Abstract
53BP1 (TP53BP1) is a chromatin-associated factor that promotes immunoglobulin class switching and DNA double-strand break (DSB) repair by non-homologous end joining. To accomplish its function in DNA repair, 53BP1 accumulates at DSB sites downstream of the RNF168 ubiquitin ligase. How ubiquitin recruits 53BP1 to break sites remains enigmatic since its relocalization involves recognition of H4 Lys20 (H4K20) methylation by its Tudor domain. Here we elucidate how 53BP1 is recruited to the chromatin that flanks DSB sites. We show that 53BP1 recognizes mono-nucleosomes containing dimethylated H4K20 (H4K20me2) and H2A ubiquitylated on Lys15 (H2AK15ub), the latter being a product of RNF168 action on chromatin. 53BP1 binds to nucleosomes minimally as a dimer using its previously characterized methyl-lysine-binding Tudor domain and a C-terminal extension, termed the ubiquitylation-dependent recruitment (UDR) motif, which interacts with the epitope formed by H2AK15ub and its surrounding residues on the H2A tail. 53BP1 is therefore a bivalent histone modification reader that recognizes a histone “code” produced by DSB signaling.
DNA double-strand breaks (DSBs) elicit a cascade of protein recruitment on the chromatin surrounding DNA lesions that regulates DNA damage repair and signaling1,2. 53BP1 is an important effector of this DSB response, as it promotes repair by non-homologous end joining (NHEJ)3 by opposing DNA end resection4, the initiating step in homologous recombination (HR). In mice, 53BP1 is necessary for immunoglobulin class switching5,6 and dysfunctional telomere fusions7, two processes that rely on NHEJ. Furthermore, 53BP1 deficiency in mice leads to a near complete reversal of the phenotypes associated with loss of BRCA1, including tumorigenesis4,8. 53BP1 must accumulate on the chromatin surrounding DSBs to accomplish its functions9. At the molecular level, 53BP1 acts as a recruitment platform for RIF1, its effector protein during DSB repair by NHEJ10–13. 53BP1 accumulation at DSB sites, as monitored by formation of ionizing radiation (IR)-induced subnuclear foci, requires the recognition of histone methylation, in particular H4K20me214, by its tandem Tudor domain15 (Fig. 1a). However, the formation of 53BP1 foci also requires the RNF168 E3 ligase1,2 raising the question of how a ubiquitin ligase promotes the accumulation of a methylated-histone binding protein at sites of DNA damage. The current models of 53BP1 recruitment to DSB sites propose that H4K20 methylation is either induced or becomes available for 53BP1 binding after DNA damage16–19. For example, it has been proposed that JMJD2A and L3MBTL1, which bind to H4K20me2, are removed in a ubiquitylation-dependent manner from the chromatin surrounding DSB sites, to allow 53BP1 binding18,19. In aggregate, these models imply that increased accessibility of H4K20me2 at DSB sites might be sufficient to trigger 53BP1 recruitment.
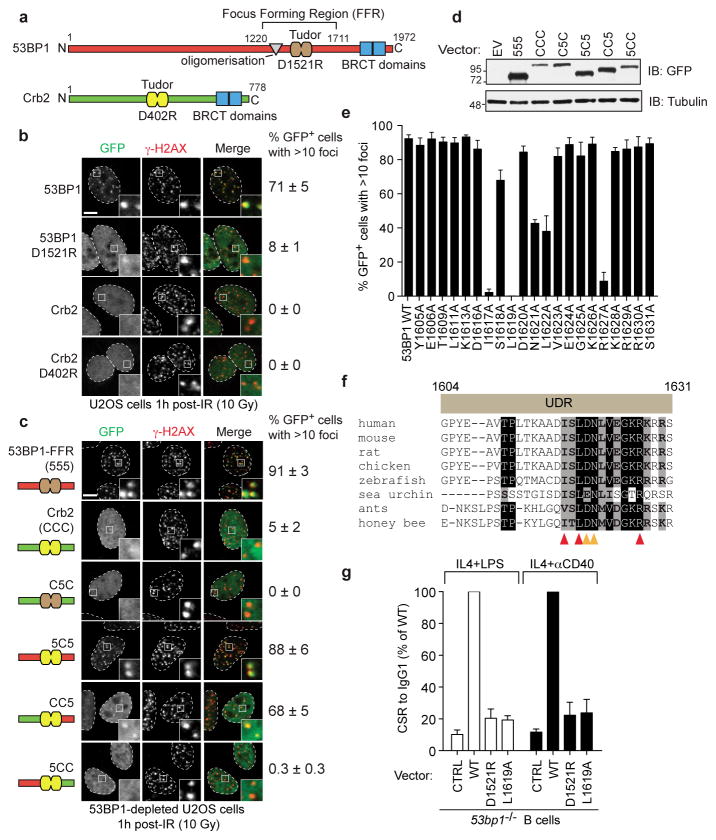
Figure 1. Identification of the 53BP1 UDR.
a, Schematic representation of 53BP1 and Crb2. b, U2OS cells transfected with GFP-53BP1 and GFP-Crb2 expression vectors were irradiated (10 Gy) and processed for GFP imaging and γ-H2AX immunofluorescence (mean ± s.e.m., N≥3). c, 53BP1-depleted U2OS cells transfected with the indicated GFP-53BP1/Crb2-derived expression vectors were irradiated (10 Gy) and processed as described in a (mean ± s.e.m., N≥3). d, Analysis of GFP fusion protein expression by immunoblotting. e, 53BP1-depleted U2OS cells transfected with vectors expressing the indicated GFP-53BP1 mutants (residues 1220-1631) were irradiated (10 Gy) and processed as in a (mean ± s.e.m., N≥3). f, Alignment of the UDR region in 53BP1 orthologs. Arrowheads highlight key UDR residues. g, Relative levels of class switching to IgG1 in 53bp1−/− murine B cells transduced with the indicated retroviruses (mean ± s.d., N≥2).
Identification of the 53BP1 UDR
We reasoned that if the above model was strictly correct, the 53BP1 ortholog from fission yeast, Crb2, should also form IR-induced foci in human cells. Indeed, Crb2 contains a tandem Tudor domain that binds to H4K20me2 (Fig. 1a)14. Crb2 accumulates at DSB sites in an H4K20me2-binding dependent manner16,20, but fission yeast does not have a recognizable RNF168 homolog, as it arose later during evolution. When expressed in human cells as a GFP fusion, Crb2 failed to form IR-induced foci whereas 53BP1 formed foci that colocalized with γ-H2AX (Fig. 1b). As expected, the accumulation of 53BP1 at DSB sites was dependent on H4K20me2 recognition since the 53BP1 D1521R mutation, which disrupts this activity of the Tudor domain, impaired the ability of 53BP1 to form IR-induced foci (Fig. 1b). The inability of Crb2 to accumulate at DSB sites in human cells was not due to a failure of Crb2 to interact with human H4K20me2 since it associated with human chromatin in a Tudor-dependent manner, as determined by fluorescence recovery after photobleaching (FRAP) (Supplementary Fig. 2a–d) and cellular subfractionation (Supplementary Fig. 2e). These experiments suggested that 53BP1 recruitment to break sites might be largely independent of an increased accessibility of H4K20me2 in damaged chromatin.
