Abstract
Fibrillins are extracellular, disulphide-rich glycoproteins that form 10–12 nm diameter microfibrils in connective tissues. They are found in the majority of higher animals, from jellyfish to humans. Fibrillin microfibrils confer properties of elasticity and strength on connective tissue and regulate growth factor availability in the extracellular matrix (ECM). Mutations in FBN1, the human gene encoding the fibrillin-1 isoform, are linked to several inherited connective tissue disorders. The fibrillin-1 N-terminus forms many functionally-important interactions, both with other fibrillin molecules and various ECM components. In particular, the first four domains, the fibrillin unique N-terminal (FUN) and three epidermal growth factor (EGF)-like domains (FUN-EGF3), are implicated in microfibril assembly and growth factor sequestration. The structure of these domains, which comprise 134 residues, is unknown. We have produced a recombinant fragment corresponding to this region of human fibrillin-1. Here, we report 1H, 13C and 15N resonance assignments of the FUN-EGF3 fragment. Assignments will facilitate structure determination, analysis of interdomain dynamics and the mapping of interaction surfaces.
Keywords: Human fibrillin-1 N-terminus, NMR resonance assignments, Extracellular matrix, Microfibril assembly, Bone morphogenetic proteins
Biological context
Fibrillin microfibrils are essential structural and regulatory components of the extracellular matrix (ECM) in connective tissues. They are found in elastic tissues, such as the skin, blood vessels and lungs, as part of elastic fibres, in which they surround an amorphous network of elastin (Sakai et al. 1986). Microfibrils are also present in non-elastic connective tissues such as the ciliary zonules of the eye and kidney glomerulus, which do not contain elastin. In both cases, microfibrils are important for strength and elasticity. Furthermore, they regulate the availability of transforming growth factor-beta (TGFβ) family signalling molecules through sequestration of their latent forms (Ramirez and Rifkin 2009). When extracted from tissue and visualised by electron or atomic force microscopy, microfibrils have a diameter of 10–12 nm and a beaded filament structure, with an average distance of ~56 nm between the beads (Sakai et al. 1991, Hanssen et al. 1998).
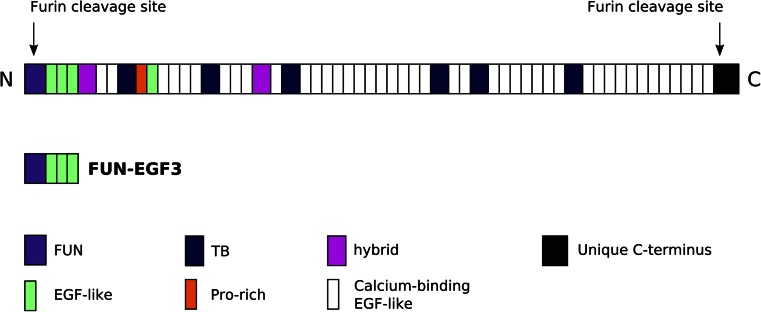
Fibrillins, the major structural components of microfibrils, are disulphide-rich glycoproteins found in most eumetazoa (Robertson et al. 2011). Their domain organisation is modular and highly conserved, with a high abundance of calcium-binding epidermal growth factor-like (cbEGF) domains, as well as several non-calcium EGF domains (Fig. 1). Arrays of cbEGF domains are interspersed by TGFβ binding-like (TB) and hybrid (hyb) domains, as well as a proline- or glycine-rich region, depending on the isoform. The structures of many of these domain types have been determined by X-ray crystallography and NMR spectroscopy (Knott et al. 1996, Yuan et al. 1997, Lee et al. 2004, Jensen et al. 2009). However, detailed structural information about the N- and C-terminal regions, including the fibrillin unique N-terminal (FUN) domain, is still lacking. High-resolution structures are important for understanding how fibrillin monomers assemble into microfibrils, the arrangements they form in microfibrils, their interactions with other ECM components and the effects of disease-causing mutations. Several inherited connective tissue disorders are linked to mutations in the human FBN1 gene, which encodes fibrillin-1: for example, Marfan syndrome (Lee et al. 1991), stiff skin syndrome (Loeys et al. 2010), acromicric and geleophysic dysplasias (Le Goff et al. 2011) and Weill-Marchesani syndrome (Faivre et al. 2003).
Fig. 1.
