subunits vary in their regional and developmental expression (see Cull-Candy et al., 2001). All of the expression clones were driven by a T7 promoter and were used to make capped RNA (cRNA) for injection into the oocytes of Xenopus laevis frogs. cRNA was prepared in-vitro using Ambion RNA transcription kits (Ambion, Inc.) according to manufacturer’s protocols. To express NMDA receptors, 2–5 ng of cRNA for each subunit was injected per oocyte. Oocytes were maintained at 18 degrees Celsius in ND96 solution (96.0 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, 1mM MgCl2, 5mM HEPES, pH 7.2–7.5) containing antibiotics (septra, amikacin, pen/strep). All voltage-clamp electrophysiology was done using oocytes 1–6 days post-injection.
Two-electrode voltage clamp electrophysiology
All oocytes were voltage clamped at −70mV at room temperature. Oocytes were gravity perfused with Mg2+ -free ND96 buffer (96.0 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, 5 mM HEPES, pH 7.2–7.5). Mg2+ was not included in the ND96 buffer because Mg2+ blocks NMDA receptors at the voltage potential used to clamp oocytes (−70mV). To reduce nonspecific absorption of peptide, bovine serum albumin (BSA) was added to ND96 buffer at a final concentration of 0.1mg/ml. To elicit current from oocytes expressing NMDA receptors, one-second pulses of gravity-perfused agonist solution were administered at intervals of 60s, 90s, or 120s, depending on the rate of receptor recovery from desensitization. Agonist solution was comprised of glutamate and co-agonist glycine suspended in Mg2+ -free ND96 buffer at final concentrations of 200 µM and 20 µM, respectively. Buffer was perfused continuously over the oocytes between agonist pulses, except during equilibration periods. During equilibration period, buffer flow was halted for either 5 or 10 minutes to create a static bath for application of either peptide (suspended in ND96 buffer at various concentrations), or control solution (ND96 buffer alone). The length of equilibration period for each peptide was equal to or greater than the time necessary to achieve maximal current inhibition at a given concentration. The effect of a peptide on NMDA receptor-mediated current was determined by measuring the amplitude of the first agonist-elicited current pulse immediately following the equilibration period as a percentage of the amplitude of the baseline current (agonist-elicited current immediately preceding equilibration period). Data acquisition was automated by a virtual instrument made by Doju Yoshikami of the University of Utah. Concentration-response curves were generated using Prism software (GraphPad Software, Inc.), using the following equation, where nH is the Hill coefficient and IC50 is the concentration of peptide causing half-maximal block: % Response = 100/{1=([peptide]/IC50)nH}.
Molecular modeling
Conantokin-T (con-T) and con-G sequences were aligned to con-Br and its chimera con-R/Br using the CLUSTALW (35) web-based program (Table S1, Supplemental Information). Since the alignments with con-T had a higher percentage identity score, the con-T coordinates (PDB code-name: 1ONT) were used for homology modeling. Modeling of con-R and con-Br/R chimera was precluded due to a lack of an appropriate conantokin model containing a disulfide bridge. γ-carboxyglutamate residues were replaced with glutamic acid residues using PyMOL (36) program. PSIPRED (37), protein secondary structure prediction program, was employed to verify the homology modeling results. To further expand the structural analysis of the modeled conantokins, we submitted resulting coordinates to molecular dynamics (MD) simulation with the AMBER 8.0 (38) program and ff99SB AMBER force field. Conantokins were solvated with the explicit water model TIP3P (39), with the box-size extended approximately 12 Å away from the solute atoms. To neutralize the charge of the system, a magnesium cation was added using the LEaP program from the AmberTools package. To monitor conformational changes for all amino acid residues we used the DSSP method (40) from the Ptraj program. The energy minimization procedure involved 5000 steps of the steepest descent (41) (SD) method and 15000 steps of conjugate gradient (42) (CG) method. To heat up the system, 50 ps MD simulation was carried out. The initial temperature was 100K and the final was 298K. Neither restraints nor constrains were applied during the heating step. Next, 1.25 ns MD simulation with the 2.0 fs time step was performed. The temperature was kept at 298K with Berendsen coupling (43) and 10Å cut-off applied. The SHAKE algorithm (44) was used on hydrogen atoms with tolerance of 0.00005 Å.
Results
Analysis of a clone encoding a conantokin from C. brettinghami
A specimen of Conus brettinghami collected by trawlers from around Manila Bay, Philippines was used to construct a cDNA library. Conus brettinghami is conventionally regarded as a form or subspecies of Conus sulcatus: in this view, Conus sulcatus is regarded as a variable species distributed broadly in the IndoPacific region from Japan to the Bay of Bengal in South India, and east to Fiji. Conus sulcatus is a somewhat confusing taxon. However, the Manila Bay populations are not typical Conus sulcatus, but rather the form known by most taxonomists as Conus sulcatus brettinghami, which has a narrower and smoother shell than the typical form (see Fig 1). Although the standard treatise on cone snail taxonomy (45) regards Conus sulcatus brettinghami as merely a form of Conus sulcatus, more recent data suggests that it is a different species from typical Conus sulcatus (B. Olivera and P. Bouchet, manuscript in preparation). The conantokin peptide described below was characterized from a cDNA clone from C. brettinghami; no independent assessment of cDNAs from typical Conus sulcatus has been carried out to date. Although there is no molecular phylogenetic data on this species complex available at the present time, we regard C. brettinghami as a distinct species from C. sulcatus.
Figure 1.
Forms of Conus sulcatus and Conus brettinghami, the species that produces conantokin-Br. The shells on the extreme left and right are Conus sulcatus; the two middle shells are generally called Conus sulcatus brettinghami; two varieties are shown. As is discussed in the text, we regard Conus brettinghami as a distinct species, not as a subspecies or form of C. sulcatus. The shell on the extreme left is a variant of the typical form. Venom ducts of several specimens of Conus sulcatus brettinghami from Manila Bay were pooled — this was the source of the cDNA clone that yielded the conantokin sequence. The specimens resembled the shell that is second from left.
A cDNA clone encoding a peptide precursor highly homologous to previously identified sequences in the conantokin gene superfamily was identified, as described under Methods. The amino acid sequence predicted from the encoded open reading frame of the clone (Fig 2) is compared to the sequences of three previously characterized conantokin precursors. The high degree of homology is consistent with the gene belonging to the conantokin superfamily.
