Abstract
The fatty acid and retinol-binding (FAR) proteins are a family of unusual helix-rich lipid binding proteins found exclusively in nematodes, and are secreted by a range of parasites of humans, animals and plants. Na-FAR-1 is from the parasitic nematode Necator americanus, an intestinal blood-feeding parasite of humans. Sequence-specific 1H, 13C and 15N resonance assignments have been obtained for the recombinant 170 amino acid protein, using three-dimensional triple-resonance heteronuclear magnetic resonance experiments. Backbone assignments have been obtained for 99.3 % of the non-proline HN/N pairs (146 out of 147). The amide resonance of T45 was not observed, probably due to rapid exchange with solvent water. A total of 96.9 % of backbone resonances were identified, while 97.7 % assignment of amino acid sidechain protons is complete. All Hα(166), Hβ(250) and Hγ(160) and 98.4 % of the Hδ (126 out of 128) atoms were assigned. In addition, 99.4 % Cα (154 out of 155) and 99.3 % Cβ (143 out of 144) resonances have been assigned. No resonances were observed for the NHn groups of R93 NεHε, arginine, Nη1H2, Nη2H2, histidine Nδ1Hδ1, Nε1Hε1 and lysine Nζ3H3. Na-FAR-1 has a similar overall arrangement of α-helices to Ce-FAR-7 of the free-living Caeorhabditis elegans, but with an extra C-terminal helix.
Keywords: Parasitic nematode, Necator americanus, Fatty-acid and retinol-binding protein, Na-FAR-1, NMR
Biological context
Over 500 million people living in tropical and subtropical regions are infected with human hookworms, Necator americanus being the most prevalent. The infective larvae burrow into skin and develop to adulthood in the intestine, where they feed on blood and tissue. The infection causes anaemia and growth stunting, the effects being most severe in children and women of child bearing age (Hotez et al. 2004).
Fatty acid and retinol-binding (FAR) proteins are α-helix-rich proteins of around 20 kDa that are produced at different life cycle stages of nematodes and have been observed to be secreted by adult parasites (Basavaraju et al. 2003; Kennedy et al. 1997; Prior et al. 2001). It is hypothesized that FARs may play a role in host-parasite interaction and pathogenesis by sequestering or delivering lipid mediators, thereby affecting the local tissue environment of the host, and compromising immune and inflammatory defences (Bradley et al. 2001). FAR proteins are already used as diagnostic tools (Burbelo et al. 2009), and have also been shown to induce immunity against one parasite infection (Fairfax et al. 2009). They are, therefore, attractive potential targets for drug or vaccine development because they have no structural counterparts in mammals (Basavaraju et al. 2003).
Na-FAR-1 has been identified in a transcriptomic survey of N. americanus (Daub et al. 2000), and we here report resonance assignments of its bacterial recombinant form. The only structure of a FAR currently available is a crystal structure of Ce-FAR-7 of the free living nematode Caenorhabditis elegans (Jordanova et al. 2009). This FAR has seven α-helixes and comprises two binding sites that could accommodate different types of ligands. We find that Na-FAR-1 has a similar overall arrangement of regular secondary structure elements, but with an extra helical region at the C-terminus.
Methods and experiments
Recombinant Na-FAR-1 was expressed in BL21 (λDE3) Escherichia coli cells using [13C, 15N]-labelled M9 minimal medium. The His-tagged fusion protein was purified by nickel affinity and gel filtration chromatographies. The removal of copurifying ligands from the bacterial expression system was achieved by reverse-phase high performance liquid chromatography (RP-HPLC) with a C8 stationary phase and water/acetonitrile/trifluoroacetic acid mobile phase. The protein was refolded in aqueous buffer and concentrated to approximately 0.5 mM in 50 mM sodium phosphate pH 7.2, 50 mM NaCl. D2O was added to a final concentration of 10 % (v/v) prior to data acquisition. All spectra were recorded at 311 K on a Bruker Avance 600 MHz spectrometer equipped with a TCI cryoprobe.
Sequence-specific resonance assignment of the Na-FAR-1 backbone was accomplished with the aid of 2D 15N-HSQC, 3D HNCACB, 3D CBCA(CO)NH (Muhandiram and Kay 1994), 3D HNCO (Kay et al. 1994) and 3D HNCACO spectra. The majority of aliphatic sidechain carbon and proton resonances were located by navigating from the backbone data using 3D (H)C(CO)NH-TOCSY, 3D HBHA(CBCA)NH (Wang et al. 1994) and 3D H(C)(CO)NH-TOCSY spectra (Grzesiek and Bax 1992). Remaining aliphatic resonances were identified using 3D HcCH-TOCSY and 3D hCCH-TOCSY (Kay et al. 1993), this latter proving particularly useful for assignment of lysine sidechain CδHδ and CεHε groups that were too overlapped in both 1H and 13C dimensions to be resolved in other experiments. A proportion of aromatic sidechain 13C/1H signals (histidine Hδ1, tyrosine Hδ,ε and phenylalanine Hδ,ε) were assigned using 2D HBCBCGCDHD and 2D HBCBCGCDCEHE spectra (Yamazaki et al. 1993) and the remainder were identified from the 13C-edited [1H,1H]-NOESY spectrum. NMR spectra were processed using AZARA (http://www.bio.cam.ac.uk/azara) and analysed with CCPN analysis (Vranken et al. 2005).
