Abstract
As-p18 is produced and secreted by larvae of the parasitic nematode Ascaris suum as they develop within their eggs. The protein is a member of the fatty acid binding protein (FABP) family found in a wide range of eukaryotes, but is distinctive in that it is secreted from the synthesizing cell and has predicted additional structural features not previously seen in other FABPs. As-p18 and similar proteins found only in nematodes have therefore been designated ‘nemFABPs’. Sequence-specific 1H, 13C and 15N resonance assignments were established for the 155 amino acid recombinant protein (18.3 kDa) in complex with oleic acid, using a series of three-dimensional triple-resonance heteronuclear NMR experiments. The secondary structure of As-p18 is predicted to be very similar to other FABPs, but the protein has extended loops that have not been observed in other FABPs whose structures have so far been solved.
Keywords: As-p18, Fatty acid binding protein, nemFABP, Nematode, Parasite, Ascaris suum
Biological context
As-p18 is a developmentally-regulated fatty acid binding protein found in the perivitelline fluid surrounding the infective third stage larva (L3) of the parasitic nematode Ascaris suum, which is closely related to the large intestinal roundworm of humans, Ascaris lumbricoides. Once fully developed, the L3 undergoes developmental arrest, persisting for several years until ingestion by the host, but little is known about the biochemical and physiological basis for such long term survival. One of the most abundant components in the fluid that surrounds the developing larva is the fatty acid binding protein, As-p18 (Mei et al. 1997). As-p18 displays significant sequence similarity to intracellular lipid binding proteins of vertebrates and other metazoan phyla, the best understood being the fatty acid binding proteins (FABPs) of humans. But, only in nematodes are they secreted from the synthesizing cell (Mei et al. 1997; Plenefisch et al. 2000). Modelling studies predicted that the protein conforms well to a ten β-stranded, two α-helix FABP, but that there are modifications to the binding site which could relate to ligand specificity. Moreover, unusual extra loops were predicted to extend from the surface of the protein that might be involved in previously unsuspected protein–protein or protein-membrane interactions related to the unusual location and function of the protein (Kennedy et al. 2000).
Phylogenetic analysis reveals that As-p18, together with potential homologues and paralogues found in Caenorhabditis elegans, comprise a distinct subclass (here named ‘nemFABPs’) within the wider FABP family that are possibly unique to nematodes (Plenefisch et al. 2000). The developmental control of As-p18’s expression, and the presence of a working secretory leader peptide encoded in the protein’s mRNA, also distinguish these nematode proteins from other FABPs. The unique features of As-p18 may reflect adaptation to a specific function within the egg related to the remarkable environmental resilience of the larva (Kennedy and Harnett 2001). Importantly, the perivitelline fluid around the developing larvae of the causative agents of lymphatic filariasis of humans (elephantiasis) also has an As-p18-like protein (Michalski et al. 2002). As a further approach to understanding the functions of this and other nemFABPs, structural studies of As-p18 were carried out on a 15N, 13C isotopically-labelled sample using solution NMR techniques. Here we report complete sequence-specific resonance assignments for As-p18 complexed with oleic acid.
Methods and experiments
Recombinant As-p18 (Mei et al. 1997) was expressed in BL21 (λDE3) Escherichia coli cells using [13C,15N]-labelled M9 minimal medium. The His-tagged fusion protein was purified by nickel chelate chromatography, followed by a gel filtration chromatography as a polishing step. The removal of contaminating ligands derived from the bacterial expression system was achieved by reverse-phase (RP) chromatography with a C8 stationary phase and water/acetonitrile/trifluoroacetic acid mobile phase, followed by refolding in aqueous buffer. The protein was concentrated to approximately 0.5 mM in 20 mM sodium phosphate pH 7.20, and D2O was added to a final concentration of 5 % (v/v) prior to data acquisition. The protein was saturated with oleic acid by adding a small volume of a concentrated solution of oleic acid in deuterated ethanol. All spectra were recorded at 298 K on a Bruker Avance spectrometer at 600 MHz equipped with a TCI cryoprobe. Proton chemical shifts were referenced relative to the H2O offset frequency and heteronuclear chemical shifts calculated from the proton reference (Wishart et al. 1995).