Figure 2.
Con-Br peptide precursor and predicted mature toxin sequences, compared to previously characterized conantokin precursors. In the mature toxin region, Glu residues either known or predicted to be post-translationally modified to Gla are indicated by γ.
As a member of the conantokin superfamily, most glutamate residues in the mature toxin region are predicted to be post-translationally-modified to γ-carboxyglutamate. The glutamate residue at position 12, however, was predicted not to be post-translationally modified based on the pattern of γ-carboxylation of previously characterized conantokins purified from venom (i.e. a Gla every 3–4 residues in the N-terminal-to-middle of the mature peptide). It has been shown for con-G in NMR studies that this pattern results in the alignment of gla residues on one face of the peptide in a linear fashion to form a calcium binding motif allowing for stabilization of the alpha helix conformation (46). Analysis of the conantokin-T sequence purified from venom suggests that glu residues in positions that do not adhere to this pattern are not modified to gla; for instance, the glu at position 16 (1 AA from the nearest gla) is not post-translationally modified in the native sequence (20). The predicted sequence of con-Br was chemically synthesized as described under Methods.
The mature toxin sequences of the venom-purified conantokins con-G and con-R are shown in Fig. 2 and Table 1 for comparison. Unlike any other conantokin characterized thus far, the sequence of the C. brettinghami clone predicts an aspartate at sequence position 2 rather than the canonical (non-post-translationally-modified) glutamate; the conserved glutamate at this position was found to be critical for NMDA receptor activity in other conantokins (47, 48).
Table 1.
Comparison of predicted mature con-Br sequence with other conantokins
| Conus Species | Conantokin | Amino-Acid Sequence | Ref. |
|---|---|---|---|
| C. brettinghami | Con-Br | GD γγ YS K FI γ RER γ AGRLDLSKFP^ | This work. |
| C. purpurascens | Con-P | GE γγ HS K YQ γ CLR γ IRVNKVQQγC(^) | Gowd et al., 2008 |
| C. parius | Con-Pr1 | GE Dγ YA γ GIR γ YQL I HGKI^ | Teichert et al., 2007 |
| C. parius | Con-Pr2 | DE Oγ YA γ AIR γ YQL K YGKI^ | Teichert et al., 2007 |
| C. parius | Con-Pr3 | GE Oγ VA K WA γ GLR γ KASSN* | Teichert et al., 2007 |
| C. radiatus | Con-R | GE γγ VA K MAA γ LAR γ NIAKGCKVNCYP^ | White et al., 2000 |
| C. lynceus | Con-L | GE γγ VA K MAA γ LAR γ DAVN* | Jimenez et al., 2002 |
| C. tulipa | Con-T | GE γγ YQ K ML γ NLR γ AEVKKNA* | Haack et al., 1990 |
| C. geographus | Con-G | GE γγ LQ γ NQ γ LIR γ KSN* | McIntosh et al., 1984 |
COOH
CONH2
Functional characterization of conantokin-Br
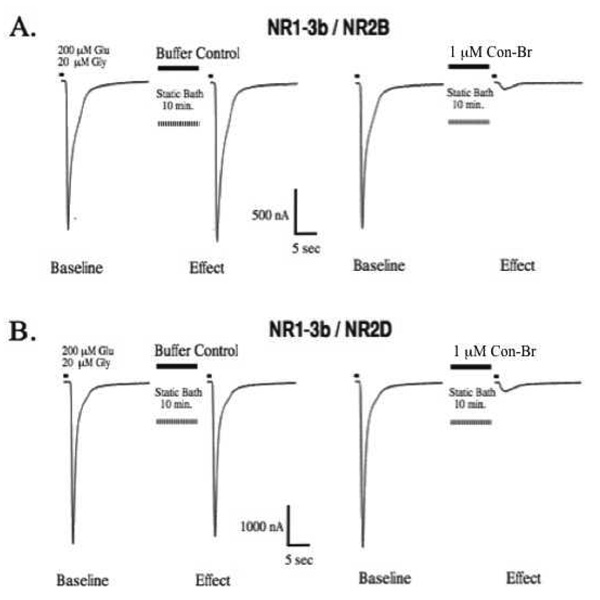
The amino acid sequence of con-Br is clearly homologous with other members of the conantokin family, which are NMDA receptor antagonists. To assess its effects on NMDA receptors, we used two-electrode voltage-clamp electrophysiology to measure current traces from Xenopus oocytes heterologously expressing various combinations of NR1-3b and NR2 subunits. Con-Br was applied to oocytes and allowed to equilibrate for 10 min. The effect of the peptide was determined by measuring the agonist-elicited current in the absence (as a control) and presence of the peptide.
Con-Br (1 µM) blocked most of the current in the NR1-3b/NR2B subtype (Fig 3A), confirming its activity as an NMDA receptor antagonist. Strikingly, a similar level of current inhibition was observed at the same concentration for the NR1-3b/NR2D subtype (Fig. 3b), suggesting con-Br is the most potent conantokin active on NR2D-containing NMDA receptors of any characterized thus far.
Figure 3.
NMDA receptor current traces in the presence and absence of con-Br. (A) Left: NR2B current elicited by agonist pulse before and after buffer control. Right: Baseline NR2B response followed by current elicited following a 10 min application of 1µM con-Br. (B) Effects of buffer control (left) and 1µM con-Br (right) on NR2D.
Concentration-response curves for each of the NR2 subunits (A-D) separately co-expressed with the NR1-3b, subunit were determined (see Table 2 and Fig 4A); con-Br had the greatest inhibitory effect on the NR2B subunit, and the order of potency for con-Br was NR2B>NR2D>NR2A>NR2C.
Table 2.