Domain organisation of human fibrillin-1. Fibrillins have two furin-cleavage sites—one at the N-terminus preceding the FUN domain and one in the unique C-terminal domain. The fragment under study (FUN-EGF3), which begins after the first furin recognition site, is highlighted
The fibrillin N-terminus is functionally versatile, interacting with many ECM components. It has been suggested that an important step in microfibril assembly is the interaction between the N- and C-terminal regions of fibrillin monomers. Several studies have demonstrated such interactions (Lin et al. 2002, Marson et al. 2005, Hubmacher et al. 2008). The minimal regions needed for the N–C terminal interaction have been identified: the four N-terminal domains (FUN-EGF3), starting after the furin proteolytic cleavage site, and the last three cbEGF domains (cbEGF41–cbEGF43) at the C-terminus (El-Hallous et al. 2007, Hubmacher et al. 2008). The FUN-EGF3 region is also a minimal interaction site for the prodomains of bone morphogenetic proteins-5 (Sengle et al. 2011) and -7 (Sengle et al. 2008). As a first step towards structural analysis of this region, a recombinant FUN-EGF3 fragment was produced. 1H, 13C and 15N resonance assignments for the protein are reported here.
Methods and experiments
Protein expression, purification and refolding
Protein expression and purification was carried out as described previously (Knott et al. 1996) with modifications. Escherichia coli BL21 cells, also containing the pREP4 plasmid (Qiagen) encoding the lacI repressor, were transformed with the pQE-30 expression vector (Qiagen) containing the human fibrillin-1 FUN-EGF3 sequence. The native fibrillin sequence started at residue R45 (numbered according to Pereira et al. 1993), the first residue after the furin proteolytic cleavage site, and finished at Q178, the residue after the last cysteine in the EGF3 domain. An N-terminal His6 tag was present for purification, followed by a factor Xa protease recognition site (I-E-G-R) to allow His6 tag removal and an S–A spacer before the start of the FUN-EGF3 sequence. 15N/13C-double labelled protein was produced by growing cells in M9 medium containing 0.1 % (w/v) 15NH4Cl and 0.5 % (w/v) 13C-glucose (Cambridge Isotope Laboratories), with the addition of 100 μg/ml ampicillin and 25 μg/ml kanamycin. 50 ml of starter culture, grown in labelled medium at 37 °C for ~18 h, was added to 0.6 l of medium. Cells were grown to OD600 ~0.8 and expression was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 2 mM. Cells were incubated at 28 °C for at least 6 h, harvested by centrifugation and stored at −80 °C until protein purification.
Cells were initially lysed in buffer containing 50 mM sodium phosphate pH 7.4, 6 M guanidinium chloride and 5 mM 2-mercaptoethanol (lysis buffer). Insoluble material was pelleted by ultracentrifugation (40,000 rpm; Beckmann 60Ti rotor; 25 °C; 30 min). The insoluble material was resuspended in lysis buffer. 2-mercaptoethanol was added to the suspension (concentration increase of 0.3 mM) and the pH was raised to ~9.0 with the addition of NaOH, followed by vigorous mixing by vortexing. The pH was then lowered to pH ~7.4 using HCl and the solution was centrifuged again. Cleared lysate was loaded onto fast flow chelating Sepharose (GE Healthcare) pre-loaded with Ni2+ and pre-equilibrated with lysis buffer. The column was then washed extensively with lysis buffer. Protein was eluted from the column in lysis buffer also containing 50 mM ethylenediaminetetraacetic acid (EDTA) and 100 mM dithiothreitol (DTT). Tris–HCl pH 8.3 was added to the eluted protein at a final concentration of 100 mM and additional DTT to final concentration of 200 mM. Following incubation at room temperature for 1 h, the mixture was adjusted to pH ~3.0 and dialysed overnight against 0.1 % (v/v) trifluoracetic acid (TFA). Protein was then desalted by reverse phase high performance liquid chromatography (HPLC).
Purified, reduced protein was refolded under the following conditions: 100 mM Tris–HCl pH 8.3, 3 mM l-cysteine, 0.3 mM l-cystine, 50 % (v/v) glycerol, 0.2 mg/ml protein at 4 °C for ~72 h. The refolding mixture was acidified to pH ~3.0 and dialysed against 0.1 % (v/v) TFA for at least 5 h. Protein was then concentrated by ultrafiltration and purified by HPLC. The His6 tag was removed through treatment with factor Xa (Novagen) under the conditions: 20 mM 2-(N-Morpholino) ethanesulfonic acid pH 5.8, 100 mM NaCl, ~4 mg/ml protein, 1 unit factor Xa/mg protein. Protein was further purified by cation exchange fast liquid protein chromatography and HPLC. The final product was analysed by denaturing SDS-PAGE in the presence or absence of 5 % (v/v) 2-mercaptoethanol, as well as by electrospray ionisation mass spectrometry (observed mass of unlabelled protein is 14,082 Da).