These observations provided an opportunity to map the region that endows 53BP1 with the ability to accumulate at DSB sites in an RNF168-dependent manner. We refer to this putative region as the ubiquitylation-dependent recruitment (UDR) motif. We thus prepared various chimeras between Crb2 and the minimal focus-forming region (FFR) of 53BP1, which consists of the Tudor domain flanked by an N-terminal oligomerization region and a C-terminal extension21,22 (i.e. 53BP1 residues 1220-1711; Fig. 1a). We separated the 53BP1FFR and Crb2 into 3 regions that were swapped between the two proteins, in various combinations. The chimeras prepared are illustrated in Fig. 1c and, to facilitate the identification of the chimeras, segments were labeled “5” if derived from 53BP1 and “C” if derived from Crb2. Since 53BP1 can oligomerize21, all experiments were carried out in cells depleted of endogenous 53BP1.
The domain swapping experiments first confirmed that the Crb2 Tudor can recognize H4K20me2 in human chromatin since the Crb2 Tudor domain inserted into the 53BP1FFR, supported localization to break sites (Fig. 1cd). Secondly, introduction of the sequence immediately C-terminal of the 53BP1 Tudor domain into Crb2 (CC5 chimera) produced a protein that accumulated into IR-induced foci that colocalized with γ-H2AX (Fig. 1cd). Importantly, the accumulation of the CC5 chimera at DSB sites was dependent on RNF168 (Supplementary Fig. 3ab), strongly suggesting that sequences C-terminal of the 53BP1 Tudor compose the UDR. We further narrowed down the UDR to the region between residues 1604 and 1631 (Supplementary Fig. 3c–e).
Next, we performed alanine-scanning mutagenesis of the UDR, in the context of 53BP11220-1631, to identify residues that participate in the recruitment of 53BP1 to DNA damage sites. These studies identified 5 residues: I1617, L1619, N1621, L1622 and R1627, whose mutation to alanine disrupts 53BP1 recruitment to DSB sites, with the I1617A and L1619A mutations having the strongest impact (Fig. 1e and Supplementary Fig. 4). The importance of L1619 and L1622 in 53BP1 recruitment to break sites was observed previously21. Introduction of these five mutations in the context of full-length 53BP1 also impaired IR-induced focus formation, confirming their importance for accumulation at DSB sites (Supplementary Fig. 5). The 5 residues important for the activity of the UDR are clustered in a 12-amino acid residue segment that is highly conserved amongst 53BP1 orthologs in organisms that have a recognizable RNF8 pathway (Fig. 1f).
The UDR is required for 53BP1 function
RIF1 is the 53BP1 effector during DSB repair10–13. We therefore examined the contribution of the UDR in promoting RIF1 IR-induced focus formation. We tested 8 UDR mutations introduced in an siRNA-resistant 53BP1 vector: the 5 mutations that affect 53BP1 recruitment and 3 others (K1613A, D1616A and E1624A) that do not. We also included in these assays 53BP1 and 53BP1D1521R, our positive and negative controls, respectively. We observed that the mutations that impaired 53BP1 accumulation at DSB sites also abrogated RIF1 foci after IR (Supplementary Fig. 5). These results suggested that the UDR is critical for the function of 53BP1 in the DSB response. In further support of this observation, reconstitution of 53bp1−/− murine B cells with either the D1521R or L1619A mutants, failed to restore class switch recombination (CSR) from IgM to IgG1, whereas reintroduction of wild type 53BP1 restored CSR (Fig. 1g and Supplementary Fig. 6). Furthermore, the UDR-defective L1619A mutant was unable to restore either resistance to IR-induced DSBs in DT40 53bp1−/− cells, or to restrict HR in BRCA1 and 53BP1 co-depleted cells (Supplementary Fig. 7). Together, these results indicate that the UDR is necessary for the biological functions of 53BP1.
53BP1 binds to ubiquitylated nucleosomes
Next, we sought to determine the mechanism by which the UDR promotes 53BP1 recruitment to DSB sites. We first considered that the UDR might increase the affinity of 53BP1 for H4K20me2 due to its location, i.e. apposed to the Tudor domain. We expressed GST-53BP1 fusion proteins consisting of the tandem Tudor domain with (Tudor-UDR) or without (Tudor) the UDR region and examined binding to a H4K20me2-derived peptide in pulldown assays. We observed that both proteins interacted equally well with H4K20me2 in a manner that required the D1521 residue (Fig. 2a). The L1619A mutation, which abolishes UDR activity, had no effect on H4K20me2 binding (Fig. 2a), indicating that the UDR does not impact recognition of H4K20me2, at least in the context of a peptide.
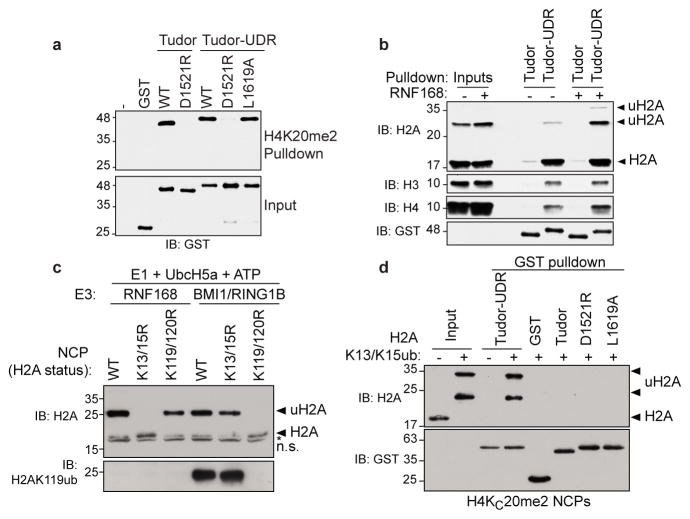
Figure 2. 53BP1 binds to ubiquitylated nucleosomes.
a, Streptavidin pulldowns of the indicated GST fusion proteins with a biotinylated H4K20me2 peptide. b, Chromatin from HEK293 cells expressing Flag-RNF168 (+) or not (−) were subjected to pulldown assays with the indicated GST fusion proteins. c, Ubiquitylation of the indicated NCPs by RNF168 and BMI1-RING1B. n.s. non-specific band. d, Pulldown assays of NCPs containing H4KC20me2 with the indicated GST fusion proteins. NCPs were ubiquitylated with RNF168 as the E3 (+); a reaction without E1 (−) was used as negative control.
