Abstract
Do intellectual property (IP) rights on existing technologies hinder subsequent innovation? Using newly-collected data on the sequencing of the human genome by the public Human Genome Project and the private firm Celera, this paper estimates the impact of Celera’s gene-level IP on subsequent scientific research and product development. Genes initially sequenced by Celera were held with IP for up to two years, but moved into the public domain once re-sequenced by the public effort. Across a range of empirical specifications, I find evidence that Celera’s IP led to reductions in subsequent scientific research and product development on the order of 20 to 30 percent. Taken together, these results suggest that Celera’s short-term IP had persistent negative effects on subsequent innovation relative to a counterfactual of Celera genes having always been in the public domain.
Innovation is a key driver of economic growth, but competitive markets may under-incentivize innovation due to the public good nature of ideas (Nelson, 1959; Arrow, 1962). Intellectual property (IP) rights, such as patents and copyrights, aim to incentivize innovation by allowing firms to capture a higher share of the social returns to their research investments. Traditional evaluations of the effectiveness of IP have focused on whether the prospect of obtaining IP rights stimulates the development of new technologies. However, in many markets technological change is cumulative, in the sense that product development results from several steps of invention and research. In markets where innovation is cumulative, the overall effectiveness of IP in promoting innovation also depends on a second, less studied question: do IP rights on existing technologies hinder subsequent innovation? The contribution of this paper is to provide empirical evidence on this second question by investigating how one form of IP on the human genome influenced subsequent scientific research and product development.
To fix ideas, suppose the firm Celera holds IP on a human gene, and that Pfizer discovers a genetic diagnostic test based on Celera’s gene. Will Celera’s IP discourage Pfizer from developing this test? In a perfect contracting environment with no transaction costs, Celera and Pfizer would negotiate a licensing agreement such that cumulative research is not hindered. However, transaction costs may cause licensing negotiations to break down, deterring some socially desirable research. While the theoretical literature on this question is well-developed (Scotchmer, 1991; Green and Scotchmer, 1995; Bessen, 2004), empirical evidence is scarce.
The empirical context analyzed in this paper is the sequencing of the human genome by the public Human Genome Project and the private firm Celera. The public effort began in 1990, and required that all sequenced genes be placed in the public domain. Celera’s effort began in 1999, and ended in 2001 when Celera disclosed an incomplete draft genome. The public effort continued, and by 2003 had sequenced all genes in Celera’s 2001 draft. Between 2001 and 2003, Celera used a contract law-based form of IP to protect genes sequenced by Celera but not yet sequenced by the public effort. This IP enabled Celera to sell its data for substantial fees, and required firms to negotiate licensing agreements with Celera for any resulting commercial discoveries - even though it was publicly known at the time that all of Celera’s genes would be sequenced by the public effort, and thus be in the public domain, by 2003. Figure 1 summarizes a timeline of these key events.
Figure 1.
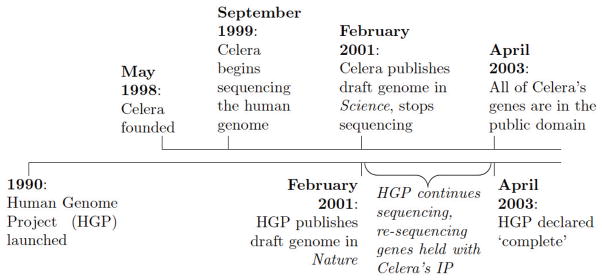
Timeline of Key Events
Notes: This figure summarizes the key events analyzed in this paper. For details, see Collins and Galas (1993), Venter et al. (1998), Venter (2000), Lander et al. (2001), Venter et al. (2001), and Wade (2003).
How did Celera’s gene-level IP influence subsequent scientific research and product development? To investigate this question, I construct a new dataset that tracks the timing of gene sequencing and Celera’s IP across the human genome, linked to gene-level measures of scientific research and product development. Specifically, I trace cumulative innovation by compiling data on links between genes and phenotypes, which are observable traits or characteristics. For example, the link between variation on the HTT gene and Huntington’s disease represents a genotype-phenotype link. For each gene, I collect data on publications investigating genotype-phenotype links, on successfully generated knowledge about genotype-phenotype links, and on the development of gene-based diagnostic tests available to consumers.1
A simple cross-tabulation illustrates how this data can be used to investigate how Celera’s IP influenced subsequent innovation. Table 1 compares subsequent innovation outcomes for Celera genes relative to non-Celera genes sequenced in the same year. Taken at face value, these data suggest that Celera’s IP led to economically and statistically significant reductions in subsequent scientific research and product development. Celera genes had an average of 1.2 publications by 2009, relative to 2.1 publications for non-Celera genes sequenced in the same year. About 3 percent of Celera genes were used in a gene-based diagnostic test as of 2009, relative to 5.4 percent of non-Celera genes sequenced in the same year.
Table 1.
Innovation Outcomes for Celera & non-Celera Genes Sequenced in 2001
| (1) Celera mean | (2) Non-Celera mean | (3) difference [(1)-(2)] | (4) p-value of difference | |
|---|---|---|---|---|
| publications in 2001–2009 | 1.239 | 2.116 | −0.877 | [0.000] |
| 1(known, uncertain phenotype) | 0.401 | 0.563 | −0.162 | [0.000] |
| 1(known, certain phenotype) | 0.046 | 0.073 | −0.027 | [0.000] |
| 1(used in any diagnostic test) | 0.030 | 0.054 | −0.024 | [0.000] |
|
| ||||
| N | 1,682 | 2,851 | ||
Notes: This table compares subsequent innovation outcomes for Celera genes relative to non-Celera genes sequenced in the same year. Gene-level observations. Sample in Column (1) includes all Celera genes; sample in Column (2) includes all non-Celera genes sequenced in 2001. The p-value reported in Column (4) is from a t-test for a difference in mean outcomes across Column (1) and Column (2). See text and online appendix for more detailed data and variable descriptions.
These simple differences in means could reflect a negative effect of Celera’s IP on subsequent research, or could reflect that Celera’s genes had lower inherent potential for follow-on research. Because the institutional context suggests that selection bias could be a concern, I present estimates from two additional empirical tests that directly address selection. First, I restrict attention to within-gene variation in IP and test whether the removal of Celera’s IP increased subsequent innovation. Second, I limit the sample to Celera genes and test for a link between the amount of time a gene was held with Celera’s IP and subsequent innovation. These additional empirical tests appear to eliminate selection bias in the following sense: within the sample of ~1,600 Celera genes, proxies for the ex ante expected value of a gene do not predict the timing of when genes were re-sequenced by the public effort. The estimates from these two additional empirical tests are roughly the same magnitude as the estimates from the simple cross-tabulation: Celera’s IP appears to have generated economically and statistically significant reductions in subsequent scientific research and product development, on the order of 20 to 30 percent.
This analysis does not evaluate the overall welfare consequences of Celera’s entry, which may have been spurred by the prospect of obtaining IP. If Celera’s entry spurred faster sequencing of the human genome, the overall timing of genome-related innovation likely shifted earlier in time, which would have had welfare gains even if Celera’s IP in isolation hindered innovation. Rather, these results suggest that, holding Celera’s entry constant, an alternative lump-sum reward mechanism may have had social benefits relative to Celera’s chosen form of IP.2
This paper joins a handful of recent papers documenting evidence that IP may hinder subsequent innovation (Murray and Stern, 2007; Huang and Murray, 2009; Murray et al., 2008). Of particular note is the work of Murray et al. (2008), who analyze short-term IP the firm DuPont held on certain types of genetically engineered mice. Using a dataset of matched mouse-article pairs, the authors document that the removal of IP increased citations to scientific papers on affected mice by 20 to 40 percent relative to papers on unaffected mice - very similar to the magnitude of my estimates.3
The paper proceeds as follows. Section 1 provides a brief scientific background and a description of my data construction. Section 2 describes the empirical context, and Section 3 presents the empirical results. Section 4 links the empirical results back to previous theoretical and empirical analyses, in an attempt to interpret which types of transaction costs were likely most important in this context. Section 5 concludes.