Approximate IC50 values of con-Br, con-R, and analogs
| Peptide | NR2A IC50
(µM) |
NR2B IC50
(µM) |
NR2C IC50
(µM) |
NR2D IC50
(µM) |
NR2D/ NR2B IC50 Ratio |
|---|---|---|---|---|---|
| Con-Br | .68 | 0.14 | 4.9 | .31 | 2.2 |
| Con-R | .53 | 0.35 | ~10 | >10 | >30 |
| Br/R | -- | 2 | -- | 5.5 | 2.75 |
| R/Br | -- | 3.7 | -- | >100* | >30* |
| Br [R 5–6] | -- | 5.2 | -- | >100 | >20 |
| R [Br 5–6] | -- | 0.40 | -- | 3.1 | 7.8 |
| Br [Y5V] | -- | 3.3 | -- | >100 | >30 |
| R [V5Y] | -- | 0.35 | -- | 3.4 | 9.7 |
Predicted values based on curve fit (see methods)
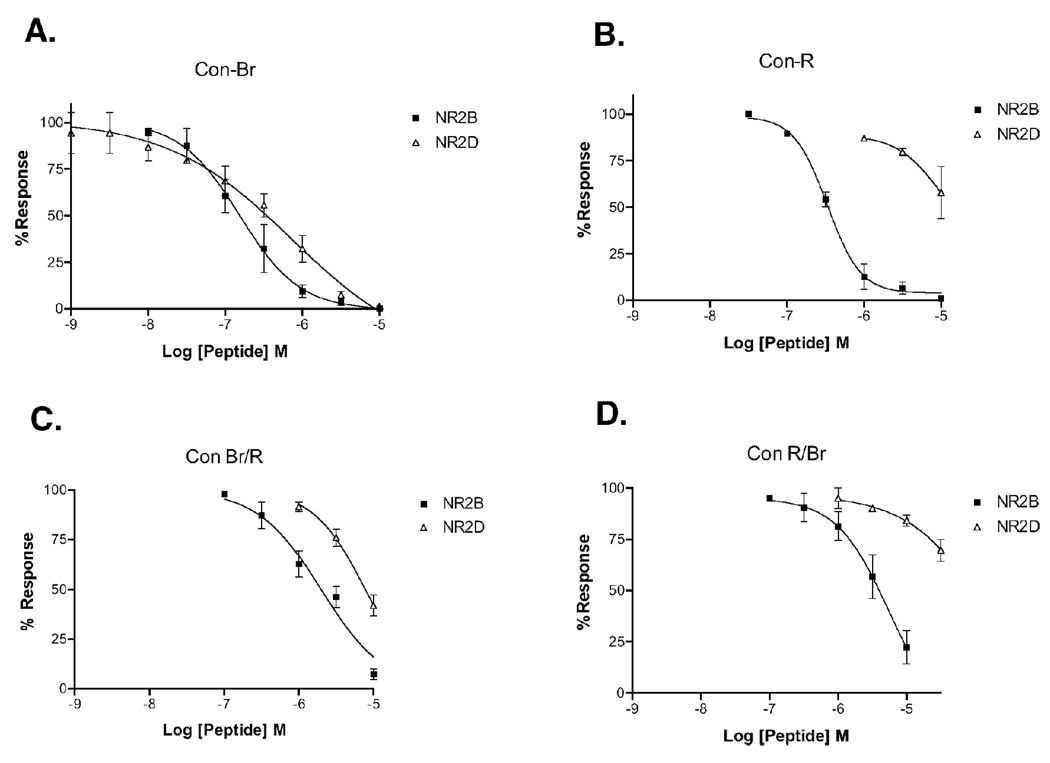
Figure 4.
NR2B and NR2D discrimination of con-Br, con-R, and chimeras. (A) Concentration-response curves for con-Br on NR1-3b/NR2B and NR1-3b/NR2D, showing weak discrimination between the two subtypes. (B) Con-R concentration-response curves for NR2B and NR2D, showing high selectivity toward NR2B. (C) Br/R concentration-response curves for NR2B and NR2D. (D) R/Br concentration-response curves for NR2B and NR2D. R/Br and Br/R sequences are shown in Table 3.
Localization of determinants for subtype selectivity
Previous studies have demonstrated that, for NMDA receptors containing NR2A and NR2B subunits, the structural determinants of subtype selectivity in conantokin-G and conantokin-T can be localized to discrete amino acid residues within the peptides (30, 49). To determine whether the unique subtype selectivity profile of con-Br can be attributed to key localized amino acid residues, we synthesized chimeric analogs of con-Br, which weakly discriminates between NR2B and NR2D, and conantokin-R (con-R), which discriminates strongly against NR2D in favor of NR2B (28). Two chimeric analogs were synthesized with the N-terminal end of one peptide attached to the C-terminal end of the other peptide with a crossover point at the third Gla residue (Gla 10 in con-Br and Gla 11 in con-R). Thus, con-Br/R had the N-terminal residues of con-Br and the C-terminal residues of con-R and con-R/Br, the N-terminal amino acids of con-R and the C-terminal amino acids of con-Br. The analogs were tested for subtype selectivity on NR1-3b/NR2B and NR1-3b/NR2D receptors expressed in Xenopus oocytes (Fig 4 and Table 2). Parent peptides were used as controls. Because previously published results for the con-R activity on NR2D were obtained using different methods (28), a concentration response curve for con-R was generated to obtain an IC50 value for the NR1-3b/NR2D subtype using the present protocol (see Table 2).
The concentration-response curves for the chimeric and parent peptides are shown in Fig 4. As expected, con-R discriminated heavily against the NR2D subunit in favor of NR2B (Fig. 4B), whereas con-Br did not (Fig. 4A). Con-R/Br showed selectivity that favors NR2B, in a similar fashion to con-R (Fig. 4D). Con-Br/R discriminated much more weakly between the NR2B and NR2D, more closely resembling the selectivity of con-Br (Fig. 4C). Interestingly, both analogs were far less potent on both receptor subtypes than the parent peptides (Table 2). The IC50 ratios for NR2D/NR2B of con-Br and con-Br/R are approximately equal (~2–3), while the corresponding ratios of con-R and con-R/Br are also similar (>30). These results suggest that one or more N-terminal residues in con-Br are responsible for the lack of discrimination between NR2B and NR2D.