An alternative function of the UDR might be that it promotes the interaction of 53BP1 with chromatin. To test this possibility, we prepared polynucleosome-enriched extracts obtained from micrococcal nuclease digestion of human chromatin. Since RNF168 overexpression can trigger 53BP1 accumulation on chromatin23, even in the absence of RNF824, we prepared a set of extracts from cells that either overexpress RNF16825, or that were transfected with an empty vector. RNF168 was recently shown to catalyze a new histone mark, H2AK13/K15 mono-ubiquitylation (H2AK13/K15ub)26,27 and thus we sought to test whether 53BP1 could potentially bind to nucleosomes containing RNF168-ubiquitylated H2A. Immunoblotting of H2A showed that the global levels of H2A ubiquitylation (uH2A) did not greatly change following RNF168 overexpression (Fig. 2b) because H2AK119ub, which is catalyzed by E3s such as BMI1-RING1B28 is much more abundant than H2AK13ub or K15ub. These extracts were then subjected to GST pulldown assays using either the 53BP1 Tudor domain or the extended Tudor-UDR module. In the absence of exogenous RNF168 expression, we observed a UDR-dependent interaction between 53BP1 and histone H2A, H3 and H4 (Fig. 2b). However, in the presence of RNF168, we observed a striking increase in the retrieval of mono- and di-ubiquitylated H2A by the Tudor-UDR protein (Fig. 2b). Together, these results suggested that the UDR may stimulate two modes of interaction between 53BP1 and nucleosomes: one mode that is independent of histone ubiquitylation, and which may reflect the constitutive interaction of 53BP1 with chromatin; and a second mode of interaction that is dependent on H2A ubiquitylation by RNF168, and which may represent the interaction that leads to 53BP1 accumulation at DSB sites.
53BP1 recognizes H2AK15ub
Since the above experiments were carried out with polynucleosomes, the interactions observed could be the product of avidity between the dimeric 53BP1 fusion protein and the multimeric nucleosomal arrays. Therefore, we tested whether we could detect binding between 53BP1 and fully recombinant monomeric nucleosome core particles (NCPs) (Supplementary Fig. 8a). We used an N-terminal fragment of RNF168 in ubiquitylation reactions with UbcH5, which recapitulated H2AK13/K15 ubiquitylation26 (Fig. 2c and Supplementary Fig. 8b–d). To generate H2AK119ub, we used a recombinant BMI1-RING1B complex as the E3 (Fig. 2c and Supplementary Fig. 8bc). For histone methylation, we produced methyl-lysine analog versions of H4K20me2 (H4KC20me2) and H3K9me2 (H3KC9me2)29 prior to octamer and nucleosome assembly.
We first assembled H4KC20me2-containing NCPs and subjected them to ubiquitylation reactions in the presence of RNF168 to produce H2AK13/K15ub (Fig. 2d). As control, we also carried out reactions without E1 and, as expected, no H2A ubiquitylation was detected (Fig. 2d). The products of these two reactions were used in GST-pulldown assays with various fusion proteins derived from 53BP1. We observed a striking, ubiquitylation-dependent interaction between the 53BP1 Tudor-UDR fusion and NCPs (Fig. 2d) that was not seen with the GST protein alone, the 53BP1 Tudor domain alone, the D1521R mutant or, finally, the L1619A mutant that disrupts UDR function. Together, these data indicate that the 53BP1 Tudor-UDR module promotes binding to methylated and ubiquitylated mono-nucleosomes in a manner that involves the same residues that are necessary for 53BP1 accumulation at DSB sites.
Next, we examined whether the binding of 53BP1 to NCPs was specific to H4K20me2 and H2AK13/K15ub. We assembled a series of NCPs that contained H4KC20me2, H3KC9me2 or their unmodified lysine counterparts. These NCPs were then used in ubiquitylation reactions with RNF168 to produce H2AK13/K15ub-containing NCPs. When these NCPs were interrogated for binding to the 53BP1 Tudor-UDR module, we observed a specific interaction between 53BP1 and the NCPs containing H4KC20me2 (and H2AK13/K15ub) but not those containing H3KC9me2 or the unmodified H4K20 (Fig. 3a and Supplementary Fig. 9a). Next, we employed H4KC20me2-containing NCPs and used them as substrates in ubiquitylation reactions with RNF168 or BMI1-RING1B. Both reactions produced similar levels of mono-ubiquitylated H2A (Fig. 3b) but when they were used in GST pulldown assays with the 53BP1 Tudor-UDR module, we only observed an interaction with the RNF168-ubiquitylated NCPs. Importantly, we excluded the possibility that the binding was due to the presence of di-ubiquitylated H2A in the RNF168 sample (Supplementary Fig. 9b). From these results, we conclude that the interaction between 53BP1 and nucleosomes requires the presence of both H4K20me2 and H2AK13/K15ub.
Figure 3. 53BP1 is a bivalent reader of the H4K20me2 and H2AK15ub histone marks.
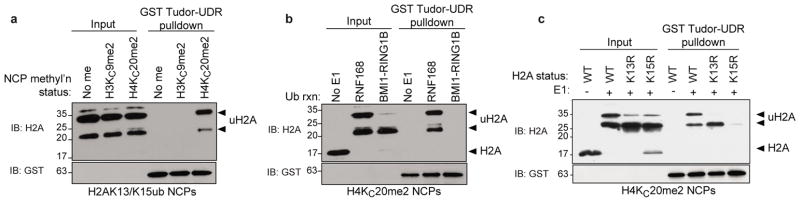
a, Pulldown assays of RNF168-ubiquitylated NCPs containing unmethylated histone H4 and H3 (No me), H4KC20me2 or H3KC9me2 with GST-Tudor-UDR. b, Pulldown assays of NCPs ubiquitylated with the indicated E3s by GST-Tudor-UDR. A reaction without E1 (No E1) acts as negative control. c, GST-Tudor-UDR pulldown assays of the indicated NCPs ubiquitylated with RNF168 (+); a reaction lacking E1 (−) was used as negative control.