1 Preliminaries: Scientific primer and data construction
This section provides a brief scientific background and describes my data construction. An appendix discusses my data construction in more detail.
1.1 Scientific primer
In classical genetics, a gene is defined as a unit of inheritance. Physically, a gene is defined as a stretch of deoxyribonucleic acid (DNA), which itself is a sequence comprised of four nucleotide bases - adenine (A), cytosine (C), guanine (G), and thymine (T).4 ‘Sequencing the genome’ refers to the process of determining the exact order of these nucleotide bases in the entire set of hereditary information for a given organism.
Genes affect health by generating proteins, which carry out functions in the human body. More precisely, genes code for messenger ribonucleic acids (mRNAs): a gene’s sequence of nucleotide bases dictates the sequence of nucleotide bases in a generated mRNA. In turn, mRNAs code for proteins: an mRNA’s sequence of nucleotide bases dictates what protein is generated. The intermediate step - mRNA - is important because a gene can code multiple proteins by coding multiple mRNAs. Reflecting this, sequencing the genome involved sequencing mRNAs.
Proteins induce variation in observable traits or characteristics of an organism, known as phenotypes. Known genotype-phenotype links can be combined with sequenced genes to form the basis for genetic tests. A gene can be involved in multiple genotype-phenotype links, and a genotype-phenotype link can involve more than one gene.
I use genes as my unit of analysis. Genes are stable scientific units, whereas the number of known mRNAs and known genotype-phenotype links in part reflects the amount of research invested in a given gene. Table 2 presents summary statistics on the gene-level data. My data include 27,882 currently known genes. The median length of a gene is around 17,000 nucleotide bases (Scherer, 2008).
Table 2.
Summary Statistics for Gene-Level Data
| mean | median | standard deviation | minimum | maximum | |
|---|---|---|---|---|---|
| Panel A: Sequencing & Celera’s IP | |||||
| year sequence disclosed | 2002.962 | 2001 | 3.551 | 1999 | 2009 |
| 1(Celera gene) | 0.060 | 0 | 0.238 | 0 | 1 |
|
| |||||
| Panel B: Outcome variables | |||||
| publications in 2001–2009 | 2.197 | 0 | 9.133 | 0 | 231 |
| 1(known, uncertain phenotype) | 0.453 | 0 | 0.498 | 0 | 1 |
| 1(known, certain phenotype) | 0.081 | 0 | 0.273 | 0 | 1 |
| 1(used in any diagnostic test) | 0.060 | 0 | 0.238 | 0 | 1 |
|
| |||||
| N = 27,882 | |||||
Notes: Gene-level observations. Note that the mean year of disclosure is affected by left-censoring since a disclosure year of 1999 represents a gene sequenced in or before 1999 (1999 is the earliest year any observations appear in the RefSeq database). See text and the online appendix for more detailed data and variable descriptions.
1.2 Tracking the public and private genome sequencing efforts
I track the timing of the public sequencing effort using data from the US National Institutes of Health’s RefSeq database. I define the year a gene was sequenced as the first year any mRNA was disclosed for that gene. The median gene was sequenced in 2001 (Panel A of Table 2).5
Istrail et al. (2004) compare Celera’s 2001 draft genome with a snapshot of the public data. Building on their analysis, I am able to determine which genes were included in Celera’s 2001 draft genome and the dates at which those genes eventually appeared in the public data. I define a Celera gene as a gene for which all known mRNAs were initially sequenced by Celera.6 Of the 27,882 currently known genes, 1,682 - about 6 percent - were held with Celera’s IP for some amount of time (Panel A of Table 2). Because Celera’s draft genome was disclosed in 2001, I code Celera genes as having been sequenced in 2001.
1.3 Measuring scientific research and product development outcomes
I collect four outcome variables: three measures of scientific research, and one measure of product development. My measures of scientific research are drawn from the Online Mendelian Inheritance in Man (OMIM) database, which aims to provide a comprehensive set of genotype-phenotype records. OMIM entries are annotated with citations to published scientific papers. From these annotations, I collect data on the number of publications related to each gene in each year. I use publications from 2001 to 2009 as an outcome; on average, genes had 2 publications over that period, with a median of 0 (Panel B of Table 2).
OMIM assigns two classifications which I use as proxies for the level of scientific knowledge about genotype-phenotype links. All genes involved in at least one genotype-phenotype link classified by OMIM as meeting a high level of scientific certainty are coded as having a “known, certain phenotype.” The set of genes classified by OMIM as meeting a lower threshold for scientific certainty (including those meeting the higher threshold) are coded as having a “known, uncertain phenotype.” I observe the former measure as of 2009, and the latter measure annually. As of 2009, forty-five percent of genes have a known, uncertain phenotype link, and 8 percent have a known, certain phenotype link (Panel B of Table 2).
My measure of product development is drawn from GeneTests.org, a self-reported, voluntary listing of US and international laboratories offering genetic testing. Although not comprehensive, GeneTests.org is the most frequently referenced genetic testing directory (Uhlmann and Guttmacher, 2008). I construct an indicator for whether each gene is used in any genetic test as of 2009. As of 2009, 6 percent of genes were used in a genetic test (Panel B of Table 2).7
1.4 Data construction: An example
A brief example may help to clarify my data construction. The mRNA NM 032753.3 first appeared in RefSeq in 2001, and was never held with Celera’s IP. This is the only known mRNA for the RAX2 gene. I define RAX2 as sequenced in 2001, and never held by Celera.
OMIM references RAX2 in two genotype-phenotypes, first appearing in 2006. Both reference a 2004 publication in Human Molecular Genetics, and are classified by OMIM as known, certain phenotypes. First, RAX2 is linked to age-related macular degeneration, a medical condition arising in older adults that destroys the type of central vision needed for common tasks such as driving, facial recognition, and reading. Second, RAX2 is linked to cone-rod dystrophy, an eye disease tending to cause vision loss. I define RAX2 as having one publication in 2004; in a known, uncertain phenotype link as of 2006; and in a known, certain phenotype link as of 2009.8
GeneTests.org lists several testing facilities offering a genetic test for RAX2’s link to age-related macular degeneration (including some academic medical centers as well as the for-profit firm Quest Diagnostics). There are no listings for genetic tests for RAX2’s link to cone-rod dystrophy. I define RAX2 as being used in a diagnostic test as of 2009.
1.5 Linking scientific research and product development
The prior empirical literature investigating how IP affects subsequent innovation has been constrained to examine only publication-related outcome variables, whereas in this paper I am able to trace how IP affected the availability of commercial products. A priori, this distinction is important: if academic and public researchers face higher incentives to disclose the results of their research than do private researchers, and if IP induces an increase in the share of research done by private researchers, then observed differences in publications could in part be explained by differences in disclosure.9 However, my product development outcome - diagnostic test availability - should be invariant with respect to disclosure preferences of researchers.
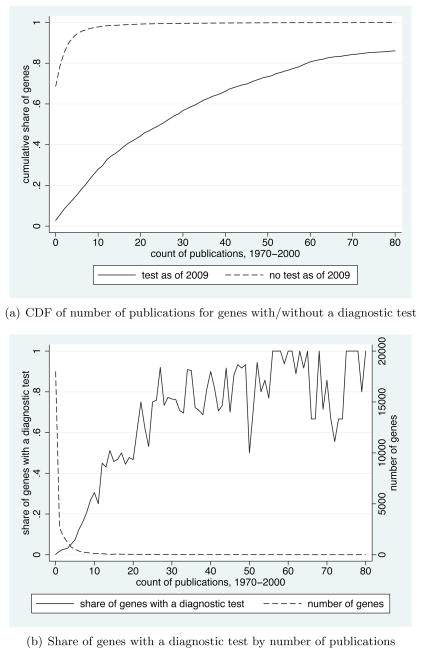
Panel A of Figure 2 presents one set of descriptive statistics illustrating that, in my data, scientific research and product development are strongly related. The dashed line (“no test as of 2009”) plots the empirical cumulative distribution function of the number of publications between 1970–2000 for genes that do not have a diagnostic test available as of 2009.10 The solid line (“test as of 2009”) plots the empirical cumulative distribution function of the number of publications between 1970–2000 for genes that do have a diagnostic test available as of 2009. Virtually all genes not used in diagnostic tests had 10 or fewer publications, whereas about 70 percent of genes used in diagnostic tests had more than 10 publications.