Localization of key amino acid residues important for subtype selectivity
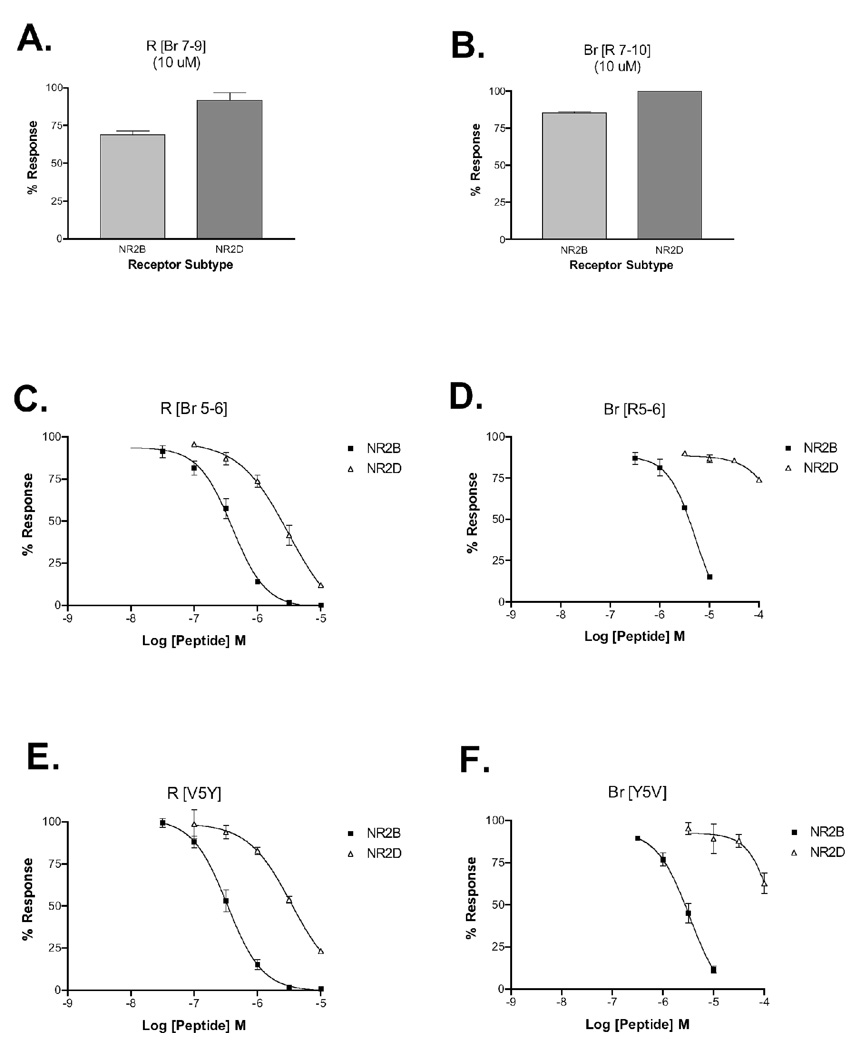
To further localize key N-terminal amino acid determinants for subtype selectivity, we synthesized six con-R and con-Br variants, in which groups of 1–4 amino acids at differing N-terminal sequence positions of one peptide were replaced with the amino acids from the corresponding regions of the other peptide (see Table 3). The subtype selectivity of these analogs with respect to NR1-3b/NR2B and NR1-3b/NR2D was assessed. Variants in which the middle amino acids were replaced (R [Br 7–9] and Br [R 7–10]) were rendered inactive or greatly diminished in potency (Fig. 5A–B). Substituting the amino acids from positions 5–6 of con-Br into the corresponding regions of con-R resulted in a peptide with similar activity to con-R with respect to NR2B but a 3-fold increase in potency for NR2D (Fig. 5C, Table 2). In the context of con-Br, replacing residues at positions 5–6 with those in con-R led to a decrease in activity on both receptor subtypes, but with a ten-fold relative increase in selectivity toward NR2B (Fig 5D, Table 2). These data suggest that one or both residues at positions 5–6 of both peptides are important for determining selectivity among these receptor subtypes.
Table 3.
Amino acid sequences of con-Br and con-R variants
| Peptide Name | Amino-Acid Sequence |
|---|---|
| Con-Br | GD γγ YS K FI γ RER γ AGRLDLSKFP |
| Con-R | GE γγ VA K MAA γ LAR γ NIAKGCKVNCYP |
| Br/R | GD γγ YS K FI γ LAR γ NIAKGCKVNCYP |
| R/Br | GE γγ VA K MAA γ RER γ AGRLDLSKFP |
| Con-Br [R7–10] | GD γγ YS K MAA γ RER γ AGRLDLSKFP |
| Con-R [Br7–9] | GE γγ VA K FI γ LAR γ NIAKGCKVNCYP |
| Con-Br [R5–6] | GD γγ VA K FI γ RER γ AGRLDLSKFP |
| Con-R [Br5–6] | GE γγ YS K MAA γ LAR γ NIAKGCKVNCYP |
| Con-Br [Y5V] | GD γγ VS K FI γ RER γ AGRLDLSKFP |
| Con-R [V5Y] | GE γγ YA K MAA γ LAR γ NIAKGCKVNCYP |
Amino acid substitutions are shown in bold
Figure 5.
Relative inhibitory potencies of con-Br and con-R variants on NR1-3b/NR2B and NR1-3b/NR2D. Con-R [Br 7–9] and con-Br [R 7–10] have greatly reduced potency on NR2B and NR2D (A–B). Con-R [Br 5–6] and con-R [V5Y] retain their potency on NR2B and discriminate less strongly against NR2D (C, E). Con-Br [R 5–6] and con-Br [Y5V] have reduced potency on both subtypes and discriminate strongly against NR2D (D, F). Sequences of con-Br and con-R variants are shown in Table 3.
Two recently characterized conantokins from C. parius (con-Pr1 and con-Pr2) with relatively high affinities for NR2D have a tyrosine at sequence position 5 (25), sharing conserved sequence homology with con-Br (see Table 1). We therefore examined the role of the con-R and con-Br amino acids at position 5 with respect to NR2B-NR2D subtype selectivity. In the context of con-R, replacing the valine at position 5 with tyrosine resulted in a peptide with an equal NR2B potency to con-R and a 3-fold increase in potency for NR2D, indicating that the amino acid at position 5 is a significant determinant of subtype selectivity seen in R [Br5–6] (Fig. 6E, Table 2). Replacing the tyrosine in con-Br with a valine resulted in a 20-fold decrease in potency for NR2B (Fig. 6F, Table 2). Despite this marked decrease in potency, however, this single amino acid substitution dramatically shifted the selectivity of con-Br toward NR2B, resulting in a NR2D/NR2B IC50 ratio similar to that seen in con-R (see Table 2). The shifts in selectivity profiles of the two position 5 variants suggest that the amino acid at this position is a key determinant of NR2B-NR2D selectivity.
Figure 6.