While RNF168 can ubiquitylate H2A K13 or K15 in vitro and in vivo26,27, we tested whether 53BP1 displayed selectivity towards K13ub or K15ub. To do so, we assembled H4KC20me2-containing NCPs with either H2AK13R or H2AK15R substitutions to leave K15 or K13, respectively, as the only residue ubiquitylated by RNF168 (Fig. 3c). To our surprise, when these ubiquitylated NCPs were used in pulldown assays, we found that the 53BP1 Tudor-UDR protein interacted specifically with NCPs containing H2AK15ub (Fig. 3c). This result indicates that 53BP1 has the striking ability to discriminate between two closely positioned ubiquitylated lysine residues on H2A.
Molecular basis of H2AK15ub selectivity
One possible cause for the 53BP1 selectivity towards H2K15ub could be the presence of sequence elements in the H2A N-terminal tail that are recognized by 53BP1. We noted that 3 mutations (K9R, A14S and R17S) were sufficient to convert the sequence surrounding K13 in the H2AK15R mutant into the sequence that normally surrounds H2AK15 (Fig. 4a). We found that the resulting mutant (H2AK15Rm3), when ubiquitylated, bound robustly to the 53BP1 Tudor-UDR module (Fig. 4a). We conclude that additional residues in the H2A N-terminus contribute to the binding of 53BP1 to H2AK15ub-containing nucleosomes.
Figure 4. Determinants of H2AK15ub recognition.
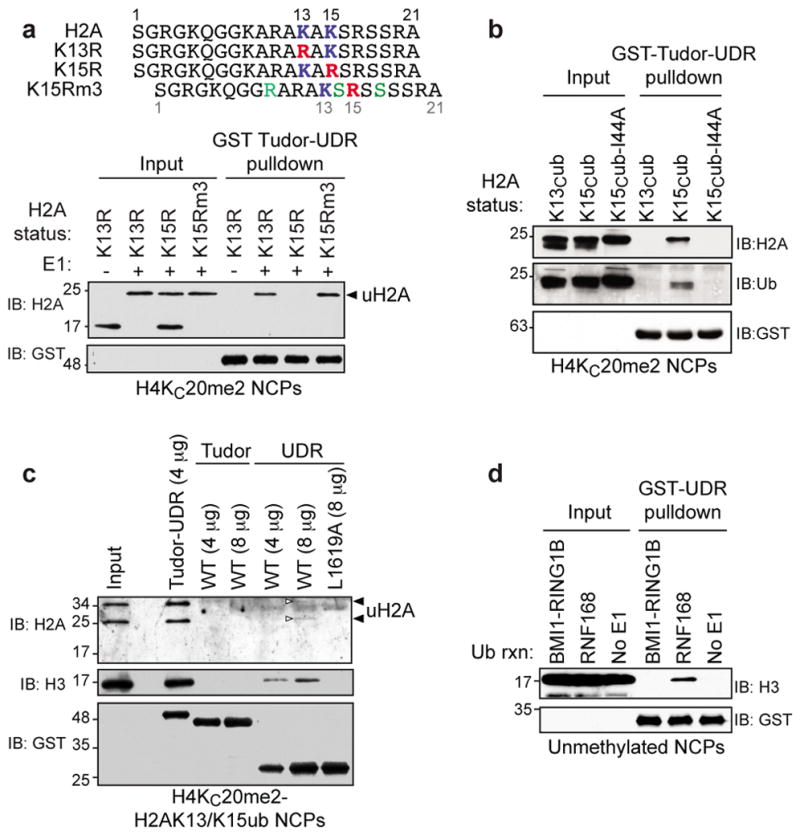
a, Upper panel: Sequence of the H2A N-termini of various mutants. Mutations are highlighted in color. Lower panel: GST-Tudor-UDR pulldown assays of RNF168-ubiquitylated NCPs (+) assembled with H4KC20me2 and the indicated H2A mutants. A reaction without E1 (−) was used as control. b, GST-Tudor-UDR pulldown assays of NPCs with the indicated H2A ubiquitylations. c, Pulldown assays of RNF168-ubiquitylated H4KC20me2-NCPs with the indicated fusion proteins. A white arrowhead highlights faint uH2A bands. d, GST-UDR pulldown assays of unmethylated NCPs that were either ubiquitylated with the indicated E3 or not (no E1).
We next investigated whether ubiquitin recognition contributed to the 53BP1-NCP interaction. Ubiquitin contains a hydrophobic patch centered on its I44 residue that contributes to most ubiquitin-dependent interactions30 and therefore we sought to test whether the ubiquitin I44 residue was important for 53BP1 recognition of H2AK15ub-NCPs. We used chemical ubiquitylation by disulfide exchange31 to prepare NCPs that contain H2A chemically ubiquitylated on H2AK13 (H2AKC13ub), H2AK15 (H2AKC15ub) and H2AK15 ubiquitylated with Ub-I44A (H2AKC15ub-I44A; Fig. 4b). Those NCPs were then used in pulldown assays with the Tudor-UDR module. As expected, we found that the interaction was selective for H2AKC15ub (Fig. 4b). However, H2AKC15ub-I44A containing NCPs were unable to be retrieved by 53BP1 (Fig. 4b), suggesting that ubiquitin recognition participates in the interaction of 53BP1 with H2AK15ub.
In aggregate, our results support a model where the Tudor-UDR module comprises two histone modification-binding domains: the Tudor domain that binds H4K20me2 and the UDR, which may interact directly with H2AK15ub. In support of this model, the transfer of the 53BP1 UDR onto the Crb2 Tudor domain endowed it with the ability to robustly bind to RNF168-ubiquitylated and H4K20me2-containing NCPs (Supplementary Fig. 10a–d). This observation prompted us to produce the isolated UDR, as a GST fusion protein, and to assess its binding properties. As expected, the isolated UDR did not bind to H4K20me2 (Supplementary Fig. 10e). However, when H2AK13/K15ub-containing NCPs were incubated with increasing amounts of the isolated UDR, we observed a dose-dependent retrieval of NCPs in a manner that required the critical L1619 residue (Fig. 4c). This weak binding of the UDR to NCPs was specific to RNF168-dependent ubiquitylation (Fig. 4d) and, as expected, was independent of H4K20me2 given the absence of the Tudor domain (Fig. 4d and Supplementary Fig. 11a). Interestingly, we failed to detect either an interaction between the UDR and free ubiquitin by nuclear magnetic resonance or an interaction between the UDR and a ubiquitylated H2A peptide in a pulldown experiment (Supplementary Fig. 11bc). These results indicate that the UDR recognizes H2AK15ub specifically in the context of the nucleosome.