Sequence-specific resonance assignment of the As-p18 backbone was accomplished with the aid of 2D 15N-HSQC, 3D HNCACB, 3D CBCA(CO)NH (Muhandiram and Kay 1994) 3D HNCO (Kay et al. 1994) and 3D HNCACO spectra. The majority of aliphatic sidechain carbon and proton resonances were located by navigating from the backbone data using 2D 13C-HSQC, 3D (H)C(CO)NH-TOCSY, 3D HBHA(CBCA)NH (Wang et al. 1994) and 3D H(C)(CO)NH-TOCSY spectra (Grzesiek and Bax 1992). Remaining aliphatic resonances were identified using 3D HCCH-TOCSY (Kay et al., 1993) or 3D 13C-edited [1H, 1H]-NOESY spectra. A proportion of aromatic sidechain 13C/1H signals (histidine Hδ1, tryptophan Hδ1, tyrosine Hδ,ε and phenylalanine Hδ,ε) were assigned using 2D HBCBCGCDHD and 2D HBCBCGCDCEHE spectra (Yamazaki et al. 1993) and the remainder were identified from the 13C-edited [1H, 1H]-NOESY spectrum. Methionine Cε/Hε were identified with the help of multiplicity edited CT-HSQC (Vuister and Bax 1992). NMR spectra were processed using AZARA (http://www.bio.cam.ac.uk/azara) and analysed with CCPNmr analysis (Vranken et al. 2005).
Extent of assignments and data deposition
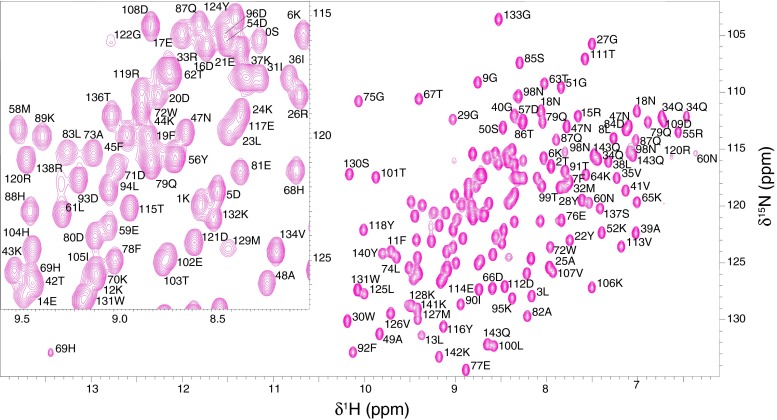
All As-p18 polypeptide backbone resonances were assigned, with the exception of the disordered N-terminal His-tag residues which are too overlapped to resolve and whose backbone amide resonances are not observed. The amide resonance of Q18, W30, D123 and Y139 were not observed, probably due to rapid exchange with solvent water. Figure 1 shows the 1H-15N HSQC spectrum with the assignments indicated. Backbone resonance assignments for the native sequence1 (excluding the flexible His-tag) have been obtained for 97.1 % of the non-proline HN/N pairs (135 out of 139), 99.3 % of the Hα (150 out of 151), 97.9 % of the Cα (140 out of 143), and 99.3 % of the Cβ (134 out of 135) atoms. In addition, assignment for 99.2 % of the Hβ and >93 % of the Hγ and Hδ resonances have been assigned. Most of the missing assignments along the residue sidechain are those of lysine CεHε groups that are too overlapped in both 1H and 13C dimensions to be resolved. No resonances were observed for the following NHn groups: arginine NεHε, Nη1H2, Nη2H2 (with the exception of R120 NεHε), histidine Nδ1Hδ1, Nε1Hε1 (with the notable exception of H69 Nδ1Hδ1 at 169.1/13.45 ppm) and lysine Nζ3H3.
Fig. 1.
Two-dimensional 1H -15N HSQC spectrum of recombinant As-p18. All assigned crosspeaks have been labelled with the single letter amino acid code and their position in the amino acid sequence. The inset shows an expanded view of the region between 1H; 8.00–9.58 and 15N; 114.40–127.00 ppm of the HSQC spectrum
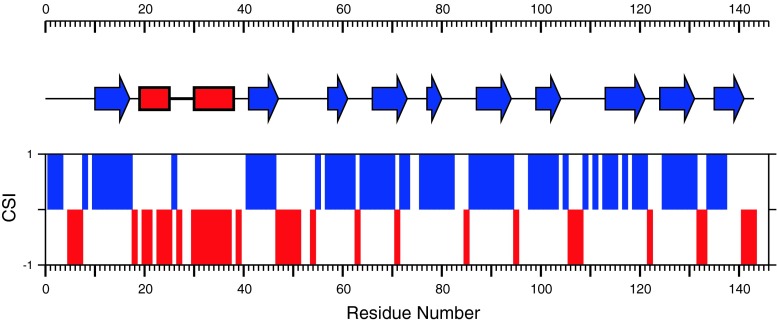
The 13C chemical-shift index (CSI; Wishart and Sykes 1994) and secondary structure analysis using DANGLE (Cheung et al. 2010) indicate the presence of two α-helices and ten β-strands (Fig. 2), and the arrangement of secondary structural elements as a function of sequence is similar to that discovered in FABPs whose structures have previously been determined (Banaszak et al. 1994). However, the chemical shift data predict an extended loop between strands βB and βC, spanning amino acids 47–55 which may be a feature of nemFABPs in general.