NMR spectroscopy
Experiments for assignment were performed using 15N-single or 15N/13C-double labelled FUN-EGF3 at a concentration of 0.5–1.5 mM in 5 % D2O/95 % H2O (v/v) at pH 5.4. NMR experiments were carried out at 298 K on three different spectrometers: home-built spectrometers with 1H-operating frequencies of 600 MHz or 750 MHz, a triple-resonance probe and GE/Omega console, or a Bruker Avance 500 MHz spectrometer with a Cryoplatform, equipped with a TCI CryoProbe. Initial sequential assignments were obtained with data from 3D 15N-edited TOCSY-HSQC, NOESY-HSQC and HSQC-NOESY-HSQC experiments acquired using 15N-labelled protein. Assignments were confirmed and extended using 3D HNCA, (H)CC(CO)NH, HNCO and HN(CA)CO experiments with 15N/13C-double labelled FUN-EGF3. Side-chain assignments were obtained using the above experiments, as well as 3D HCCH-TOCSY, 3D 13C-edited NOESY-HSQC, 2D 1H-13C(aromatic) HSQC, 2D 1H NOESY, 2D 1H DQF-COSY and 2D 1H TOCSY. NMR data were processed using NMRPipe (Delaglio et al. 1995) and analysed using CcpNmr Analysis (Vranken et al. 2005).
Assignments and data deposition
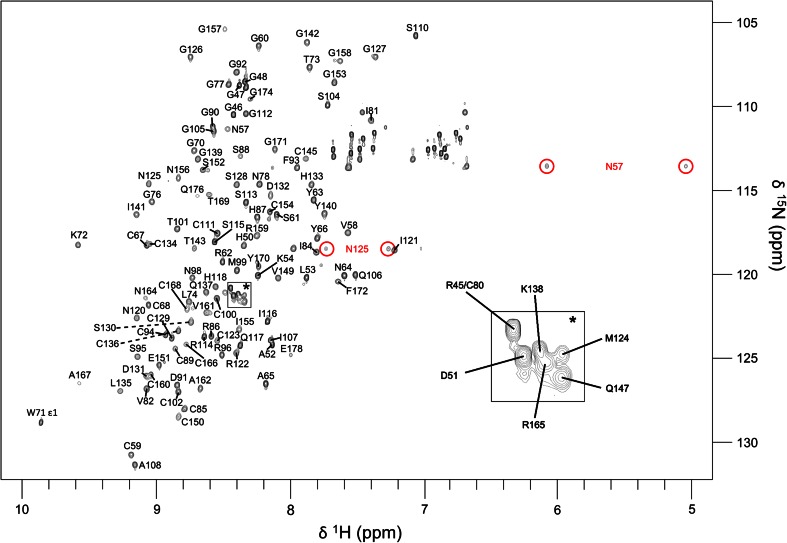
Resonances were assigned for all 134 residues in the native sequence, starting at R45. Figure 2 shows a 1H-15N HSQC spectrum of FUN-EGF3. All non-proline backbone 1HN and 15N resonances were assigned except C119 and T173. 15N assignments of the 10 proline residues were not obtained. Overall, 97.3 % of all 1H assignments were obtained, including 100 % of 1Hα, 99.0 % of 1Hβ, 100 % of 1Hγ and 97.6 % of 1Hδ. 89.9 % of 13C resonance assignments were obtained, including 100 % of 13Cα, 99.1 % of 13Cβ and 100 % of 13C′. Complete proline side-chain 1H and 13C assignments were obtained. The remaining unassigned 1H and 13C resonances are mainly in aromatic side chains.
Fig. 2.
750 MHz 1H-15N HSQC spectrum of 15N-labelled FUN-EGF3 in 5 % D2O/95 % H2O at pH 5.4 and 298 K. Peak assignments for backbone amides and the W71 Nε1-Hε1 peak are shown. The NH2 side-chain peaks of N57 and N125 are also indicated by red circles. Assignments are not shown for overlapping peaks. The boxed region marked * is annotated separately on the lower right-hand side
Several unusual chemical shifts were measured, most likely arising from the proximity of these nuclei to aromatic rings. The 1Hδ2 side-chain resonances of N57 are upfield-shifted, with chemical shifts of 5.05 and 6.09 ppm (Fig. 2). NOESY data indicate that the side chain of N57 in the FUN domain is close to the W71 aromatic ring, also in the FUN domain (data not shown). The 1Hβ resonance of T101 in the EGF1 domain is also upfield-shifted (2.76 ppm)—the atom is likely to lie above the plane of the aromatic ring of F93 (data not shown). In addition, the 15Nδ resonance of N125 in the EGF2 domain is downfield-shifted, with a chemical shift of 118.35 ppm (Fig. 2).