Discussion
In response to DSBs, ATM signaling triggers a first wave of chromatin ubiquitylation that is dependent on RNF82. The role of RNF8, and its E2 UBC13, is to trigger the recruitment of RNF168 to DSB sites where it catalyzes H2AK13/K15 mono-ubiquitylation. Together, our work identifies 53BP1 as a bivalent histone modification reader that recognizes nucleosomes modified with H4K20me2 and the DNA damage-inducible H2AK15ub mark (Supplementary Fig. 1). We propose that the engagement of H4K20me2 by the Tudor domain positions the UDR in the correct orientation to contact the epitope formed by H2AK15ub, a scenario supported by modeling (Supplementary Fig. 12). In the Supplementary Data section, we also present evidence that 53BP1 recognizes nucleosomes minimally as a dimer (Supplementary Figs. 13–14). Together, these observations suggest that 53BP1 may alter nucleosomal array structure either by acting akin to a wheel-clamp if a dimer engages a mono-nucleosome (Supplementary Fig. 1), or by acting as a ubiquitylation-dependent nucleosome crosslinker if it can bridge adjacent nucleosomes (Supplementary Fig. 1). These plausible binding modes may be central to the function of 53BP1 as an inhibitor of end-resection.
Our experiments also identify the first site-specific reader of histone ubiquitylation. 53BP1 is likely to be one of many readers that interpret the various histone ubiquitylation marks identified to date. Proteins such as ASH2L (for H2BK120ub)32, RNF168 and RNF169 (for H2AK13/K15ub)25 are prime candidates for ubiquitin mark readers. RNF169 presents an attractive case since it acts as a competitive inhibitor of 53BP125,33. These observations and the identification of 53BP1 as an H2AK15ub reader further emphasize the need to decipher the chromatin modification landscape, its regulation and its interpretation, at sites of DNA damage.
METHODS
Cell culture and plasmid transfection
Human cell culture media were supplemented with 10% fetal bovine serum (FBS). U-2-OS (U2OS) cells were cultured in McCoy’s medium (Gibco). HEK293T and HeLa DR-GFP cells were cultured in DMEM (Gibco). HCT116 Flp-In T-REx Flag and Flag-RNF168 stably transfected cell lines were cultured in DMEM supplemented with 250 μg/mL hygromycin B and 5 μg/mL blasticidin. CT116 Flp-In T-REx Flag and Flag-RNF168 stably transfected cell lines were described previously25, U2OS and HEK293T cells were purchased from ATCC and HeLa DR-GFP cells were a gift from the laboratory of Roger Greenberg. All cell lines were tested negative for mycoplasma contamination. To induce protein expression in these cell lines, 5 μg/mL doxycycline was added to the culture medium for 24 h. DT40 cells were obtained from the laboratory of Dongyi Xu and grown at 39.5 °C, 5% CO2 in RPMI 1640 medium (Gibco) supplemented with 10% fetal calf serum, 1% chicken serum and 0.1 mM β-mercaptoethanol. Plasmid transfections were generally carried out using Lipofectamine 2000 Transfection Reagent (Invitrogen) or Effectene (Qiagen).
Unless stated otherwise, for microscopy experiments, cells were fixed 1 h post-irradiation (10 Gy). The DNA was also counter-stained with DAPI (not shown) and used to trace the outline of the nuclei.
Retroviral restitution of 53BP1 in B cells
Class switching to IgG1 was assayed in 53bp1−/− murine primary B cells complemented with 53BP1 (1-1711) and mutants thereof by retroviral delivery. Mature B lymphocytes were isolated from the spleens of 2 males and 1 female 8–15 week old 53bp1−/− C57BL/6 strain 129-Trp53bp1tm1Jc/J mice3 by depletion of CD43+ cells using CD43 microbeads (Miltenyi Biotech) according to the manufacturer’s instructions. Mice were obtained from Jackson laboratories. Purified B cells were resuspended at a concentration of 106 cells/mL in the presence of 50 ng/mL IL-4 (Preprotech) and 25 μg/mL LPS (Sigma-Aldrich) or 1 μg/mL agonist anti-CD40 (BD) to allow B cell proliferation/activation. Retroviral particles were collected from the supernatant of Plat-E packaging cells35 transfected with 10 μg of the different retroviral pMX constructs. Retroviral supernatants were passed through a 0.45 μm filter and then ultracentrifuged at 20,000 x g at 25 °C for 90 min through a 20% sucrose layer to obtain purified virus. B cells were subsequently infected with the retroviral concentrate in the presence of 8 μg/mL Polybrene (Sigma-Aldrich) and 20 mM HEPES, pH 7.5 by plate centrifugation. The B cell medium was subsequently changed and replaced with fresh RPMI medium supplemented with 50 ng/mL IL-4 and 25 μg/mL LPS or 1 μg/mL agonist anti-CD40 to induce class switching to IgG1. CSR was analyzed 3 days post-infection by flow cytometry as described previously11. Experiments with 53bp1−/− mice (Trp53bp1tm1Jc/tm1Jc) were carried out according to regulatory standards and were approved by the Mount Sinai Hospital animal care committee (Protocol AUP#0200a).
Chromatin pulldown
HEK293 chromatin-enriched extracts (CEE) were prepared essentially as described25. Chromatin pulldowns were performed with 2.5 μg of recombinant GST-tagged proteins immobilized on glutathione sepharose 4B (GE Healthcare) in chromatin pulldown buffer (CPB: 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM DTT) for 1 h at 4 °C. Pulldowns were then carried out by mixing 125 μg of CEE isolated from cells stably expressing Flag-RNF168 in a final volume of 1.5 mL for 3 h at 4°C. Pulldowns were then washed 4 times with 1 mL of CPB and eluted in 2X Laemmli SDS-PAGE sample buffer for analysis by immunoblotting.
NCP reconstitution
Recombinant histones were purified essentially as described36,37. Briefly, after preparation of inclusion bodies, the histones were purified under denaturing conditions on either a HiPrep 16/60 Sephacryl S-300 HR (GE Healthcare) size exclusion column or a 5 mL HiTrap™ SP HP (GE Healthcare) cation exchange column. Fractions containing the purified histones were pooled and extensively dialyzed into water and 2 mM β-mercaptoethanol before lyophilization. His6-G76C ubiquitin and His6-I44A/G76C ubiquitin to be installed on H2A K13C or K15C were purified over a Ni-NTA column and the His-tag was removed with TEV protease. DTT was added to the ubiquitin to reduce any oxidized bonds and then quickly buffer exchanged into degassed water (with no DTT) using a PD-10 column (GE Healthcare). The reduced ubiquitin was immediately snap frozen and lyophilized to dryness. Octamers were refolded by mixing the four histones in equimolar ratios, followed by dialysis into 2 M NaCl, and then purified on a Superdex 200 10/300 GL size exclusion column (GE Healthcare). Nucleosome core particles (NCPs) were reconstituted as described36. The 151 base pair DNA used to wrap the mono-nucleosomes was obtained from an EcoRV digest of 32x601 DNA plasmid (a gift from Cheryl Arrowsmith).