Extent of assignments and data deposition
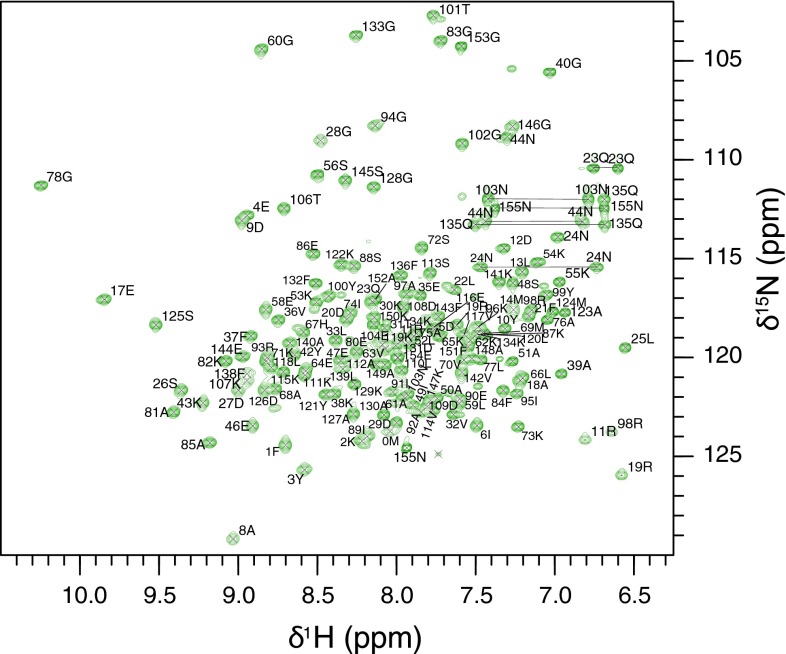
All Na-FAR-1 polypeptide backbone resonances were assigned, with the exception of the 15 His-tag N-terminal residues (MGSSHHHHHHSSGHM). Excluding the His-tag residues, backbone resonance assignments have been obtained for 99.3 % of the non-proline HN/N pairs (146 out of 147). The amide resonance of T45 was not observed, probably due to rapid exchange with solvent water. Figure 1 shows the 1H–15N HSQC spectrum with the assigned crosspeaks. A total of 96.9 % of backbone resonances were identified, while 97.7 % assignment of amino acid sidechain protons is complete. All Hα(166), Hβ(250) and Hγ(160) and 98.4 % of the Hδ (126 out of 128) atoms were assigned. In addition, 99.4 % Cα (154 out of 155) and 99.3 % Cβ (143 out of 144) resonances have been assigned. No resonances were observed for the following NHn groups: R93 NεHε, arginine, Nη1H2, Nη2H2, histidine Nδ1Hδ1, Nε1Hε1 and lysine Nζ3H3.
Fig. 1.
Two-dimensional 1H–15N HSQC spectrum of recombinant Na-FAR-1 showing the backbone amide resonance assignments. Crosspeaks have been labelled with the single letter amino acid code along with the native sequence specific number
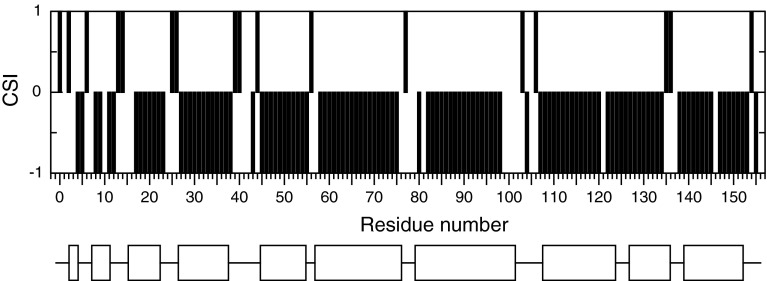
Chemical shift index (CSI) (Wishart and Sykes 1994) and DANGLE secondary structure analysis (Cheung et al. 2010) of Na-FAR-1 revealed a α-helix pattern consistent with the previously reported C. elegans FAR protein, with an additional α-helix segment at the C-terminus (Fig. 2).
Fig. 2.
Na-FAR-1 CSI consensus values (bars) and secondary structure elements (bottom) alignment. Negative CSI indicate α-helical segments which are shown as boxes below
The 1H, 13C and 15N chemical shift assignments have been deposited with the BioMagResBank database (http://www.bmrb.wisc.edu), accession number 18637.