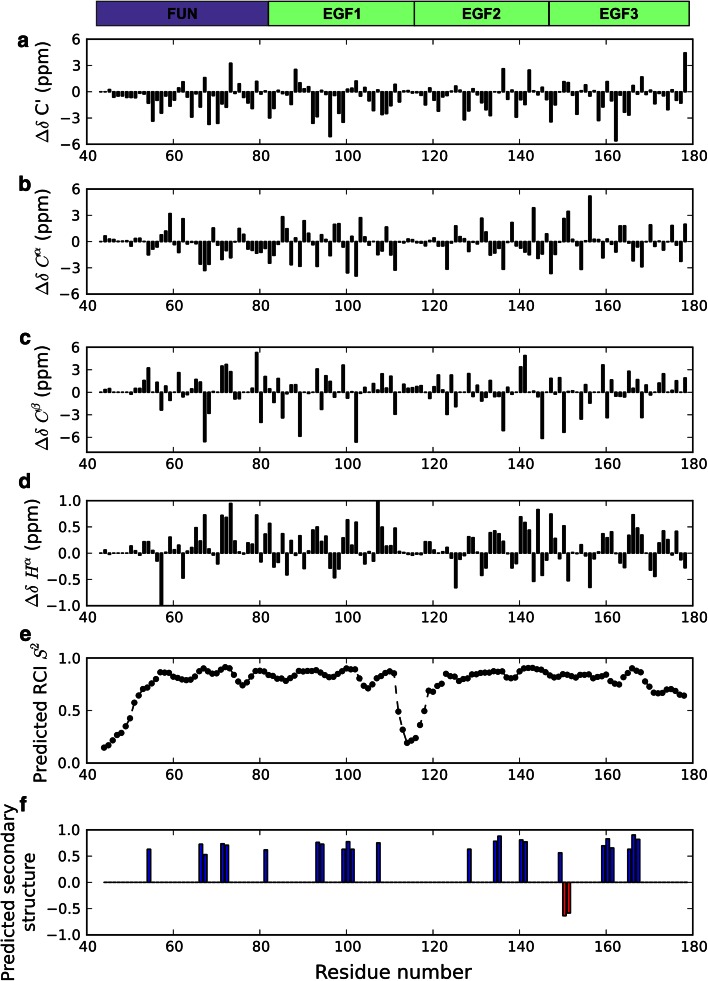
Secondary chemical shifts (Fig. 3a-d) indicate that the majority of residues are in structured regions of FUN-EGF3, as the values differ from random coil chemical shifts. They were used to make predictions about the structure and dynamics of the protein. Figure 3e shows a plot of predicted order parameters (S 2), on the basis of the random coil index (Berjanskii and Wishart 2005, Shen et al. 2009, Shen and Bax 2012), against residue number. The predictions identify several potentially flexible regions in the structure: firstly, the N-terminal residues (R45-G55) have predicted S 2 < 0.8; secondly, residues in the linker sequence between the last cysteine of the EGF1 domain and first cysteine of EGF2 (G112-H118) all have predicted S 2 < 0.5. Thirdly, residues at the C-terminus of the EGF3 domain, after the penultimate cysteine (T169-E178) may be more flexible, all having predicted S 2 < 0.8. In the full-length fibrillin molecule this loop may pack against the hyb1 domain, which follows the EGF3 domain in the amino acid sequence. There is also variability in the predicted S 2 value in other regions of the structure-residues with lower values may lie in flexible loops.
Fig. 3.