Histone labeling
The installation of a dimethyl-lysine analog (di-MLA) at the mutated cysteine of the H4K20C protein or H3K9C (C110A) protein was done exactly as described29. The di-MLA installation was confirmed by mass spectrometry for the H3KC9me2 protein, and by mass spectrometry and immunoblotting against H4K20me2 for the H4KC20me2 full-length protein. Once labeled, the H4KC20me2 and H3KC9me2 proteins were refolded into octamers as previously described. Installation of a modified wild-type or I44A mutant ubiquitin was achieved by disulfide-directed conjugation. H2A K15C or K13C were conjugated with G76C ubiquitin or I44A/G76C ubiquitin via disulfide exchange prior to octamer refolding. Specifically, the cysteine on the histone was activated by dissolving 5 mg of lyophilized H2A in 1 mL of water containing 5 mM TCEP. Then 5 mg of DTNP (2,2′-dithiobis(5-nitropyridine); Sigma Aldrich) dissolved in 2 mL of acetic acid was added to the histone and this mixture was agitated at room temperature overnight. The activated histone reaction was then dialyzed extensively against water before purification by S75 10/300 gel filtration (GE Healthcare) in conjugation reaction buffer. The conjugation reaction was set up in 6 M Guanidinium-HCl, 50 mM Tris-HCl pH 6.9 at room temperature with a 2:1 ratio of lyophilized ubiquitin to degassed activated histone. After 1 h, the completion of the reaction was confirmed by mass spectrometry. Unconjugated ubiquitin and any oxidized ubiquitin-ubiquitin conjugates were removed in subsequent gel filtration runs.
In vitro ubiquitylation of the NCP
Nucleosomes were ubiquitylated by incubating 2.5 μg recombinant mononucleosomes with 30 nM E1, 1.5 μM UbcH5a, 4 μM RNF168 (1-113) or BMI1-RING1B complex, 22 μM ubiquitin or methylated ubiquitin (Boston Biochem), and 3.33 mM ATP in a buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 μM ZnOAc and 1 mM DTT at 30 °C for 2 h.
NCP pulldown assays
NCP pulldowns were done in a total volume of 100 μL by using 15–20 μL ubiquitylation reaction, 4 or 8 μg GST- or MBP-protein coupled to glutathione sepharose 4B (GE Healthcare) or amlyose resin (New England Biolabs), respectively, in the same buffer as the peptide pulldowns, except containing 0.1% BSA. Pulldown reactions were incubated for 2 h at 4 °C. Pulldowns were then washed 3 times with 0.5 mL of the pulldown buffer plus 0.1% BSA and eluted in 2X Laemmli SDS-PAGE sample buffer for analysis by immunoblotting.
Plasmids
The GFP-53BP1 expression vector (DDp1910) resistant to siRNA 53BP1 #1 (ThermoFisher D-003548-01) was described previously11. The GFP was swapped for mCherry using the KpnI-AscI sites to generate pcDNA5-mCherry-FRT/TO-53BP1 (DDp2005). The 53BP1 deletion vectors (consisting of residues 1220-1711, 1220-1631, 1484-1603, 1484-1631 or 1604-1631) were created by inserting PCR-amplified fragments (derived from DDp1910) into the NotI and ApaI sites of pcDNA5-GFP-FRT/TO; EcoRI and NotI sites of pcDNA5-Flag-FRT/TO-DmrA (DDp1911) and pcDNA5-HA-FRT/TO-DmrC (DDp1912); in the BamHI and EcoRI sites of a modified pETM-30-02 vector in which the ORF of GST was inserted between the hexahistidine tag and the TEV cleavage site or between the BamHI and PstI sites of pMAL-c2X (New England Biolabs). Mammalian expression vectors for the components of the heterodimerization system were generated by PCR amplification of DmrA (FKBP12) and DmrC (FRB) from pLVX-Het-2 and pLVX-Het-1 (iDimerize™ Inducible Heterodimer System, Clonetech), respectively, and by ligation into the BamHI and EcoRI sites of pcDNA5-Flag-FRT/TO and pcDNA5-HA-FRT/TO (DDp1915). The GFP-Crb2 and GFP-CC (i.e. residues 1-507 of Crb2) vectors (DDp1913 and DDp1914, respectively) were constructed by inserting PCR-amplified fragments of Crb2 into the NotI and ApaI sites of pcDNA5-GFP-FRT/TO or pcDNA5-GFP-NLS-FRT/TO (DDp1916). The source of the Crb2 coding sequence was the pJK148-Crb2 plasmid (gift from Li-Lin Du)38. Chimeras of 53BP1 and Crb2 were obtained by annealing overlapping PCR fragments (where 555 = aa 1220-1483, 1484-1603, 1604-1711 and CCC = aa 1-357, 358-507, 507-708). The annealed fragments were then ligated into the NotI and ApaI sites of pcDNA5-GFP-NLS-FRT/TO (DDp1916). The GST-Crb2 Tudor domain (i.e. residues 358-507 of Crb2) alone or fused to 53BP1 UDR (residues 1604-1631), yielding GST-Tudor(C) and GST-Tudor(C)-UDR(5) vectors, respectively were constructed by inserting PCR-amplified sequences into the BamHI and EcoRI sites of a modified pETM-30-02 described above. Bacterial expression vectors for histones (His6-human H2A in pET15b, His6-human H2B in pET15b, Xenopus laevis H3 in pET3d and X. laevis H4 in pET3a) were obtained from Cheryl Arrowsmith. The RNF168 bacterial expression vector (residues 1-113; DDp1878) was obtained by PCR amplification of the DDp110925 and cloned into pPROEX HTa (Invitrogen) using the BamHI and SpeI sites. The BM11-His6 (residues 1-108) bacterial expression vector (DDp1886) was obtained by PCR amplification of pGEX-4T1-BMI1(1-108) and cloned into pET24b(+) using NdeI and XhoI sites. The RING1B (residue 1-116) bacterial expression vector (DDp1887) was obtained by PCR amplification of pET28-MHL-RINGb(1-120) and cloned into pGEX-6P-1 using the BamHI and NotI sites. pGEX-4T1-BMI1(1-108) and pET28-MHL-RINGB (1-120) were gifts of Yufeng Tong. The retroviral vector pMX-53BP1 (1-1711) and its D1521R derivative were gifts of André Nussenzweig9. All mutations were introduced by site-directed mutagenesis using QuikChange (Stratagene) and all plasmids were sequence-verified.