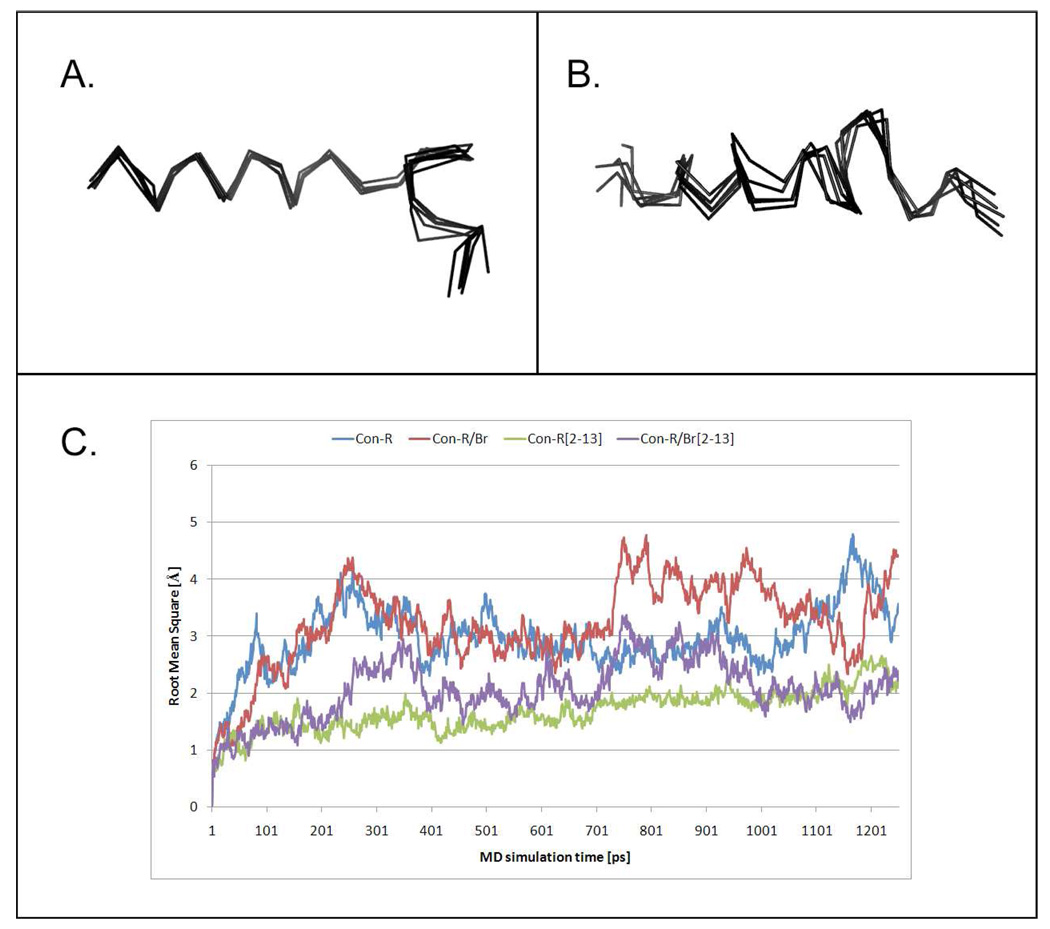
Molecular modeling of con-Br and con-R/Br. Ribbon representation of con-Br (A) and con-R/Br chimera (B). Superimposion of five averaged structures generated with frequency of 250 ps from 1.25 ns MD simulation run. C - RMS deviation for conantokins con-Br and the con-R/Br chimera calculated during the MD simulation for each residue (all atoms were included). RMS deviation was also calculated for chosen helical region, which included residues 2–13 for both peptides.
Molecular modeling
A characteristic feature of conantokins is their helical conformation stabilized by interactions of Gla residues with divalent cations (31, 46, 50). Modeling of two linear conantokins, con-Br and the con-R/Br chimera, was performed using the NMR model of con-T, as described in the Methods section. Con-Br and the chimera exhibited a substantial amount of helical conformation (Figure S1, Supplemental Information) that was further confirmed using the secondary structure prediction program, PSIPRED (37, 51) (Figure S2, Supplemental Material). To better characterize conformational properties of con-Br and con-R/Br, the initial model structures containing Mg2+ cations (used as counterions to neutralize the charge and solvated by the explicit water model) were submitted to molecular dynamics analysis. The MD results are summarized in Figure 6 and Tables S2 and S3 (Supplemental Information).
Figure 6 shows superimposition of five averaged structures of con-Br and con-R/Br generated from 1.25 ns MD run with 250 ps frequency. The results of the MD simulations confirmed that the helical structure between the residues 2–13 is dominant for both conantokins. The helical structure is particularly pronounced in con-Br between the residues 2 and 12 and for the con-R/Br chimera between the residues 2–13 with three additional residues (17–19) forming a short helical fragment. From the superimposition we are able to observe that the conformation of the con-R/Br chimera is somewhat less stable in an aqueous environment than con-Br. On the other hand, the structure of con-Br is very stable during the MD simulation. In both cases the helical motif is present between residues 2–13. To measure the stability of the conformations, we calculated the RMS value during MD simulations for both conantokins as well as for the selected helical region between residues 2–13 (Figure 6C). It is notable that the calculated values of the RMSD are lower for calculation of the helical region, suggesting that the helices stabilize the structure of the peptides.
Discussion
In this work, we characterized con-Br, a conantokin peptide from a species not previously analyzed, Conus brettinghami. A comparison of the sequence of con-Br to known conantokins is shown in Table 1. One functional difference between conantokin-Br and previously characterized conantokins is the relatively high potency for NMDA receptors that have an NR2D subunit. All conantokins characterized so far have a preference for NR2B; presumably the relevant target in the fish prey of these piscivorous species is an NMDA receptor complex with a subunit similar to the mammalian NR2B subtypes. However, the relative affinities for other NMDA receptor subtypes vary, and conantokin-Br is noteworthy in the relatively high potency for NR2D containing NMDA receptor complexes. In contrast, con-R has low potency (>10um) for the NR2D subtype.