Analysis of FUN-EGF3 chemical shifts. Domains, defined by exon boundaries in the FBN1 gene (Pereira et al. 1993), are indicated by coloured boxes above the plots. a 13C′ secondary chemical shifts plotted as a function of residue number. b 13Cα secondary chemical shifts. c 13Cβ secondary chemical shifts d 1Hα secondary chemical shifts. e Random coil index (RCI) order parameters (S 2) predicted from chemical shifts, plotted as a function of residue number. Predictions were made using the TALOS+ and MICS servers (Shen et al. 2009, Shen and Bax 2012), using the method of Berjanskii and Wishart (2005). f Secondary structure prediction by TALOS+/MICS. Bars indicate the predicted secondary structure adopted by each residue, with the height showing the prediction probability. Positive values and blue bars indicate extended (β-strand) structure, while negative values and red bars indicate helical (α-helix) structure. Residues without bars are predicted to occur in loops, turns or regions with random-coil structure
Chemical shift-based predictions by TALOS+ (Shen et al. 2009) and MICS (Shen and Bax 2012) suggest that there are no long, continuous regions of regular secondary structure in FUN-EGF3 (Fig. 3f). However, there are several stretches of two to three residues with predicted β-sheet-like structure. These mainly coincide with the expected locations of β-hairpin structures, which are typically found in EGF-like domains (Bork et al. 1996). For example, residues R159-V161 and R165-A167 in the EGF3 domain are expected to form a two-stranded β-sheet. Only two residues, C150 and E151 in the EGF3 domain, are predicted to adopt α-helical secondary structure. EGF-like domains typically have only small regions of regular secondary structure, but are stabilised by the presence of three disulphide bonds. The FUN domain, which has an unknown structure, is also predicted to contain little secondary structure, with short β-sheet-like elements. It has four cysteine residues, which may form two disulphide bonds. In future, structure determination of the FUN-EGF3 fragment and characterisation of its dynamics will shed further light on the properties on the FUN domain and the interdomain interfaces.
The chemical shift assignments for FUN-EGF3 have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 18843.
Acknowledgments
DAY acknowledges a Wellcome Trust-funded PhD studentship and IBR acknowledges a BBSRC-funded PhD studentship. SAJ, PAH and CR acknowledge Wellcome Trust grant 086613. CR also acknowledges Wellcome Trust grant 079440.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Contributor Information
Penny A. Handford, Phone: +1-44-1865-613260, FAX: +44 1865 613201, Email: penny.handford@bioch.ox.ac.uk
Christina Redfield, Phone: +1-44-1865-275330, FAX: +44 1865 613201, Email: christina.redfield@bioch.ox.ac.uk.
References
- Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc. 2005;127(43):14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- Bork P, Downing A, Kieffer B, Campbell I. Structure and distribution of modules in extracellular proteins. Q Rev Biophys. 1996;29(2):119–167. doi: 10.1017/s0033583500005783. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- El-Hallous E, Sasaki T, Hubmacher D, Getie M, Tiedemann K, Brinckmann J, Bätge B, Davis EC, Reinhardt DP. Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adaptor function to tropoelastin. J Biol Chem. 2007;282(12):8935–8946. doi: 10.1074/jbc.M608204200. [DOI] [PubMed] [Google Scholar]
- Faivre L, Gorlin RJ, Wirtz MK, Godfrey M, Dagoneau N, Samples JR, Le Merrer M, Collod-Beroud G, Boileau C, Munnich A, Cormier-Daire V. In frame fibrillin-1 gene deletion in autosomal dominant Weill-Marchesani syndrome. J Med Genet. 2003;40(1):34–36. doi: 10.1136/jmg.40.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen E, Franc S, Garrone R. Atomic force microscopy and modeling of natural elastic fibrillin polymers. Biol Cell. 1998;90(3):223. doi: 10.1016/s0248-4900(98)80018-x. [DOI] [PubMed] [Google Scholar]