Figure 2.
Documenting the Link Between Scientific Research & Product Development
Notes: These figures provide two sets of descriptive statistics that document the link between scientific research and product development in my data. In sub-figure (a), the dashed line plots the empirical cumulative distribution function of the number of publications between 1970-2000 for genes that do not have a diagnostic test available as of 2009, and the solid line plots the empirical cumulative distribution function of the number of publications between 1970-2000 for genes that do have a diagnostic test available as of 2009. In sub-figure (b), the dashed line plots the distribution of genes by the number of publications between 1970-2000, and the solid line plots the share of genes with a diagnostic test as of 2009 at each number of publications. See text and online appendix for more detailed data and variable descriptions.
Panel B of Figure 2 presents an alternative set of descriptive statistics illustrating a similar point. The dashed line plots the distribution of genes, ordered by the number of publications between 1970–2000; consistent with the summary statistics presented in Table 2, most genes have zero publications. The solid line plots the share of genes with a diagnostic test for each number of publications. The share of genes with a diagnostic test increases roughly monotonically with the number of publications between 0 and around 30 publications, and then continues to increase more slowly above that point. These data suggest that the number of publications is strongly related to the probability that a diagnostic test is available.
2 Empirical context
This section briefly reviews the institutional context relevant for the empirical analysis.11
2.1 Timeline of sequencing efforts
The public sequencing effort - the Human Genome Project - was launched in 1990, and originally aimed to finish sequencing the entire genome by 2005 (Collins and Galas, 1993). In May 1998, Celera - a new firm led by scientist Craig Venter - formed with the intention to sequence the entire human genome within three years (Venter et al., 1998). The public effort subsequently announced a revised plan to complete its sequencing efforts by 2003 (Collins et al., 1998), and to release an earlier “draft” sequence of the human genome (Pennisi, 1999).12 Departing from its previous goal of producing a near-perfect sequence, the aim of this draft sequence was to place most of the genome in the public domain as soon as possible. The two efforts jointly published draft genomes in February 2001, the public effort in Nature (Lander et al., 2001) and Celera in Science (Venter et al., 2001).13 Celera’s sequencing effort stopped with this publication, whereas the public effort continued and was declared complete in April 2003 (Wade, 2003).
2.2 Intellectual property: The Bermuda rules and Celera’s IP
As of 1996, genes sequenced by the public effort were covered by the “Bermuda rules,” requiring data to be posted on an open-access website within twenty-four hours of sequencing.14 The stated goal was “…to encourage research and development and to maximize [the data’s] benefit to society.”15 Eisenberg (2000) argues the Bermuda rules also aimed to discourage gene patenting.
Between 2001 and 2003, Celera used a contract law-based form of IP to protect genes that had been sequenced by Celera but not yet sequenced by the public effort. Celera’s IP had several key features.16 First, Celera’s data were ‘disclosed’ in 2001 (Venter et al., 2001), in the sense that any individual could view data on the assembled genome through Celera’s website, or by obtaining a free data DVD from the company.17 Academic researchers were free to use Celera’s data for non-commercial research and academic publications. Second, by placing restrictions on redistribution, Celera was able to sell its data to larger institutions - including pharmaceutical companies, universities, and research institutes. Although the terms of specific deals were private, Service (2001) reports that pharmaceutical companies were paying between $5 million and $15 million a year, whereas universities and nonprofit research organizations were paying between $7,500 and $15,000 for each lab given access to the data. Third, any researcher wanting to use the data for commercial purposes was required to negotiate a licensing agreement with Celera. Celera was able to charge these data access and licensing fees even though all available accounts suggest it was publicly known in 2001 that all of Celera’s genes would be re-sequenced by the public effort, and thus move into the public domain, by 2003. Shreeve (2005) quotes Craig Venter as saying: “Amgen, Novartis, and now Pharmacia Upjohn have signed up knowing damn well the data was going to be in the public domain in two years anyways. They didn’t want to wait for it.” In addition to this short-term IP, Shreeve (2005) documents that Celera was actively pursuing gene patent applications for genes in its database; ex post most of these applications were not granted patents, but given the contemporaneous and subsequent policy uncertainty surrounding gene patenting it is difficult to know what researchers’ expectations were at the time.18 Beyond database sales and licensing revenues, Celera’s business model also included in-house research and profits from genes granted patents (Service, 2001). Celera eventually grew into a healthcare firm that develops and manufactures gene-based technologies.
2.3 Sequencing strategies: Implications for selection into Celera’s IP
The public sequencing effort was a large consortium that, for the purposes of this paper, can be conceptualized as two distinct efforts. A ‘targeted’ effort focused on sequencing genes with known medical value, such as the gene linked to Huntington’s disease. A ‘large-scale’ effort focused on the same type of large-scale sequencing undertaken by Celera. Large-scale sequencing by both Celera and the public effort relied on the shotgun sequencing method, in which DNA is randomly broken up into small segments that are sequenced and re-assembled (Lander et al., 2001). From an empirical perspective, shotgun sequencing should have introduced some effectively random variation in whether genes were initially sequenced by Celera or by the public effort.
The vast majority of genes were sequenced under the large-scale public effort, which started in mid-1999 (Lander et al., 2001). However, because the targeted public effort focused on sequencing genes that had high ex ante expected medical value, we expect that Celera genes - on average - will be negatively selected in the sense of having a lower inherent potential for follow-on research. This selection is important, because it implies that the simple cross-tabulation presented in Table 1 could be misleading. This concern motivates the construction of empirical measures that proxy for the ex ante expected value of each gene in order to empirically investigate patterns of selection in my data.
Based on discussions with scientists, one reasonable proxy for the ex ante expected value of a gene is the number of scientific papers published about the gene before it was sequenced. For example, a long scientific literature has documented evidence that Huntington’s disease has a genetic basis. Many of these papers were published prior to the development of gene sequencing techniques, and the evidence from these papers likely led scientists to target the sequencing of genes related to Huntington’s disease more than genes related to conditions that were less well-understood.
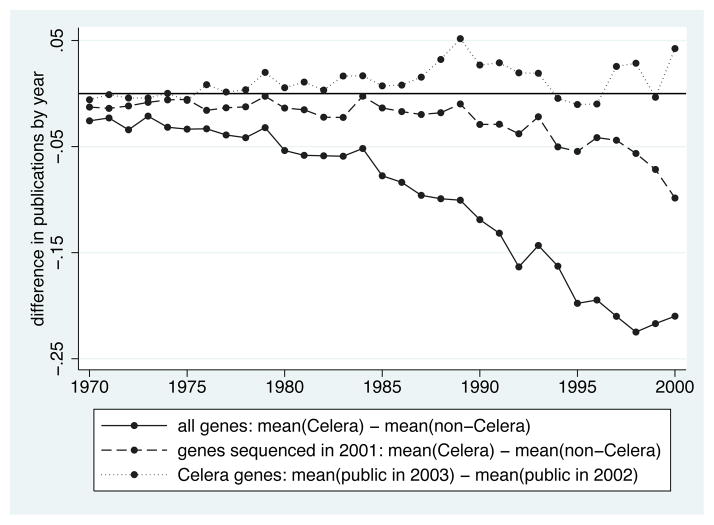
Figure 3 uses data on the number of OMIM publications about a gene from 1970 to 2000 to investigate selection into Celera’s IP. The solid line (“all genes”) plots the difference in mean publications on Celera genes and mean publications on non-Celera genes in each year from 1970 to 2000. All observations are less than zero, providing empirical evidence consistent with the type of selection I described: genes initially sequenced by the public effort had higher ex ante expected value than genes initially sequenced by Celera. One formal test for such selection is to use an ordinary-least-squares model to predict the gene-level “celera” indicator as a function of count variables for publications in each year from 1970–2000. In the full sample, the p-value from an F-test for joint significance is less than 0.001.
Figure 3.