We used the differential affinities of conantokin-Br and conantokin-R for the NR2D subunit to evaluate chimeras and analogs of conantokin-Br and conantokin-R for activity on NMDA receptors containing the NR2B and NR2D subunits. When chimeras were made of conantokin-Br and conantokin-R, both of the resulting chimeras had a significantly lowered potency for both NMDA receptor subtypes, compared to the native parent peptides. However, even at the lower affinities, the ratio between NR2B and NR2D differed between the chimeras; only the chimera with the N-terminal half of con-Br showed a comparable potency for NR2D and NR2B, while the chimera with the N-terminal half of con-R showed a ~30-fold lower potency for NR2D compared to NR2B. These results suggest that despite the fall in potency for both receptor subtypes, there were determinants for differential binding to NR2D in the N-terminal region. A residue switch at position 5 between con-Br and con-R was particularly revealing. In (V5Y) con-R, the potency of the peptide for the NR2B subtype was not significantly perturbed, but a higher potency for NR2D subtype was observed. This result suggests that the presence of tyrosine in con-Br contributes to its higher potency for the NR2D subtype, although there may be other determinants that further improve the potency for this subtype that remain to be identified. To this end, we also synthesized a chimeric peptide to test the role of Asp at position 2 in con-Br, the other likely determinant of selectivity; however, the data suggest that this is not a significant determinant of NR2D selectivity (not shown). The reciprocal analog, (Y5V) con-Br had very poor relative potency for NR2D, thus confirming that position 5 is an important determinant of NR2D selectivity -- however, this single residue substitution also lowered the potency for NR2B approximately 20-fold, suggesting that it had lower binding energy for shared sites in both the NR2B and NR2D subtypes. Interestingly, the amino acid at position 5 has been shown to be essential for activity in conantokin-T (48) and is a key determinant of NR2A-NR2B selectivity in conantokin-G (49). Furthermore, all native conantokins that have a tyrosine at position 5 have a relatively higher potency for NR2D (see Teichert et al., 2007). Thus, the residue at this position may be partially predictive of the subunit selectivity of a given conantokin.
From the data included in this work, it cannot be directly ruled out that the C-terminal disulfide loop in con-R analogues may confer properties that govern peptide activity and subunit selectivity. Several lines of evidence suggest this possibility is highly unlikely, however. First, earlier structure function studies of con-R using circular dichroism analysis and [3H]MK-801 binding displacement assays suggest that structure and NMDA receptor activity are virtually unchanged in C-terminal truncation variants comprising the first 17 amino acids of con-R which exclude the C-terminal disulfide bond (47, 52). Secondly, unpublished data from our recent characterization of con-P, which contains an unusually long disulfide loop, (24) shows similar activity and subunit selectivity in both reduced alkylated and oxidized forms, indicating that the disulfide bond plays little or no role in the NMDA antagonist activity of the peptide. Finally, extensive structure-activity studies of con-G and con-T, in addition to con-R (reviewed in 31), and the data in this work, suggest that structural properties important for peptide activity are conferred by the N-terminal amino acids of the peptide.
From the discovery of the first conantokins and the presence of γ-carboxyglutamate residues in these peptides, combined with the similarity in the spacing of the Gla residues to the Gla domains of mammalian blood-clotting factors, it was immediately hypothesized that these peptides assumed a helical structure stabilized by Gla residues chelated to Ca2+ or other divalent cations. This has been strongly supported by a variety of NMR studies (31, 46, 50, 53). Molecular modeling and MD studies on con-Br and con-R/Br confirmed substantial amount of the α-helical conformation in the N-terminal parts of the peptides. The data obtained here also suggest that the conantokin framework of Gla residues should not be regarded as a rigid helical scaffold from which side chains are displayed that can interact with partner residues on their specific NMDA receptor targets. Such a model works reasonably well for the (V5Y) con-R analog: the substitution of Tyr for Val clearly increases the potency of conantokin-R for the NR2D subunit, without a large effect on the potency for the NR2B subunit. However, in most other chimeras or substitutions, the significant decrease in potency observed for the different NMDA subunits tested indicate that the potency of conantokins for particular NMDA receptors is a more subtle function of the entire structure; the non-reciprocal nature of the complementary substitutions at position 5 of con-R and con-Br in terms of NR2B potency underline this general perception.
Although differences in selectivity uncovered so far are modest, the lack of subtype selective NMDA receptor ligands makes the development and further exploration of the conantokin family highly desirable. The conantokins are widespread across Conus and clearly, the concerted discovery strategy used for other families of Conus peptides (29, 54) gives grounds for optimism that a broader survey of the conantokin gene family across the entire phylogenetic range of cone snails may yield novel conantokin peptides and their derivatives that will prove useful for identifying additional determinants of subtype selectivity. The first step in being able to efficiently scan a library of natural sequences for potentially novel-targeting specificity is to have information regarding which residues may be selectivity determinants; the work that we have described in this report is a significant step in this important direction.
It is highly desirable to continue developing NR2D specific ligands. The NR2D subunit confers a number of physiologically distinct properties to NMDA receptor complexes, including low sensitivity to Mg2+ block, resistance to H+ block, and prolonged deactivation kinetics (reviewed in 12), which may make neurons expressing this subunit particularly vulnerable to excitotoxicity. In addition, expression of the NR2D subunit is more regionally restricted within the brain than NR2A or NR2B subunits, with expression primarily localized to the diencephalons and midbrain (34), and an enrichment of expression in the substantia nigra and striatum (55, 56). Thus, the development of an NR2D-specific ligand may help to evaluate the role of this NMDA receptor subtype in a number of pathological conditions affecting these regions, such as Parkinson’s disease.
Supplementary Material
Acknowledgements
This work was supported by GM48677 from the National Institute of General Medical Science (to Baldomero M. Olivera). Vernon Dean Twede was funded by a student fellowship from the Neuroscience Training Grant NIH NIDCD 5T32 DC008553. Rajmund Kaźmierkiewicz acknowledges support from fund DS/B50A-4-162-8.
We would like to thank Tiffany Han and Aleksandra Walewska for their assistance of with some of the bioassays and oxidation reactions.