- Hubmacher D, El-Hallous EI, Nelea V, Kaartinen MT, Lee ER, Reinhardt DP. Biogenesis of extracellular microfibrils: multimerization of the fibrillin-1 C terminus into bead-like structures enables self-assembly. Proc Natl Acad Sci USA. 2008;105(18):6548. doi: 10.1073/pnas.0706335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SA, Iqbal S, Lowe ED, Redfield C, Handford PA. Structure and interdomain interactions of a hybrid domain: a disulphide-rich module of the fibrillin/LTBP superfamily of matrix proteins. Structure. 2009;17(5):759. doi: 10.1016/j.str.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V, Downing AK, Cardy CM, Handford P. Calcium binding properties of an epidermal growth factor-like domain pair from human fibrillin-1. J Mol Biol. 1996;255(1):22. doi: 10.1006/jmbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Le Goff C, Mahaut C, Wang LW, Allali S, Abhyankar A, Jensen S, Zylberberg L, Collod-Beroud G, Bonnet D, Alanay Y, Brady AF, Cordier MP, Devriendt K, Genevieve D, Kiper P, Kitoh H, Krakow D, Lynch SA, Le Merrer M, Mégarbane A, Mortier G, Odent S, Polak M, Rohrbach M, Sillence D, Stolte-Dijkstra I, Superti-Furga A, Rimoin DL, Topouchian V, Unger S, Zabel B, Bole-Feysot C, Nitschke P, Handford P, Casanova JL, Boileau C, Apte SS, Munnich A, Cormier-Daire V. Mutations in the TGFβ binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am J Hum Genet. 2011;89(1):7–14. doi: 10.1016/j.ajhg.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Godfrey M, Vitale E, Hori H, Mattei MG, Sarfarazi M, Tsipouras P, Ramirez F, Hollister DW. Linkage of Marfan syndrome and a phenotypically related disorder to two different fibrillin genes. Nature. 1991;352(6333):330–334. doi: 10.1038/352330a0. [DOI] [PubMed] [Google Scholar]
- Lee SS, Knott V, Jovanovic J, Harlos K, Grimes JM, Choulier L, Mardon HJ, Stuart DI, Handford PA. Structure of the integrin binding fragment from fibrillin-1 gives new insights into microfibril organization. Structure. 2004;12(4):717. doi: 10.1016/j.str.2004.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Tiedemann K, Vollbrandt T, Peters H, Batge B, Brinckmann J, Reinhardt DP. Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J Biol Chem. 2002;277(52):50795. doi: 10.1074/jbc.M210611200. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Gerber EE, Riegert-Johnson D, Iqbal S, Whiteman P, McConnell V, Chillakuri CR, Macaya D, Coucke PJ, De Paepe A, Judge DP, Wigley F, Davis EC, Mardon HJ, Handford P, Keene DR, Sakai LY, Dietz HC. Mutations in fibrillin-1 cause congenital scleroderma: stiff skin syndrome. Sci Transl Med. 2010;2(23):23ra20. doi: 10.1126/scitranslmed.3000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Rock MJ, Cain SA, Freeman LJ, Morgan A, Mellody K, Shuttleworth CA, Baldock C, Kielty CM. Homotypic fibrillin-1 interactions in microfibril assembly. J Biol Chem. 2005;280(6):5013. doi: 10.1074/jbc.M409029200. [DOI] [PubMed] [Google Scholar]
- Pereira L, D’Alessio M, Ramirez F, Lynch JR, Sykes B, Pangilinan T, Bonadio J. Genomic organization of the sequence coding for fibrillin, the defective gene product in Marfan syndrome. Hum Mol Genet. 1993;2(7):961. doi: 10.1093/hmg/2.7.961. [DOI] [PubMed] [Google Scholar]
- Ramirez F, Rifkin DB (2009) Extracellular microfibrils: contextual platforms for TGFbeta and BMP signaling. Curr Opin Cell Biol [DOI] [PMC free article] [PubMed]
- Robertson I, Jensen S, Handford P. TB domain proteins: evolutionary insights into the multifaceted roles of fibrillins and LTBPs. Biochem J. 2011;433(2):263–276. doi: 10.1042/BJ20101320. [DOI] [PubMed] [Google Scholar]
- Sakai L, Keene D, Engvall E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J Cell Biol. 1986;103(6 Pt 1):2499–2509. doi: 10.1083/jcb.103.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai LY, Keene DR, Glanville RW, Bachinger HP. Purification and partial characterization of fibrillin, a cysteine-rich structural component of connective tissue microfibrils. J Biol Chem. 1991;266(22):14763. [PubMed] [Google Scholar]
- Sengle G, Charbonneau NL, Ono RN, Sasaki T, Alvarez J, Keene DR, Bächinger HP, Sakai LY. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J Biol Chem. 2008;283(20):13874–13888. doi: 10.1074/jbc.M707820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengle G, Ono RN, Sasaki T, Sakai LY. Prodomains of transforming growth factor beta (TGFbeta) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. J Biol Chem. 2011;286(7):5087–5099. doi: 10.1074/jbc.M110.188615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. Identification of helix capping and beta-turn motifs from NMR chemical shifts. J Biomol NMR. 2012;52(3):211–232. doi: 10.1007/s10858-012-9602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken W, Boucher W, Stevens T, Fogh R, Pajon A, Llinas M, Ulrich E, Markley J, Ionides J, Laue E. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Yuan X, Downing A, Knott V, Handford P. Solution structure of the transforming growth factor beta-binding protein-like module, a domain associated with matrix fibrils. EMBO J. 1997;16(22):6659–6666. doi: 10.1093/emboj/16.22.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]