Investigating Selection into Celera’s IP
Notes: This figure provides three sets of descriptive statistics investigating the selection of genes into Celera’s intellectual property (IP). The solid line (“all genes”) plots the difference in mean publications on Celera genes and mean publications on non-Celera genes in each year from 1970 to 2000. The dashed line (“genes sequenced in 2001”) plots the difference in mean publications on Celera genes and mean publications on non-Celera genes that were sequenced in 2001 in each year from 1970 to 2000. The dotted line (“Celera genes”) plots the difference in mean publications on Celera genes resequenced in 2003 and mean publications on Celera genes resequenced in 2002 in each year from 1970 to 2000. See text and online appendix for more detailed data and variable descriptions.
Unfortunately, I do not observe which genes were sequenced by the targeted public effort, so I cannot directly exclude those genes from the sample. As an alternative, I limit the comparison group of non-Celera genes to those sequenced in 2001 (the year Celera’s draft genome was disclosed). Because the number of genes sequenced under the targeted public effort was likely small in 2001 relative to the number of genes sequenced under the large-scale effort, selection should be reduced in this sample. The dashed line in Figure 3 (“genes sequenced in 2001”) suggests this sample restriction reduces but does not eliminate observed selection. In this restricted sample, the p-value from an F-test for joint significance is 0.033. Selection will thus be a concern in my cross-section estimates; this motivates two additional empirical tests that address selection.
My second and third empirical tests rely on variation in the timing of when Celera genes were resequenced by the public effort (either 2002 or 2003). The dotted line in Figure 3 (“Celera genes”) plots the difference in mean publications on Celera genes resequenced in 2003 and mean publications on Celera genes resequenced in 2002, in each year from 1970 to 2000. Here, I find no evidence of selection: predicting a gene-level indicator for being resequenced in 2003 as a function of these count variables for publications in each year, the p-value from an F-test for their joint significance is 0.169. This result suggests that, post-2001, the public effort was either not targeting or not successfully targeting the resequencing of more valuable Celera genes. This evidence supports the validity of my second and third empirical tests.
3 Empirical results
3.1 Cross-section estimates
The basic comparison underlying my cross-section estimates can be a presented in a simple cross-tabulation. Table 1 compares subsequent innovation outcomes for Celera genes and for non-Celera genes sequenced in 2001. These data suggest Celera genes have lower levels of subsequent scientific research and product development relative to non-Celera genes sequenced in the same year. For example, about 3 percent of Celera genes were used in a gene-based diagnostic test in 2009, relative to 5.4 percent of non-Celera genes sequenced in the same year.
The concern with interpreting these simple differences in means is that they could reflect a negative effect of Celera’s IP on subsequent research, or could instead reflect selection if Celera’s genes had lower inherent potential for follow-on research. In order to address this concern, Table 3 formalizes the basic comparison from Table 1 in a regression framework that allows me to investigate the robustness of these patterns. For gene g, I estimate the following:
Table 3.
Cross-Section Estimates: Impact of Celera’s IP on Innovation Outcomes
| (1) | (2) | (3) | |
|---|---|---|---|
| Panel A: publications in 2001–2009 | |||
| mean = 2.197 | |||
| celera | −0.877 (0.177)*** | −0.328 (0.099)*** | −0.264 (0.107)*** |
|
| |||
| Panel B: 1(known, uncertain phenotype) | |||
| mean = 0.453 | |||
| celera | −0.162 (0.015)*** | −0.158 (0.015)*** | −0.128 (0.017)*** |
|
| |||
| Panel C: 1(known, certain phenotype) | |||
| mean = 0.081 | |||
| celera | −0.027 (0.007)*** | −0.017 (0.006)*** | −0.014 (0.007)** |
|
| |||
| Panel D: 1(used in any diagnostic test) | |||
| mean = 0.060 | |||
| celera | −0.023 (0.006)*** | −0.014 (0.005)*** | −0.013 (0.006)** |
|
| |||
| indicator variables for year of disclosure | yes | yes | yes |
| number of publications in each year 1970–2000 | no | yes | yes |
| detailed cytogenetic & molecular covariates | no | no | yes |
|
| |||
| N | 27,882 | 27,882 | 16,485 |
Notes: Gene-level observations. All estimates are from ordinary-least-squares (OLS) models. Samples in Columns (1) and (2) include all genes; sample in Column (3) includes all genes with non-missing cytogenetic and molecular covariates. Robust standard errors shown in parentheses.
p< 0.10;
p< 0.05;
p< 0.01.
“Celera”: 0/1, =1 for a Celera gene. Indicator variables for year of disclosure: 0/1 indicator variables for the year sequence for the gene was disclosed. Number of publications in each year 1970–2000: count variables for the number of publications on each gene in each year from 1970 to 2000. Detailed cytogenetic & molecular covariates: 0/1 indicator variables for the chromosome (1–22, X, or Y) and arm (p or q) on which a gene is located; continuous variables for region, band, subband, start base pair, and end base pair; and 0/1 indicator variables for the orientation of the gene on the genome assembly (plus or minus). See text and the online appendix for more detailed data and variable descriptions.
The coefficient on the “celera” variable is the main estimate of interest. I show estimates from ordinary-least-squares (OLS) models, and report heteroskedasticity-robust standard errors.19
Column (1) of Table 3 replicates the cross-tabulation results on the full sample, controlling for year of disclosure but no other covariates.20 Column (2) of Table 3 includes a set of count variables for the number of publications on each gene in each year from 1970 to 2000. We saw in Section 2.3 that Celera genes looked less valuable than non-Celera genes based on these proxies for ex ante expected value. As expected, including these variables as covariates reduces the magnitudes of the point estimates. Column (3) limits the sample to genes with non-missing data on location variables (n = 16, 485), and investigates whether the estimates are sensitive to conditioning on detailed location variables. This robustness check addresses the possibility that scientists may have targeted their sequencing efforts based on a gene’s ex ante known location on the genome. For example, certain chromosomes (such as chromosome 19) were estimated to be more “gene-rich” than others, and scientists may have targeted the sequencing of such chromosomes. To test for this possibility, I collect detailed variables on both types of gene location descriptors used by geneticists (cytogenetic location and molecular location). The estimates in Column (3) are quite similar in magnitude to the estimates in Column (2).
For brevity, I focus on interpreting the magnitudes of the point estimates in Column (1). The estimate in Panel A implies Celera genes had about 0.88 fewer publications from 2001 to 2009, a decline on the order of 40% of the mean number of publications for genes over that time period. The estimate in Panel B implies a 16 percentage point reduction in the probability of having a known, uncertain phenotype link, a decline on the order of 35% relative to the sample mean. The estimate in Panel C implies a 2.7 percentage point reduction in the probability of having a known, certain phenotype link, a decline on the order of 33% of the sample mean. The estimate in Panel D implies a 2.3 percentage point reduction in the probability of a gene being used in any currently available diagnostic test, a decline on the order of 38% of the sample mean. One way to interpret the magnitudes of these differences in means is the following: if Celera genes had counterfactually had the same rate of subsequent innovation as non-Celera genes, there would have been 1,400 additional publications between 2001 and 2009, and 40 additional diagnostic tests as of 2009.21 The magnitudes of the estimated coefficients decline in magnitude when I add additional control variables, and the estimates in Columns (2) and (3) are on the order of 20 to 30 percent of the sample means.
Of course, despite my attempts to control for selection, the lingering concern is that these estimates could be driven by non-random selection into Celera’s IP. Sections 3.2 and 3.3 present results from my second and third empirical tests, which address selection more directly.
3.2 Panel estimates
My second empirical test investigates whether the the removal of Celera’s IP affected within-gene flow measures of subsequent innovation. For gene-year gy, I estimate the following:
The “celera” variable is now an indicator for whether gene g had been sequenced only by Celera as of that year. This “celera” variable varies within genes over time, and a transition from 1 to 0 represents the removal of Celera’s IP from a given gene. Year fixed effects control for year-specific shocks that are common across genes, such as annual changes in the level of research funding available from public sector agencies. Gene fixed effects control for time-invariant differences across genes, such as a gene’s inherent commercial potential. I limit the years in the sample to 2001–2009, focusing on the time period in which all Celera genes had been sequenced, but vary in their IP status over time.22 I show estimates from OLS models and report heteroskedasticity-robust standard errors clustered at the gene level.