The abbreviations used are
- NMDA
N-methyl-D-aspartate
- AMPA
alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid
- con
conantokin
- Gla
γ, γ-carboxyglutamate
- ACN
acetonitrile
- HPLC
high performance liquid chromatography
- MD
molecular dynamics
- RMS
root mean square
- RMSD
root mean square deviation
- DSSP
define secondary structure of protein
- PDB
protein data bank
Footnotes
Supplemental Information
Three supplemental tables summarizing homology modeling experiments and two figures showing homology models and secondary structure prediction results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Coomans HE, Moolenbeek RG, Wils E. Alphabetical revision of the (sub)species in recent Conidae 5. baccatus to byssinus, including Conus brettinghami nomen novum . Basteria. 1982;46(1/4):3–67. [Google Scholar]
- 2.Simeone TA, Sanchez RM, Rho JM. Molecular biology and ontogeny of glutamate receptors in the mammalian central nervous system. J Child Neurol. 2004;19:343–360. doi: 10.1177/088307380401900507. discussion 361. [DOI] [PubMed] [Google Scholar]
- 3.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 4.Hille B. Ion Channels of Excitable Membranes. Third Edition. Sunderland, MA: Sinauer Associates, Inc; 2001. [Google Scholar]
- 5.Hollmann M, Heinemann S. Cloned glutamate receptors. Ann. Rev. Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 6.Meldrum BS. The role of glutamate in epilepsy and other CNS disorders. Neurology. 1994;44:S14–S23. [PubMed] [Google Scholar]
- 7.Ulas J, Weihmuller FB, Brunner LC, Joyce JN, Marshall JF, Cotman CW. Selective increase of NMDA-sensitive glutamate binding in the striatum of Parkinson's disease, Alzheimer's disease, and mixed Parkinson's disease/Alzheimer's disease patients: an autoradiographic study. J Neurosci. 1994;14:6317–6324. doi: 10.1523/JNEUROSCI.14-11-06317.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozawa S, Kamiya H, Tsuzuki K. Glutamate receptors in the mammalian central nervous system. Prog Neurobiol. 1998;54:581–618. doi: 10.1016/s0301-0082(97)00085-3. [DOI] [PubMed] [Google Scholar]
- 9.Childers WE, Jr, Baudy RB. N-methyl-D-aspartate antagonists and neuropathic pain: the search for relief. J Med Chem. 2007;50:2557–2562. doi: 10.1021/jm060728b. [DOI] [PubMed] [Google Scholar]
- 10.Bisaga A, Popik P, Bespalov AY, Danysz W. Therapeutic potential of NMDA receptor antagonists in the treatment of alcohol and substance use disorders. Expert Opin Investig Drugs. 2000;9:2233–2248. doi: 10.1517/13543784.9.10.2233. [DOI] [PubMed] [Google Scholar]
- 11.Bisaga A, Popik P. In search of a new pharmacological treatment for drug and alcohol addiction: N-methyl-D-aspartate (NMDA) antagonists. Drug Alcohol Depend. 2000;59:1–15. doi: 10.1016/s0376-8716(99)00107-6. [DOI] [PubMed] [Google Scholar]
- 12.Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr. Opin. Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 13.Cavara NA, Hollmann M. Shuffling the deck anew: how NR3 tweaks NMDA receptor function. Mol Neurobiol. 2008;38:16–26. doi: 10.1007/s12035-008-8029-9. [DOI] [PubMed] [Google Scholar]
- 14.Schuler T, Mesic I, Madry C, Bartholomaus I, Laube B. Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in N-methyl-D-aspartate receptor assembly. J Biol Chem. 2008;283:37–46. doi: 10.1074/jbc.M703539200. [DOI] [PubMed] [Google Scholar]
- 15.Layer RT, Wagstaff JD, White HS. Conantokins: peptide antagonists of NMDA receptors. Curr Med Chem. 2004;11:3073–3084. doi: 10.2174/0929867043363901. [DOI] [PubMed] [Google Scholar]
- 16.Micu I, Jiang Q, Coderre E, Ridsdale A, Zhang L, Woulfe J, Yin X, Trapp BD, McRory JE, Rehak R, Zamponi GW, Wang W, Stys PK. NMDA receptors mediate calcium accumulation in myelin during chemical ischaemia. Nature. 2006;439:988–992. doi: 10.1038/nature04474. [DOI] [PubMed] [Google Scholar]
- 17.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 18.Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, Stewart CL, Morgan JI, Connor JA, Curran T. Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron. 1994;13:325–338. doi: 10.1016/0896-6273(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 19.Estrada G, Villegas E, Corzo G. Spider venoms: a rich source of acylpolyamines and peptides as new leads for CNS drugs. Nat Prod Rep. 2007;24:145–161. doi: 10.1039/b603083c. [DOI] [PubMed] [Google Scholar]
- 20.Haack JA, Rivier J, Parks TN, Mena EE, Cruz LJ, Olivera BM. Conantokin T: a g-carboxyglutamate-containing peptide with N-methyl-D-aspartate antagonist activity. J. Biol. Chem. 1990;265:6025–6029. [PubMed] [Google Scholar]
- 21.Hammerland LG, Olivera BM, Yoshikami D. Conantokin-G selectively inhibits NMDA-induced currents in Xenopus oocytes injected with mouse brain mRNA. Eur. J. Pharmacol. 1992;226:239–244. doi: 10.1016/0922-4106(92)90067-6. [DOI] [PubMed] [Google Scholar]
- 22.Mena EE, Gullak MF, Pagnozzi MJ, Richter KE, Rivier J, Cruz LJ, Oivera BM. Conantokin-G: a novel peptide antagonist to the N-methyl-D-aspartate acid (NMDA) receptor. Neurosci. Lett. 1990;118:241–244. doi: 10.1016/0304-3940(90)90637-o. [DOI] [PubMed] [Google Scholar]
- 23.Donevan SD, McCabe RT. Conantokin-G is an NR2B-selective competitive antagonist of N-methyl-D-aspartate receptors. Mol. Pharmacol. 2000;58:614–623. doi: 10.1124/mol.58.3.614. [DOI] [PubMed] [Google Scholar]
- 24.Gowd KH, Twede V, Watkins M, Krishnan KS, Teichert RW, Bulaj G, Olivera BM. Conantokin-P, an unusual conantokin with a long disulfide loop. Toxicon. 2008;52:203–213. doi: 10.1016/j.toxicon.2008.04.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teichert RW, Jimenez EC, Twede V, Watkins M, Hollmann M, Bulaj G, Olivera BM. Novel conantokins from Conus parius venom are specific antagonists of N-methyl-D-aspartate receptors. J Biol Chem. 2007;282:36905–36913. doi: 10.1074/jbc.M706611200. [DOI] [PubMed] [Google Scholar]