Fig. 2.
Alignment of CSI consensus values (bars) with the predicted secondary structure elements of As-p18. Positive and negative CSI values indicate β-strand and α-helix segments, respectively. The secondary structure prediction, was calculated using DANGLE for assigned Cα, Cβ, Hα, C’ and NH. Blue arrows and red rectangles represent β-strands and α-helical stretches, respectively
The 1H, 13C and 15N chemical shift assignments have been deposited with the BioMagResBank database (http://www.bmrb.wisc.edu), accession number 18632.
Acknowledgments
This work was supported by the National Research Council of Argentina (CONICET), and The Wellcome Trust (UK) grant number 083625 to MWK, AC, BOS and BC. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The expression vector for As-p18 was kindly provided by Rick Komuniecki, University of Toledo, Ohio, USA.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Footnotes
Native sequence spans residues K1-Q143. Negative numbering accounts for the twelve residues in the His-tag at the N-terminus of recombinant As-p18.
Contributor Information
Malcolm W. Kennedy, Email: malcolm.kennedy@glasgow.ac.uk
Brian O. Smith, Email: Brian.Smith@glasgow.ac.uk
References
- Banaszak L, Winter N, Xu ZH, Bernlohr DA, Cowan S, Jones TA. Lipid-binding proteins- a family of fatty-acid and retinoid transport proteins. Adv Protein Chem. 1994;45:89–1511. doi: 10.1016/S0065-3233(08)60639-7. [DOI] [PubMed] [Google Scholar]
- Cheung MS, Maguire ML, Stevens TJ, Broadhurst RW. DANGLE: a Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J Magn Reson. 2010;202(2):223–233. doi: 10.1016/j.jmr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31-kDa protein. J Magn Reson. 1992;96(2):432–440. [Google Scholar]
- Kay LE, Xu GY, Singer AU, Muhandiram DR, FormanKay JD. A gradient enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J Magn Reson (B) 1993;101:333–337. doi: 10.1006/jmrb.1993.1053. [DOI] [Google Scholar]
- Kay LE, Xu GY, Yamazaki T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J Magn Reson (A) 1994;109(1):129–133. doi: 10.1006/jmra.1994.1145. [DOI] [Google Scholar]
- Kennedy MW, Harnett W. Parasitic nematodes: molecular biology, biochemistry, and immunology. New York, Wallingford: CABI Publishing; 2001. [Google Scholar]
- Kennedy MW, Scott JC, Lo SJ, Beauchamp J, McManus DP. Sj-FABPc fatty acid binding protein of the human blood fluke Schistosoma japonicum: structural and functional characterisation and unusual solvent exposure of a portal-proximal tryptophan residue. Biochem J. 2000;349:377–384. doi: 10.1042/0264-6021:3490377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei B, Kennedy MW, Beauchamp J, Komuniecki PR, Komuniecki R. Secretion of a novel, developmentally regulated fatty acid-binding protein into the perivitelline fluid of the parasitic nematode, Ascaris suum. J Biol Chem. 1997;272(15):9933–9941. doi: 10.1074/jbc.272.15.9933. [DOI] [PubMed] [Google Scholar]
- Michalski ML, Monsey JD, Cistola DP, Weil GJ. An embryo-associated fatty acid-binding protein in the filarial nematode Brugia malayi. Mol Biochem Parasitol. 2002;124(1–2):1–10. doi: 10.1016/S0166-6851(02)00081-6. [DOI] [PubMed] [Google Scholar]
- Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J Magn Reson (B) 1994;103:203–216. doi: 10.1006/jmrb.1994.1032. [DOI] [Google Scholar]
- Plenefisch J, Xiao H, Mei B, Geng J, Komuniecki PR, Komuniecki R. Secretion of a novel class of iFABPs in nematodes: coordinate use of the Ascaris/Caenorhabditis model systems. Mol Biochem Parasitol. 2000;105(2):223–236. doi: 10.1016/S0166-6851(99)00179-6. [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Vuister GW, Bax A. Resolution enhancement and spectral editing of uniformly 13C enriched proteins by homonuclear broadband 13C decoupling. J Magn Reson. 1992;98:428–435. [Google Scholar]
- Wang AC, Lodi PJ, Qin J, Vuister GW, Gronenborn AM, Clore GM (1994) An efficient triple-resonance experiment for proton-directed sequential backbone assignment of medium-sized proteins. J Magn Reson Ser B 105(2):196–198 [DOI] [PubMed]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6(2):135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Forman-Kay JD, Kay LE. Two-dimensional NMR experiments for correlating 13C-β and 1H-δ/ε chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J Am Chem Soc. 1993;115:11054–11055. doi: 10.1021/ja00076a099. [DOI] [Google Scholar]