Table 4 presents estimates from the panel specification. Columns (1) and (2) are analogous to the cross-section specifications from Table 3: both control for year fixed effects, Column (1) includes indicator variables for the year of disclosure, and Column (2) adds count variables for the number of publications in each year from 1970 to 2000. Column (3), my preferred specification, retains the year fixed effects but replaces the time-invariant covariates with gene fixed effects.
Table 4.
Panel Estimates: Impact of Celera’s IP on Innovation Outcomes
| (1) | (2) | (3) | |
|---|---|---|---|
| Panel A: publications | |||
| mean = 0.244 | |||
| celera | −0.160 (0.017)*** | −0.121 (0.011)*** | −0.109 (0.011)*** |
|
| |||
| Panel B: 1(known, uncertain phenotype) | |||
| mean = 0.381 | |||
| celera | −0.163 (0.009)*** | −0.160 (0.008)*** | −0.083 (0.008)*** |
|
| |||
| year fixed effects | yes | yes | yes |
| indicator variables for year of disclosure | yes | yes | no |
| number of publications in each year 1970–2000 | no | yes | no |
| gene fixed effects | no | no | yes |
|
| |||
| N | 250,938 | 250,938 | 250,938 |
Notes: Gene-year-level observations. All estimates are from ordinary-least-squares (OLS) models. The sample includes all gene-years from 2001 to 2009 (27,882 genes for 9 years implies N = 250,938 total gene-year observations). Robust standard errors, clustered at the gene level, shown in parentheses.
: p< 0.10;
: p< 0.05;
: p< 0.01.
“Celera”: 0/1, =1 for a Celera gene. Indicator variables for year of disclosure: 0/1 indicator variables for the year sequence for the gene was disclosed. Number of publications in each year 1970–2000: count variables for the number of publications on each gene in each year from 1970 to 2000. See text and online appendix for more detailed data and variable descriptions.
Panel A of Table 4 reports estimates for the gene-year publications outcome. As in the cross-section specification, adding the publication variables does affect the estimated effect of Celera’s IP. In addition, replacing the time-invariant covariates with gene fixed effects further reduces the magnitude of the estimated effect. That said, the magnitudes of the coefficients in Columns (2) and (3) are broadly similar, which I interpret as suggestive evidence that the cross-section controls are at least somewhat effective in controlling for gene-specific variation in the publications outcome. In terms of magnitudes, the coefficient in Column (3) in Panel A of Table 4 suggests Celera’s IP was associated with 0.11 fewer publications per year, a decline on the order of 45% of the sample mean.
Panel B of Table 4 reports analogous estimates for the gene-year indicator for a gene having any known, uncertain phenotype link as of that year. The coefficient in Column (3) suggests Celera’s IP was associated with a 8.3 percentage point reduction in the probability that a gene had a known, uncertain phenotype link, a decline on the order of 22% of the sample mean.
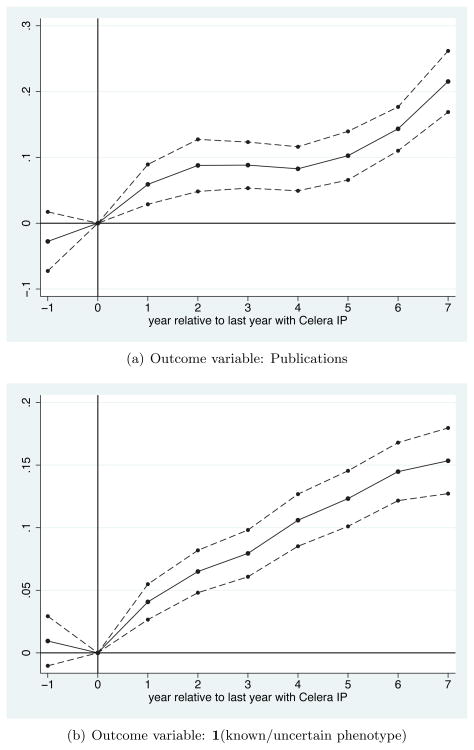
To explore the timing of the estimated effects, Figure 4 presents graphical versions of the following event study specification:
Figure 4.
Panel Estimates: Impact of Celera’s IP on Innovation Outcomes
Notes: These figures plot coefficients (and 95 percent confidence intervals) from the event study specification described in Section 3.2. On the x axes are years z relative to a “zero” relative year that marks the last year the gene was held with Celera’s IP (that is, year 1 marks the first year the gene was in the public domain). As in the specifications in Table 4, this specification is based on gene-year level observations, the coefficients are estimates from ordinary-least-squares (OLS) models, the sample includes all gene-years from 2001 to 2009, and the standard errors are robust and clustered at the gene level. See text and online appendix for more detailed data and variable descriptions.
On the x axes are years z relative to a “zero” relative year that marks the last year the gene was held with Celera’s IP (that is, year 1 marks the first year the gene was in the public domain). The dotted lines show 95 percent confidence intervals.
Panel A of Figure 4 presents results for the gene-year level publications outcome. These estimates suggest that in the first year a gene enters the public domain (t = 1 on the graph), there is a discrete level shift in the flow of publications related to that gene, which remains relatively constant through the end of my data. In theory, the panel estimates in Table 4 could have been driven by short-term shifts in the timing of when research takes place that may or may not have persistent effects on welfare. In practice, the results show no clear “bunching” of publications that would be predicted by stories in which researchers strategically wait until IP is removed to publish scientific papers.
Panel B of Figure 4 presents results for the gene-year level indicator for a gene having any known, uncertain phenotype link. This outcome increases in the first year a gene enters the public domain (t = 1 on the graph), and continues to increase through the end of my data.
3.3 Focusing on Celera genes
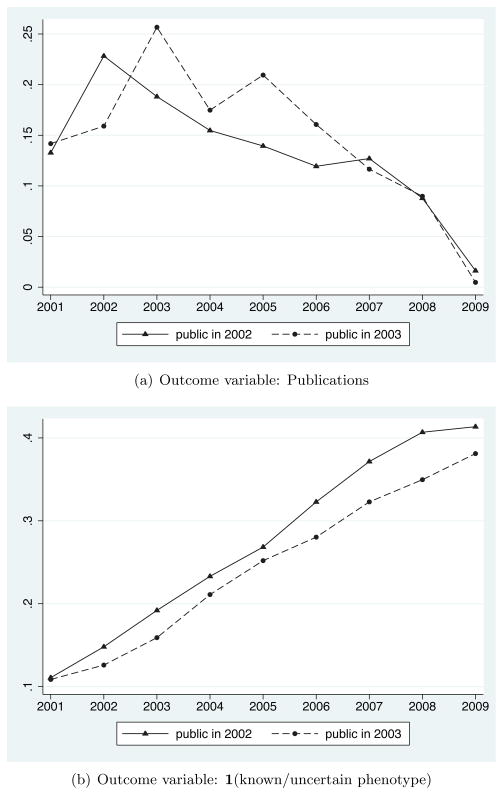
Figure 5 presents results from my third empirical test. I limit the sample to include only Celera genes, and rely solely on variation in how long genes were held with Celera’s IP - that is, whether the Celera gene was re-sequenced by the public effort in 2002 (N = 1,047; “public in 2002”) or in 2003 (N = 635; “public in 2003”). The evidence presented in Section 2.3 and Figure 3 suggests that the year in which Celera genes were re-sequenced by the public effort cannot be predicted with gene-level observables. Hence, this analysis should provide a clean test for investigating the effect of being held with Celera’s IP for one additional year.
Figure 5.
Average Innovation Outcomes for Celera Genes by Year, by Year of Re-sequencing by the Public Effort
Notes: These figures plot the descriptive statistics described in Section 3.3. Sample includes all Celera genes. Means are shown separately for Celera genes that were re-sequenced by the public effort in 2002 (N = 1,047) and for Celera genes that were re-sequenced by the public effort in 2003 (N = 635). See text and online appendix for more detailed data and variable descriptions.