Subellular fractionation
A cytoplasmic fraction (CYTO) was obtained by harvesting HEK293 cells in EBC1 buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.5% IGEPAL CA-630, 1 mM EDTA, 1 mM DTT, 1X protease inhibitors - Complete, EDTA-free; Roche). Following centrifugation at 1000 x g for 15 min at 4°C, the nuclear pellet was resuspended and periodically vortexed in EBC2 buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM CaCl2, 1X protease inhibitors - Complete, EDTA-free; Roche) over 30 min. Following centrifugation at 1000 x g for 15 min, the supernatant was harvested as the nuclear soluble fraction (NS). The remaining insoluble chromatin fraction was then solubilized by microccocal nuclease digestion for 30 min at 30 °C and centrifugation at 1000 x g for 15 min; the supernatant was harvested at the nuclear soluble fraction (CHR)
RNA interference
All siRNAs employed in this study were single duplex siRNAs purchased from ThermoFisher. RNAi transfections (40 nM) were performed using DharmaFECT 1 (ThermoFisher) or RNAiMax (Invitrogen) in a forward transfection mode, following the manufacturer’s protocol. The individual siRNA duplexes used were: 53BP1 (ThermoFisher, D-003548-01, target sequence: 5′-GAGAGCAGAUGAUCCUUUA-3′), RNF168 (ThermoFisher, D-007152-04, target sequence: 5′-GAAGAGUCGUGCCUACUGAUU-3′), BRCA1 (ThermoFisher D-003461-05, target sequence: 5′-CAGCUACCCUUCCAUCAUAUU-3′) and non-targeting control siRNA (ThermoFisher, D-001210-02, target sequence: 5′-UAAGGCUAUGAAGAGAUAC-3′). Except when stated otherwise, siRNA were transfected 48 h prior to cell processing.
Antibodies
We employed the following antibodies: mouse anti-53BP1 (clone 19, BD Biosciences), rabbit anti-53BP1 (A300-273A, Bethyl), mouse anti-γ-H2AX (clone JBW301, Millipore), rabbit anti-γ-H2AX (#2577, Cell Signaling Technologies), rabbit anti-BRCA1 (#07-434, Millipore), rabbit anti-KAP1 (A300-274A, Bethyl), goat anti-RIF1 (N20) (sc55979, Santa Cruz), mouse anti-Flag (clone M2, Sigma), mouse anti-tubulin (clone DM1A, Calbiochem), mouse anti-GFP (Roche) and rabbit anti-cyclin A (gift from M. Pagano), mouse anti-HA (F-7, sc-7392, Santa Cruz), rabbit anti-H2A (ab18255, Abcam), rabbit anti-H4 (NBP1-19404, Novus Biologicals), rabbit anti-H3 (ab1791, Abcam), rabbit anti-H2B (ab1790, Abcam), rabbit anti-H4K20me2 (9759, Cell Signaling Techologies), rabbit anti-GST (sc-459, Santa Cruz), mouse anti-MBP (E8032, NEB), rabbit anti-ubiquitin (Z0458, Dako) and mouse anti-actin (CP01, Calbiochem). Peroxidase-affiniPure goat anti-rabbit IgG (111 035 144, Jackson ImmunoResearch) and HRP-linked sheep anti-mouse IgG (NA931, GE Healthcare) were used as secondary antibodies in immunoblotting. The following antibodies were used as secondary antibodies in immunofluorescence microscopy: Alexa Fluor 488 goat anti-mouse, Alexa Fluor 488 donkey anti-mouse, Alexa Fluor 555 goat anti-mouse, Alexa Fluor 555 goat anti-rabbit, Alexa Fluor 555 donkey anti-goat, Alexa Fluor 647 donkey anti-mouse (Molecular Probes).
Fluorescence Recovery After Photobleaching (FRAP)
For FRAP experiments, cells were seeded onto 25 mm round coverslips, transferred to a Chamlide Chamber and imaged using a Quorum WaveFX Spinning Disc Confocal System (Quorum Technologies Inc., Guelph, Canada) equipped with a 63X oil objective and a temperature-controlled chamber (37 °C, 5% CO2). All images were acquired using Volocity Software (Improvision). FRAP experiments were performed 24 h after cell transfection. Five images were acquired before photobleaching a region of interest using a Photonic Instruments Mosaic (450–515 nm Ar laser, 0.6 sec) to achieve at least 60–70% of measured fluorescence loss. Images were then acquired every 0.08 sec for 20 sec. Image processing was performed using Volocity and included photobleaching and background correction. Recovery time was obtained by fitting a single exponential equation. As the image set of each sample was acquired with nonuniform time intervals, a cubic spline interpolation technique was used to resample data on a common time base.
Recombinant protein production
GST and MBP fusions proteins were produced as previously described25,34. Briefly, MBP and GST proteins expressed in Escherichia coli were purified on amylose (New England Biolabs) or glutathione sepharose 4B (GE Healthcare) resins according to the batch method described by the manufacturer and stored in 50 mM HEPES pH 7.5, 150 mM NaCl, 5% glycerol. His6-UbcH5a and His6-RNF168 (1-113) were purified on Ni-NTA agarose (Qiagen) and stored in 50 mM Tris-HCl pH 7.5, 1 mM EDTA, 10% glycerol. For NMR, the proteins were further purified by gel filtration in NMR buffer (S75 10/300 HiPrep Superdex, GE Healthcare). The pET24b(+)-BMI1 and pGEX-6p-1-RING1B expression plasmids were co-transformed and the proteins were purified as a complex as described39.
Peptides
The H4K20me2 peptide (H4K20me2: biotin-YGKGGAKRHR-K(me2)-VLRD) was purchased from BioBasic and the H2AK15ub peptide (biotin-spacer-ARAKAK(Ub)SRSSR; Spacer = 8-amino-3,6-dioxaoctanoic acid) was purchased from Lifesensors Inc.
Peptide pulldowns were performed by incubating 2.5 μM MBP or GST-tagged 53BP1 proteins with 25 μM of the indicated biotinylated histone H4-derived peptide in peptide pulldown buffer (PPB) (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.05% NP-40, 1% BSA). After 2 h at 4 °C, 10 μL of the pulldown reaction mixtuere was removed as input control and 10 μL of streptavidin-Dynabeads (Dynal) were added to the pulldown mixture and incubated for an additional 30 min at 4 °C. The Dynabeads were then washed twice with 750 μL PPB, twice with 750 μL of 50 mM Tris-HCl pH 8.0, 150 mM NaCl and were then eluted in 25 μL 2X Laemmli SDS-PAGE sample buffer for analysis by immunoblotting.