- 26.Jimenez EC, Donevan SD, Walker C, Zhou L-M, Nielsen J, Cruz LJ, Armstrong H, White HS, Olivera BM. Conantokin-L, a new NMDA receptor antagonist: determinants for anticonvulsant potency. Epilepsy Res. 2002;51:73–80. doi: 10.1016/s0920-1211(02)00101-8. [DOI] [PubMed] [Google Scholar]
- 27.McIntosh JM, Olivera BM, Cruz LJ, Gray WR. g-Carboxyglutamate in a neuroactive toxin. J. Biol. Chem. 1984;259:14343–14346. [PubMed] [Google Scholar]
- 28.White HS, McCabe RT, Armstrong H, Donevan S, Cruz LJ, Abogadie FC, Torres J, Rivier JE, Paarman I, Hollmann M, Olivera BM. In vitro and in vivo characterization of conantokin-R, a selective NMDA antagonist isolated from the venom of the fish-hunting snail Conus radiatus . J. Pharmacol. Exp. Therap. 2000;292:425–432. [PubMed] [Google Scholar]
- 29.Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: a model for concerted pharmacological discovery. Mol Interv. 2007;7:251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- 30.Sheng Z, Dai Q, Prorok M, Castellino FJ. Subtype-selective antagonism of N-methyl-D-aspartate receptor ion channels by synthetic conantokin peptides. Neuropharmacology. 2007;53:145–156. doi: 10.1016/j.neuropharm.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prorok M, Warder SE, Blandl T, Castellino FJ. Calcium binding properties of synthetic gamma-carboxyglutamic acid-containing marine cone snail "sleeper" peptides, conantokin-G and conantokin-T. Biochemistry. 1996;35:16528–16534. doi: 10.1021/bi9621122. [DOI] [PubMed] [Google Scholar]
- 32.Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. Constant and hypervariable regions in conotoxin propeptides. EMBO J. 1990;1:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green BR, Bulaj G. Oxidative folding of conotoxins in immobilized systems. Protein Pept Lett. 2006;13:67–70. doi: 10.2174/092986606774502162. [DOI] [PubMed] [Google Scholar]
- 34.Laurie DJ, Bartke I, Schoepfer R, Naujoks K, Seeburg PH. Regional, developmental and interspecies expression of the four NMDAR2 subunits, examined using monoclonal antibodies. Brain Res Mol Brain Res. 1997;51:23–32. doi: 10.1016/s0169-328x(97)00206-4. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 37.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 38.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorgensen WL, Chandrasekhar J, Madura J, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:10. [Google Scholar]
- 40.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:61. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 41.Arfken G. Mathematical Methods for Physicists. 3. Orlando, FL: Academic Press; 1985. The Method of Steepest Descents; pp. 428–443. [Google Scholar]
- 42.Fletcher R, M RC. Function minimization by conjugate gradients. Computer Journal. 1964;7:6. [Google Scholar]
- 43.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola AJRH. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:7. [Google Scholar]
- 44.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:15. [Google Scholar]
- 45.Röckel D, Korn W, Kohn AJ. Manual of the Living Conidae. Vol. I. Wiesbaden, Germany: Indo-Pacific Region, Verlag Christa Hemmen; 1995. [Google Scholar]
- 46.Rigby AC, Baleja JD, Li L, Pedersen LG, Furie BC, Furie B. Role of gamma-carboxyglutamic acid in the calcium-induced structural transition of conantokin G, a conotoxin from the marine snail Conus geographus. Biochemistry. 1997;36:15677–15684. doi: 10.1021/bi9718550. [DOI] [PubMed] [Google Scholar]
- 47.Blandl T, Zajicek J, Prorok M, Castellino FJ. Sequence requirements for the N-methyl-D-aspartate receptor antagonist activity of conantokin-R. J Biol Chem. 2001;276:7391–7396. doi: 10.1074/jbc.M006648200. [DOI] [PubMed] [Google Scholar]
- 48.Warder SE, Blandl T, Klein RC, Castellino FJ, Prorok M. Amino acid determinants for NMDA receptor inhibition by conantokin-T. J Neurochem. 2001;77:812–822. doi: 10.1046/j.1471-4159.2001.00281.x. [DOI] [PubMed] [Google Scholar]
- 49.Klein RC, Prorok M, Galdzicki Z, Castellino FJ. The amino acid residue at sequence position 5 in the conantokin peptides partially governs subunit-selective antagonism of recombinant N-methyl-D-aspartate receptors. J. Biol. Chem. 2001;276:26860–26867. doi: 10.1074/jbc.M102428200. [DOI] [PubMed] [Google Scholar]
- 50.Chen Z, Blandl T, Prorok M, Warder SE, Li L, Zhu Y, Pedersen LG, Ni F, Castellino FJ. Conformational changes in conantokin-G induced upon binding of calcium and magnesium as revealed by NMR structural analysis. J. Biol. Chem. 1998;273:16248–16258. doi: 10.1074/jbc.273.26.16248. [DOI] [PubMed] [Google Scholar]
- 51.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:2. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 52.Blandl T, Warder SE, Prorok M, Castellino FJ. Structure-function relationships ofthe NMDA receptor antagonist peptide, conantokin-R. FEBS Lett. 2000;470:139–146. doi: 10.1016/s0014-5793(00)01309-0. [DOI] [PubMed] [Google Scholar]
- 53.Skjaebaek N, Nielsen KJ, Lewis RJ, Alewood P, Craik DJ. Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy. J. Biol. Chem. 1997;272:2291–2299. doi: 10.1074/jbc.272.4.2291. [DOI] [PubMed] [Google Scholar]
- 54.Olivera BM, Quik M, Vincler M, McIntosh JM. Subtype-selective conopeptides targeted to nicotinic receptors: Concerted discovery and biomedical applications. Channels (Austin) 2. 2008 doi: 10.4161/chan.2.2.6276. [DOI] [PubMed] [Google Scholar]
- 55.Counihan TJ, Landwehrmeyer GB, Standaert DG, Kosinski CM, Scherzer CR, Daggett LP, Velicelebi G, Young AB, Penney JB., Jr Expression of N-methyl-D-aspartate receptor subunit mRNA in the human brain: mesencephalic dopaminergic neurons. J Comp Neurol. 1998;390:91–101. [PubMed] [Google Scholar]
- 56.Kuppenbender KD, Standaert DG, Feuerstein TJ, Penney JB, Jr, Young AB, Landwehrmeyer GB. Expression of NMDA receptor subunit mRNAs in neurochemically identified projection and interneurons in the human striatum. J Comp Neurol. 2000;419:407–421. doi: 10.1002/(sici)1096-9861(20000417)419:4<407::aid-cne1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.