Figure 5 presents means by year for the two panel outcome variables. As expected, the mean levels of both outcome variables are quite similar across the “public in 2002” and “public in 2003” groups in 2001, when both sets of genes were held with Celera’s IP. Panel A shows that Celera genes re-sequenced in 2002 saw a relative uptick in publications in that year, while Celera genes re-sequenced in 2003 show a similar uptick in 2003. Flow of scientific effort into these two cohorts of genes appears to have converged over time: although the difference in means in 2002 is statistically significant at the 10 percent level, mean differences in other years are not statistically significant.
Panel B shows that Celera genes re-sequenced in 2002 saw a relative increase in the probability of having a known, uncertain genotype-phenotype link in 2002. However, rather than the “public in 2003” group catching up with their “public in 2002” counterparts one year later, the “public in 2003” group has persistently lower levels of this outcome variable through the end of my data. The difference in means is statistically significant in 2003 (at the 10 percent level), 2006 (at the 10 percent level), 2007 (at the 5 percent level), and 2008 (at the 5 percent level).
The results in Figure 5 suggest that although the flow of scientific effort into these two cohorts of genes (as measured by annual publications) converged over time, the average stock of scientific knowledge about genes in these two cohorts (as measured by having a known, uncertain phenotype) did not converge. I test this formally by estimating whether the mean of the “public in 2002” cohort in year t is statistically distinguishable from the mean of the “public in 2003” cohort in year t+1; in no year can I reject the null that those means are equal. That is, I cannot reject a model in which an additional year of Celera’s IP induces a permanent loss of one year of research. These results provide clear evidence that even very temporary forms of intellectual property - here, lasting only one year - can have persistent effects on subsequent innovation.
4 What kinds of transaction costs were relevant?
Return to the example from the introduction: suppose Pfizer discovers a gene-based diagnostic test that requires licensing one of Celera’s genes. Would Celera’s IP impede Pfizer’s research?23 The empirical evidence in this paper suggests it would, which implies that some form of transaction costs hindered licensing negotiations over Celera’s IP. In evaluating which potential sources of transaction costs were most likely relevant, both Celera’s short-term IP and the expectation that Celera was pursuing patent applications on their genes are relevant.
In a perfect contracting environment with no transaction costs, Celera and Pfizer would negotiate a licensing agreement such that cumulative research is not hindered. Consider the model of Green and Scotchmer (1995). Licensing agreements can occur at two stages: ex ante, before Pfizer invests in the diagnostic test, or ex post, after Pfizer has invested in the test. The key distinction is whether Pfizer has sunk its research costs at the time of the licensing negotiation. The Green and Scotchmer (1995) framework delivers a strong prediction that ex ante licenses are optimal and will always be negotiated. When negotiating ex ante, Pfizer has a credible threat not to invest unless Celera is willing to share a positive fraction of the diagnostic test profits. When negotiating ex post, Pfizer has diminished bargaining power and faces a potential holdup problem.
Despite this strong theoretical prediction, transaction costs may prevent ex ante licensing agreements from being successfully negotiated. For example, the Green and Scotchmer (1995) framework assumes symmetric information, but in practice Celera may not have known Pfizer’s cost of developing its gene-based diagnostic test. Bessen (2004) explores the implications of this type of private information in the Green and Scotchmer (1995) framework, showing that private information may cause negotiations to break down, deterring some socially desirable research.
Empirically, only a small share of licensing agreements appear to be set ex ante. Anand and Khanna (2000) document that in SIC28 (chemicals and pharmaceuticals), only 23% of licensing agreements were set ex ante. Consistent with this data, Celera’s data access agreement (Science Online, 2001), Celera’s DVD user agreement, and my informal discussions with academic and commercial researchers all suggested Celera’s licensing agreements were frequently if not always negotiated ex post rather than ex ante.
Celera could have avoided transaction costs by conducting in-house research; indeed, Celera developed and manufactured several gene-based technologies. However, ideas in this market were likely scarce in the sense of Scotchmer (1991): Celera’s scientists did not know how to develop the full set of possible subsequent innovations. Taken together, this suggests that a scarcity of ideas together with asymmetric information about the costs of development may have generated a first source of transaction costs.
A second source of potential transaction costs is a version of the classic disclosure problem (Arrow, 1962), highlighted by Gallini and Wright (1990) and Gans and Stern (2000). To negotiate a licensing agreement with Celera, Pfizer had to disclose its idea. Because Celera was developing gene-based technologies, Celera had a credible threat to engage in imitative R&D. Either the expectation of Celera’s bargaining position, or the actual impact of Celera’s bargaining power in licensing negotiations, may have generated a second source of transaction costs.
A final source of potential transaction costs is uncertainty over the academic research exemption. Formally, Celera placed no restrictions on academic research. However, for at least two reasons academic researchers may have nonetheless been deterred from using Celera’s data. First, informal discussions with academic scientists suggested they faced uncertainty over some of Celera’s contractual terms. For example, one scientist I spoke with expressed uncertainty over whether the restrictions on redistribution implied she could not share Celera’s data with her graduate students. Because accessing the data required agreeing to Celera’s terms of use, perceived litigation risks may have deterred research even by academics who solely wanted to use the data for non-commercial research. Second, given that the boundary between academic and commercial research is often not clearly delineated - perhaps particularly for biomedical research (Cohen and Walsh, 2008) - the ‘exemption’ for academic research may not have been clear in practice. Celera’s sequencing took place during the biotech boom, when many academics were doing research with an eye towards commercial applications. Celera’s IP may have discouraged that type of academic research even in the absence of formal restrictions on academic publications.
5 Concluding remarks
Intellectual property (IP) is a widely-used policy lever for promoting innovation, yet relatively little is known about how IP on existing technologies affects subsequent innovation. The sequencing of the human genome provides a useful empirical context, generating variation in IP across a relatively large group of ex ante similar technologies. Across a range of empirical specifications, I find evidence that Celera’s IP led to reductions in subsequent scientific research and product development on the order of 20 to 30 percent.
A caveat to this interpretation of these results is that if innovation inputs are scarce, my estimates could reflect the substitution of innovative effort away from Celera genes towards non-Celera genes (as opposed to a net decrease in total innovation over the set of all genes).24 Looking at a broad set of academic biomedical researchers, surveys by Walsh, Cho and Cohen (2005) and Walsh, Cohen and Cho (2007) suggest some substitution is relevant: restricted access to tangible research inputs (including information, data, and software) appears to shift scientists’ research project choices. If substitution is relevant and researchers optimally choose their line of research in the absence of IP, quantifying the welfare costs of IP on cumulative innovation requires estimating the cost of distorting research towards sub-optimal projects. If more socially valuable technologies are more likely to be held with IP, these welfare costs could be substantial.
While Celera’s gene-level IP did not depend on patent protection, the evidence in this paper is related to the ongoing legal controversy surrounding patents on human genes.25 Echoing the broader debate on patents, proponents argue that gene patents incentivize investment in gene-related technologies, while opponents argue that gene patents stifle subsequent product development and restrict patients’ access to gene-related technologies.26 To the best of my knowledge, there exists no direct evidence on how gene patents have affected subsequent product development. Moreover, the overall welfare consequences of gene patents - and patents more generally - depend on the trade-off between ex ante incentives for innovation, the ex post costs of restricting patients’ access to technologies, and any potential effects of IP on subsequent innovation. From a policy perspective, the evidence in this paper informs this gene patent debate by documenting that - at least for some forms of intellectual property - getting the incentives “right” for subsequent innovators is quantitatively important for encouraging subsequent scientific research and product development.
Supplementary Material
Footnotes
I am very grateful to Wes Cohen, Joe Doyle, Dan Fetter, Matt Gentzkow, Claudia Goldin, Sam Kortum, Amanda Kowalski, Fiona Murray, Jesse Shapiro, Scott Stern, three anonymous referees, numerous seminar participants, and especially my PhD advisers David Cutler, Amy Finkelstein, and Larry Katz for comments. Several individuals from Celera, the Human Genome Project, and related institutions provided invaluable guidance, including Sam Broder, Peter Hutt, and particularly Mark Adams, David Altshuler, Bob Cook-Deegan, Eric Lander, Robert Millman, and seminar participants at the Broad Institute. David Robinson provided valuable assistance with the data collection. Financial support from NIA Grant Number T32-AG000186 to the NBER, NSF Grant Number 1151497, and the Center for American Political Studies at Harvard is gratefully acknowledged.