Gel filtration
Estimation of the GST-53BP1 Tudor-UDR and MBP-53BP1 Tudor-UDR molecular weights in solution was done by gel filtration analysis using a 24 mL S200 Superdex column (S200 10-300 GL, (GE Healthcare)) in 50 mM HEPES pH 7.5, 150 mM NaCl. Approximately 400 μg of purified GST or MBP-tagged protein was injected onto the column. The molecular weight of each sample was estimated according to the elution profile of gel filtration standard molecular weight markers (151-1901, Bio-Rad).
NMR spectroscopy
NMR data were acquired at 25 °C on a 600 MHz Bruker AVANCE III spectrometer equipped with a 1.7 mm TCI CryoProbe. Two-dimensional 1H,15N HSQC (Heteronuclear Single Quantum Coherence) spectra were collected for 0.2 mM 15N-ubiquitin in the absence or presence of GST, GST-53BP1-UDR (1604-1631) or CDC34. All NMR samples were prepared in 50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM DTT and 10% D2O. Backbone resonance assignments for human ubiquitin have been reported previously40. Residue A46 was not seen in the spectra, and therefore was excluded from analysis.
Mass spectrometry
Electrospray ionization mass spectrometry analysis was performed on an Agilent LC/MSD TOF mass spectrometer. Samples were diluted in 0.1% trifluoroacetic acid prior to analysis. Deconvolution was performed using Agilent MassHunter workstation software for the analysis of modified histones.
Supplementary Material
Acknowledgments
We are grateful to R. Szilard and S. Panier for critically reading the manuscript; to L-L. Du, A. Nussenzweig, Y. Tong and C. Arrowsmith for plasmids; to M. Cook and A. Rosebrock for centrifugal elutriation and to B. Sauriol for help with FRAP data data analysis. A.F.T. and A.O. both receive post-doctoral fellowships from the CIHR, C.E.D. is an Ontario Post-doctoral Fellow, J.K.L. receives a post-doctoral fellowship from the Leukemia and Lymphoma Society, whereas C.C.Y.L. and M.C.L. are post-doctoral fellows of the Canadian Breast Cancer Foundation (Ontario Division). D.D. is the Thomas Kierans Chair in Mechanisms of Cancer Development and a Canada Research Chair (Tier 1) in the Molecular Mechanisms of Genome Integrity. Work in the D.D. laboratory was supported by CIHR grant MOP84297 and grant GL2-01-010 from the Ontario Research Fund.
Footnotes
Supplementary Information. A Supplementary Data section detailing the importance of dimerization for the recognition of nucleosomes by 53BP1, a full Methods section, along with 14 Supplementary Figures can be found in the Supplementary Information document.
Author contributions
A.F.T. initiated the project, carried out most of the cell biological experiments, found the interaction between ubiquitylated H2A and 53BP1 and contributed to the experimental design and data interpretation. M.D.C. produced the recombinant nucleosomes. A.F.T. and M.D.C. and carried out the recombinant nucleosome pulldown studies. C.E.D. examined RIF1 focus formation, the role of the UDR in HR and helped with some immunofluorescence experiments. A.O. carried out the class switching experiments and the DT40 work. C.C.Y.L. helped with protein binding studies and generated the nucleosome model shown in Supplementary Fig. 12. M.C.L. helped with the FRAP experiments. J.K.L. carried out chemical ubiquitylation and mass spectrometry to verify histone methylation. H.H. carried out the NMR experiments. SN carried out mass spectrometry to verify H4K20 methylation. F.S. supervised H.H. D.D. supervised the project and wrote the manuscript with input from the other authors.
The authors declare no competing financial interests.
References
- 1.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Molecular cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura K, et al. Genetic dissection of vertebrate 53BP1: a major role in non-homologous end joining of DNA double strand breaks. DNA Repair (Amst) 2006;5:741–749. doi: 10.1016/j.dnarep.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. S0092-8674(10)00285-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 7.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. nature07433 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. nsmb.1831 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. S1097-2765(11)00247-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 Regulates DSB Repair Using Rif1 to Control 5′ End Resection. Science. 2013 doi: 10.1126/science.1231573. science.1231573 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escribano-Diaz C, et al. A Cell Cycle-Dependent Regulatory Circuit Composed of 53BP1-RIF1 and BRCA1-CtIP Controls DNA Repair Pathway Choice. Molecular cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Di Virgilio M, et al. Rif1 Prevents Resection of DNA Breaks and Promotes Immunoglobulin Class Switching. Science. 2013 doi: 10.1126/science.1230624. science.1230624 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman JR, et al. RIF1 Is Essential for 53BP1-Dependent Nonhomologous End Joining and Suppression of DNA Double-Strand Break Resection. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.01.002. S1097–2765(13)00003-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 16.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. S0092867404010530 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Pei H, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. nature09658 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acs K, et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. nsmb.2188 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Mallette FA, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. The EMBO journal. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura TM, Moser BA, Du LL, Russell P. Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol Cell Biol. 2005;25:10721–10730. doi: 10.1128/MCB.25.24.10721-10730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zgheib O, Pataky K, Brugger J, Halazonetis TD. An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. Molecular and cellular biology. 2009;29:1050–1058. doi: 10.1128/MCB.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pryde F, et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci. 2005;118:2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- 23.Gudjonsson T, et al. TRIP12 and UBR5 Suppress Spreading of Chromatin Ubiquitylation at Damaged Chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. S0092-8674(12)00883-5 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 25.Panier S, et al. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Molecular cell. 2012;47:383–395. doi: 10.1016/j.molcel.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 26.Mattiroli F, et al. RNF168 Ubiquitinates K13-15 on H2A/H2AX to Drive DNA Damage Signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Gatti M, et al. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 2012;11:2538–2544. doi: 10.4161/cc.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 29.Simon MD, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nature reviews. Molecular cell biology. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Ai Y, Wang J, Haracska L, Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat Chem Biol. 2010;6:270–272. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, et al. ASH2L Regulates Ubiquitylation Signaling to MLL: trans-Regulation of H3 K4 Methylation in Higher Eukaryotes. Molecular cell. 2013;49:1108–1120. doi: 10.1016/j.molcel.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poulsen M, Lukas C, Lukas J, Bekker-Jensen S, Mailand N. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J Cell Biol. 2012;197:189–199. doi: 10.1083/jcb.201109100. jcb.201109100 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juang YC, et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol Cell. 2012;45:384–397. doi: 10.1016/j.molcel.2012.01.011. S1097-2765(12)00077-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 36.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Meth Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 37.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentley ML, et al. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang AC, Grzesiek S, Tschudin R, Lodi PJ, Bax A. Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium-decoupled HA(CA)N and HA(CACO)N. J Biomol NMR. 1995;5:376–382. doi: 10.1007/BF00182281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.