Acknowledgments
This project is funded through a Wellcome Trust grant (ref. 083625) awarded to MWK, AC, BOS and BC, and by the National Research Council of Argentina (CONICET). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Contributor Information
Malcolm W. Kennedy, Email: malcolm.kennedy@glasgow.ac.uk
Brian O. Smith, Email: brian.smith@glasgow.ac.uk
References
- Basavaraju SV, Zhan B, Kennedy MW, Liu Y, Hawdon J, Hotez PJ (2003) Ac-FAR-1, a 20 kDa fatty acid- and retinol-binding protein secreted by adult Ancylostoma caninum hookworms: gene transcription pattern, ligand binding properties and structural characterisation. Mol Biochem Parasitol 126(1):63–71. doi:10.1016/S0166-6851(02)00253-0 [DOI] [PubMed]
- Bradley JE, Nirmalan N, Klager SL, Faulkner H, Kennedy MW. River blindness: a role for parasite retinoid-binding proteins in the generation of pathology? Trends Parasitol. 2001;17(10):471–475. doi: 10.1016/S1471-4922(01)02036-0. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Leahy HP, Iadarola MJ, Nutman TB. A four-antigen mixture for rapid assessment of Onchocerca volvulus infection. PLoS Negl Trop Dis. 2009;3(5):e438. doi: 10.1371/journal.pntd.0000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MS, Maguire ML, Stevens TJ, Broadhurst RW. DANGLE: a Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J Magn Reson. 2010;202(2):223–233. doi: 10.1016/j.jmr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Daub J, Loukas A, Pritchard DI, Blaxter M. A survey of genes expressed in adults of the human hookworm, Necator americanus. Parasitology. 2000;120(Pt 2):171–184. doi: 10.1017/S0031182099005375. [DOI] [PubMed] [Google Scholar]
- Fairfax KC, Vermeire JJ, Harrison LM, Bungiro RD, Grant W, Husain SZ, Cappello M (2009) Characterisation of a fatty acid and retinol binding protein orthologue from the hookworm Ancylostoma ceylanicum. Int J Parasitol 39 (14):1561–1571. doi:10.1016/j.ijpara.2009.06.005 [DOI] [PMC free article] [PubMed]
- Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein (1969) J Magn Reson. 1992;96(2):432–440. [Google Scholar]
- Hotez PJ, Brooker S, Bethony JM, Bottazzi ME, Loukas A, Xiao S. Hookworm infection. N Engl J Med. 2004;351(8):799–807. doi: 10.1056/NEJMra032492. [DOI] [PubMed] [Google Scholar]
- Jordanova R, Groves MR, Kostova E, Woltersdorf C, Liebau E, Tucker PA. Fatty acid- and retinoid-binding proteins have distinct binding pockets for the two types of cargo. J Biol Chem. 2009;284(51):35818–35826. doi: 10.1074/jbc.M109.022731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE, Xu GY, Singer AU, Muhandiram DR, Forman-kay JD. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J Magn Reson, Ser B. 1993;101(3):333–337. doi: 10.1006/jmrb.1993.1053. [DOI] [Google Scholar]
- Kay LE, Xu GY, Yamazaki T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J Magn Reson, Ser A. 1994;109(1):129–133. [Google Scholar]
- Kennedy MW, Garside LH, Goodrick LE, McDermott L, Brass A, Price NC, Kelly SM, Cooper A, Bradley JE. The Ov20 protein of the parasitic nematode Onchocerca volvulus. A structurally novel class of small helix-rich retinol-binding proteins. J Biol Chem. 1997;272(47):29442–29448. doi: 10.1074/jbc.272.47.29442. [DOI] [PubMed] [Google Scholar]
- Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional nmr experiments with improved sensitivity. J Magn Reson, Ser B. 1994;103(3):203–216. [Google Scholar]
- Prior A, Jones JT, Blok VC, Beauchamp J, McDermott L, Cooper A, Kennedy MW. A surface-associated retinol- and fatty acid-binding protein (Gp-FAR-1) from the potato cyst nematode Globodera pallida: lipid binding activities, structural analysis and expression pattern. Biochem J. 2001;356(Pt 2):387–394. doi: 10.1042/0264-6021:3560387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wang AC, Lodi PJ, Qin J, Vuister GW, Gronenborn AM, Clore GM. An efficient triple-resonance experiment for proton-directed sequential backbone assignment of medium-sized proteins. J Magn Reson B. 1994;105(2):196–198. doi: 10.1006/jmrb.1994.1123. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4(2):171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Forman-Kay JD, Kay LE. Two-dimensional NMR experiments for correlating carbon-13.beta. and proton.delta./.epsilon. chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J Am Chem Soc. 1993;115(23):11054–11055. doi: 10.1021/ja00076a099. [DOI] [Google Scholar]