Patent citations are a frequently-used measure of cumulative innovation (Jaffe, Trajtenberg and Henderson, 1993). However, by construction patent citations cannot measure cumulative innovation on non-patented technologies. The type of data constructed in this paper - measuring scientific research and product development directly, rather than via patent citations - is critical in enabling a test of how IP affects cumulative innovation.
For example, under the patent buyout mechanism discussed by Kremer (1998), the public sector (or another entity) could have paid Celera some fee to “buy out” Celera’s IP and place Celera genes in the public domain. See Kremer and Williams (2010) for further discussion of other alternative mechanisms for rewarding innovation.
Murray et al. (2008) also provide evidence, consistent with the model of Aghion, Dewatripont and Stein (2008), that IP reduces the diversity of scientific experimentation.
In recent years the exact definition of a gene has become more complicated than this basic description; for a more detailed discussion see Snyder and Gerstein (2003). However, these subtleties are not important for the empirical analysis in this paper; my use of the term “gene” follows the definitions set out in the US National Institutes of Health’s RefSeq database, which is used internationally as the standard for genome annotation.
The mean for this variable is left-censored, because 1999 is the first year coded in the RefSeq database.
The mean number of known mRNAs per gene is 1.67, and the median is 1. Thus, alternative definitions -such as the share of known mRNAs that were Celera mRNAs, or an indicator for whether any mRNA on the gene was a Celera mRNA - are identical for the majority of genes.
These tests can be developed quite quickly; Cho et al. (2003) note it may only take weeks or months to go from a research finding that a particular genetic variant is associated with a disease to a clinically validated test.
As detailed above, I only observe the “known, certain phenotype” measure as of 2009.
Of course, disclosure itself presumably has social value, and to the extent that IP induces reductions in disclosure this effect is also relevant in measuring the welfare effects of IP. Moon (2011) provides an empirical study of disclosure in the context of genetic research. Analyzing the discovery of a genotype-phenotype link in an event study framework, he shows that non-academic research organizations become less likely to publish relative to universities after the discovery of a phenotype link.
There are very few pre-1970 publications cited in the OMIM data.
For more details, see Cook-Deegan (1994), Shreeve (2005), Sulston and Ferry (2002), and Venter (2007).
Many observers attribute this scale-up to Celera’s entry (Marshall, 1998).
Celera and a few “early subscriber” firms had access to intermediate data updates during late 1999 and 2000, but my understanding is that the vast majority of Celera’s data were first released in the 2001 draft genome.
In 1996, the heads of the largest labs involved in the public effort agreed at a Bermuda-based meeting to these rules as a set of guidelines for data sequenced under the public effort. The Bermuda Rules replaced a US policy that data be made available within six months (Marshall, 2001).
These rules are described in various policy statements by the US National Human Genome Research Institute (NHGRI). Non-adherence was expected to result in black marks on future grant reviews (Marshall, 2001).
For details, see Celera’s data access agreement (Science Online, 2001), and Celera’s DVD user agreement. I am very grateful to Mike Meurer, Robert Millman (then-Chief IP Counsel at Celera from 1999–2002), and Ben Roin for discussions on Celera’s IP, but of course none of them is responsible for any errors in my descriptions.
Viewing the data or obtaining the DVD required agreeing not to commercialize or redistribute the data.
What the US Patent and Trademark Office has allowed to be covered by a “gene patent” has changed dramatically over time; see, e.g. National Academy of Sciences (2006). There has also been substantial variation over time in the judicial enforcement of existing gene patents.
As a robustness check, in Appendix Table B.1 I present estimates from proportional models that are analogous to the estimates in Table 3: quasi-maximum likelihood Poisson models for publications, and logit models for the three binary outcomes.
There are two ways to control for variation in innovation outcomes across genes as of 2009 that is a function of the year in which the genes were sequenced: limiting the sample to the set of genes sequenced in 2001, or using the full sample but including indicator variables to control for year of disclosure. Because all Celera genes were sequenced in 2001, the “celera” variable only varies in 2001. Hence, re-estimating the specification in Column (1) on the sub-sample of genes sequenced in 2001 estimates identical coefficients. I use the full sample in these robustness checks because the additional non-Celera genes are useful for identifying the covariates.
I calculated these figures using non-Celera genes sequenced in 2001 as my counterfactual, as in Table 1, as follows. Between 2001 and 2009, there were 8,118 publications on the cohort of genes sequenced in 2001, with an average of 2.116 publications for non-Celera genes and an average of 1.239 publications for Celera genes. If we assume that Celera genes had attained the non-Celera average number of publications, there would instead have been 9,592 publications. Hence, Celera’s IP led to around 1,400 fewer publications between 2001 and 2009 (9, 592–8, 118 ≈ 1, 400). Undertaking a similar calculation for diagnostic tests, the number of genes sequenced in 2001 with a diagnostic test available as of 2009 was 204, and the probability of having a diagnostic test was 0.030 for Celera genes and 0.054 for non-Celera genes. If Celera genes had attained the non-Celera average probability of being used in a diagnostic test, there would instead have been 245 diagnostic tests as of 2009. Hence, Celera’s IP led to around 40 fewer diagnostic tests as of 2009 (245 – 204 ≈ 40).
As noted in Section 2.1, Celera’s sequencing began in 1999, and its draft genome was disclosed in 2001. My understanding is that the vast majority of Celera’s data were first released in the 2001 draft genome, but I do not observe the timing of sequencing from 1999–2001. In the absence of such data, I limit my panel specification to include the years 2001–2009 since prior to 2001 I do not know whether or not Celera genes had yet been sequenced.
Beyond the references in the introduction (Scotchmer, 1991; Green and Scotchmer, 1995; Bessen, 2004), see also the discussions in Merges and Nelson (1990), Heller and Eisenberg (1998), and Shapiro (2000).
A priori, the relevance of substitution depends on whether inputs to gene-related research should be considered relatively fixed or relatively flexible.
Two recent court cases are relevant. First, Association for Molecular Pathology v. Myriad Genetics (formerly Association for Molecular Pathology v. U.S. Patent and Trademark Office) is a lawsuit challenging the validity of gene patents held by the firm Myriad Genetics related to the BRCA1 and BRCA2 genes. In November 2012, the US Supreme Court agreed to hear the AMP v. Myriad case, with a decision expected before July 2013. Second, Mayo Collaborative Services, dba Mayo Medical Laboratories, et al. v. Prometheus Laboratories Inc. was a lawsuit challenging the patentability of diagnostic tests with implications for the patentability of gene-based diagnostic tests. The US Supreme Court handed down a unanimous decision in Mayo v. Prometheus in March 2012, upholding the District court decision that declared Prometheus’s diagnostic test not patent eligible, and reversing the US Court of Appeals for the Federal Circuit.
Opponents of gene patents have also argued that such patents may be invalid on the grounds that DNA is not patentable subject matter.
References
- Aghion Philippe, Dewatripont Mathias, Stein Jeremy. Academic freedom, private-sector focus, and the process of innovation. RAND Journal of Economics. 2008;39(3):617–635. [Google Scholar]
- Anand Bharat, Khanna Tarun. The structure of licensing contracts. Journal of Industrial Economics. 2000;48(1):103–135. [Google Scholar]
- Arrow Kenneth. Economic welfare and the allocation of resources for invention. In: Nelson Richard., editor. The Rate and Direction of Inventive Activity. Princeton University Press; 1962. [Google Scholar]
- Bessen James. Holdup and licensing of cumulative innovations with private information. Economics Letters. 2004;82(3):321–326. [Google Scholar]
- Cho Mildred, Illangasekare Samantha, Weaver Meredith, Leonard Debra, Merz Jon. Effects of patents and licenses on the provision of clinical genetic testing services. Journal of Molecular Diagnostics. 2003;5(1):3–8. doi: 10.1016/S1525-1578(10)60444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Wesley, Walsh John. Real impediments to academic biomedical research. In: Lerner Josh, Stern Scott., editors. Innovation Policy and the Economy. Vol. 8. University of Chicago Press; 2008. [Google Scholar]
- Collins Francis, Galas David. A new five-year plan for the U.S. Human Genome Project. Science. 1993;262(5130):43–46. doi: 10.1126/science.8211127. [DOI] [PubMed] [Google Scholar]
- Collins Francis, Patrinos Ari, Jordan Elke, Chakravarti Aravinda, Gesteland Raymond, Walters LeRoy the members of the DOE, and NIH planning groups. New goals for the US Human Genome Project: 1998–2003. Science. 1998;282(5389):682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- Cook-Deegan Robert. The Gene Wars: Science, Politics, and the Human Genome. W. W. Norton & Company; 1994. [Google Scholar]
- Eisenberg Rebecca. Genomics in the public domain: Strategy and policy. Nature Reviews Genetics. 2000;1(1):70–74. doi: 10.1038/35049590. [DOI] [PubMed] [Google Scholar]
- Gallini Nancy, Wright Brian. Technology transfer under asymmetric information. RAND Journal of Economics. 1990;21(1):147–160. [Google Scholar]
- Gans Joshua, Stern Scott. Incumbency and R&D incentives: Licensing the gale of creative destruction. Journal of Economics & Management Strategy. 2000;9(4):485–511. [Google Scholar]
- Green Jerry, Scotchmer Suzanne. On the division of profit in sequential innovation. RAND Journal of Economics. 1995;26(1):20–33. [Google Scholar]
- Heller Michael, Eisenberg Rebecca. Can patents deter innovation? The anticommons in biomedical research. Science. 1998;280(5364):698–701. doi: 10.1126/science.280.5364.698. [DOI] [PubMed] [Google Scholar]
- Huang Kenneth, Murray Fiona. Does patent strategy shape the long-run supply of public knowledge: Evidence from human genetics. Academy of Management Journal. 2009;52(6):1198–1221. [Google Scholar]
- Istrail Sorin, et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proceedings of the National Academy of Sciences. 2004;101(7):1916–1921. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe Adam, Trajtenberg Manuel, Henderson Rebecca. Geographic localization of knowledge spillovers as evidenced by patent citations. Quarterly Journal of Economics. 1993;108(3):577–598. [Google Scholar]
- Kremer Michael. Patent buyouts: A mechanism for encouraging innovation. Quarterly Journal of Economics. 1998;113(4):1137–1167. [Google Scholar]
- Kremer Michael, Williams Heidi. Incentivizing innovation: Adding to the toolkit. In: Lerner Josh, Stern Scott., editors. Innovation Policy and the Economy. Vol. 10. University of Chicago Press; 2010. pp. 1–17. [Google Scholar]
- Lander Eric, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Marshall Eliot. NIH to produce a ‘working draft’ of the genome by 2001. Science. 1998;281(5384):1774–1775. doi: 10.1126/science.281.5384.1774. [DOI] [PubMed] [Google Scholar]
- Marshall Eliot. Bermuda Rules: Community spirit, with teeth. Science. 2001;291(5507):1192. doi: 10.1126/science.291.5507.1192. [DOI] [PubMed] [Google Scholar]
- Merges Robert, Nelson Richard. On the complex economics of patent scope. Columbia Law Review. 1990;90(4):839–916. [Google Scholar]
- Moon Seongwuk. How does the management of research impact the disclosure of knowledge? Evidence from scientific publications and patenting behavior. Economics of Innovation and New Technology. 2011;20(1):1–32. [Google Scholar]
- Murray Fiona, Stern Scott. Do formal intellectual property rights hinder the free flow of scientific knowledge? An empirical test of the anti-commons hypothesis. Journal of Economic Behavior and Organization. 2007;63(4):648–687. [Google Scholar]
- Murray Fiona, Aghion Philippe, Dewatripont Mathias, Kolev Julian, Stern Scott. Of mice and academics: Examining the effect of openness on innovation. 2008. unpublished MIT mimeo. [Google Scholar]
- National Academy of Sciences. Reaping the Benefits of Genomic and Proteomic Research: Intellectual Property Rights, Innovation, and Public Health. National Academies Press; 2006. [PubMed] [Google Scholar]
- Nelson Richard. The simple economics of basic scientific research. Journal of Political Economy. 1959;67(3):297–306. [Google Scholar]
- Pennisi Elizabeth. Human genome: Academic sequencers challenge Celera in a sprint to the finish. Science. 1999;283(5409):1822–1823. doi: 10.1126/science.283.5409.1822. [DOI] [PubMed] [Google Scholar]
- Scherer Stewart. A Short Guide to the Human Genome. Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Science Online. [last accessed 21 December 2011];Accessing the Celera human genome sequence data. 2001 http://www.sciencemag.org/feature/data/announcement/gsp.dtl.
- Scotchmer Suzanne. Standing on the shoulders of giants: Cumulative research and the patent law. Journal of Economic Perspectives. 1991;5(1):29–41. [Google Scholar]
- Service Robert. Can data banks tally profits? Science. 2001;291(5507):1203. doi: 10.1126/science.291.5507.1203. [DOI] [PubMed] [Google Scholar]
- Shapiro Carl. Navigating the patent thicket: Cross licenses, patent pools, and standard setting. In: Jaffe Adam, Lerner Josh, Stern Scott., editors. Innovation Policy and the Economy. Vol. 1. MIT Press; 2000. [Google Scholar]
- Shreeve James. The Genome War: How Craig Venter Tried to Capture the Code of Life and Save the World. Ballantine Books; 2005. [PubMed] [Google Scholar]
- Snyder Michael, Gerstein Mark. Defining genes in the genomics era. Science. 2003;300(5617):258–260. doi: 10.1126/science.1084354. [DOI] [PubMed] [Google Scholar]
- Sulston John, Ferry Georgina. The Common Thread: Science, Politics, Ethics, and the Human Genome. Corgi Books; 2002. [Google Scholar]
- Uhlmann Wendy, Guttmacher Alan. Key internet genetics resources for the clinician. Journal of the American Medical Association. 2008;299(11):1356–1358. doi: 10.1001/jama.299.11.1356. [DOI] [PubMed] [Google Scholar]
- US National Human Genome Research Institute (NHGRI), US National Institutes of Health (NIH) [last accessed 21 December 2011];NHGRI policy regarding intellectual property of human genomic sequence: Policy on availability and patenting of human genomic DNA sequence produced by NHGRI pilot projects (funded under RFA HG-95-005) 1996 http://www.genome.gov/10000926.
- Venter J Craig. Prepared statement of J Craig Venter, PhD President and Chief Scientific Officer Celera Genomics, a PE Corporation Business before the Subcommittee on Energy and Environment. U.S. House of Representatives Committee on Science; 2000. [last accessed 21 December 2011]. http://clinton4.nara.gov/WH/EOP/OSTP/html/00626_4.html. [Google Scholar]
- Venter J Craig. A Life Decoded: My Genome, My Life. Viking Adult; 2007. [Google Scholar]
- Venter J Craig, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Venter J Craig, Adams Mark, Sutton Granger, Kerlavage Anthony, Smith Hamilton, Hunkapiller Michael. Shotgun sequencing of the human genome. Science. 1998;280(5369):1540–1542. doi: 10.1126/science.280.5369.1540. [DOI] [PubMed] [Google Scholar]
- Wade Nicholas. New York Times. 2003. Apr 15, Once again, scientists say human genome is complete. [Google Scholar]
- Walsh John, Cho Charlene, Cohen Wesley. View from the bench: Patents and material transfers. Science. 2005;309(5743):2002–2003. doi: 10.1126/science.1115813. [DOI] [PubMed] [Google Scholar]
- Walsh John, Cohen Wesley, Cho Charlene. Where excludability matters: Material versus intellectual property in academic biomedical research. Research Policy. 2007;36(8):1184–1203. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.