Abstract
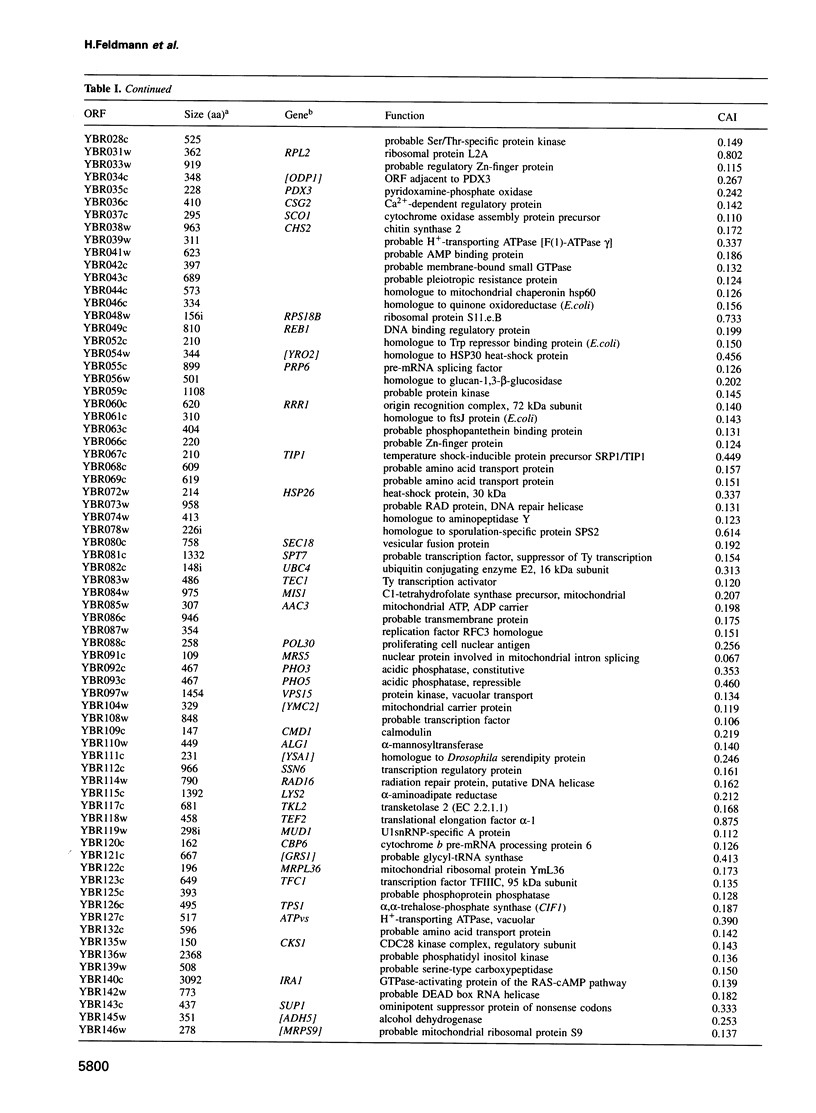
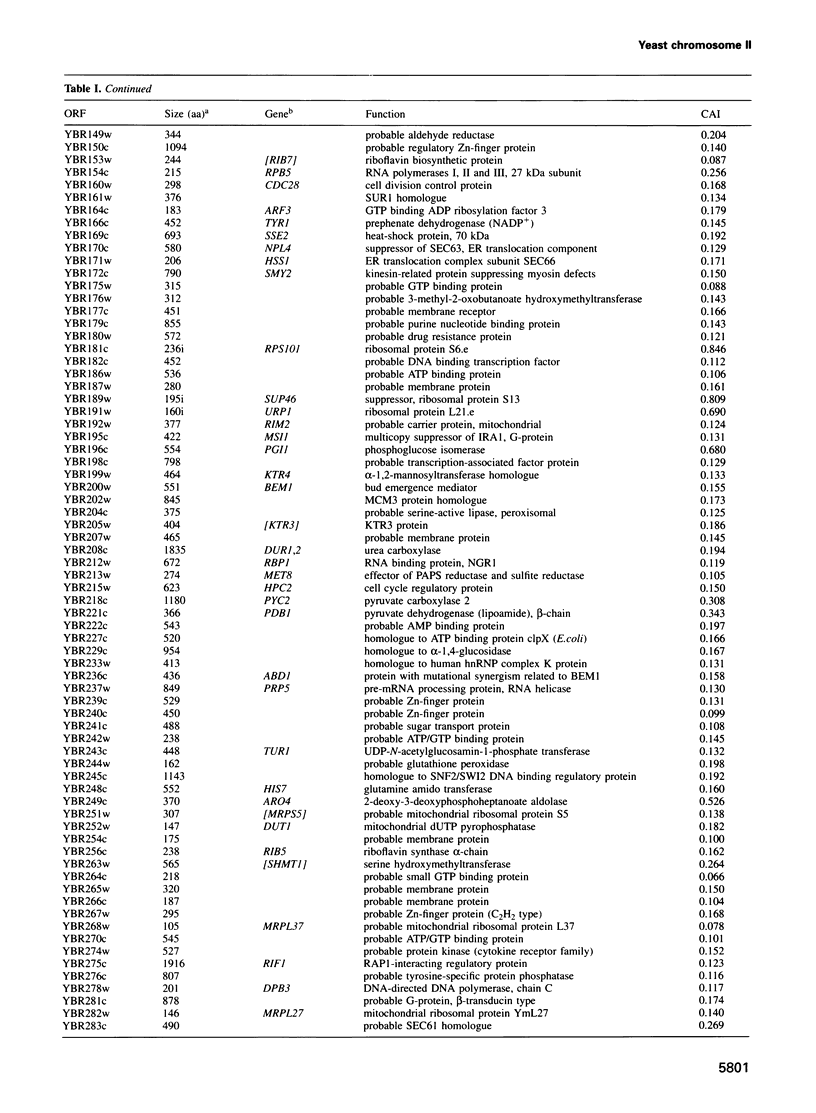
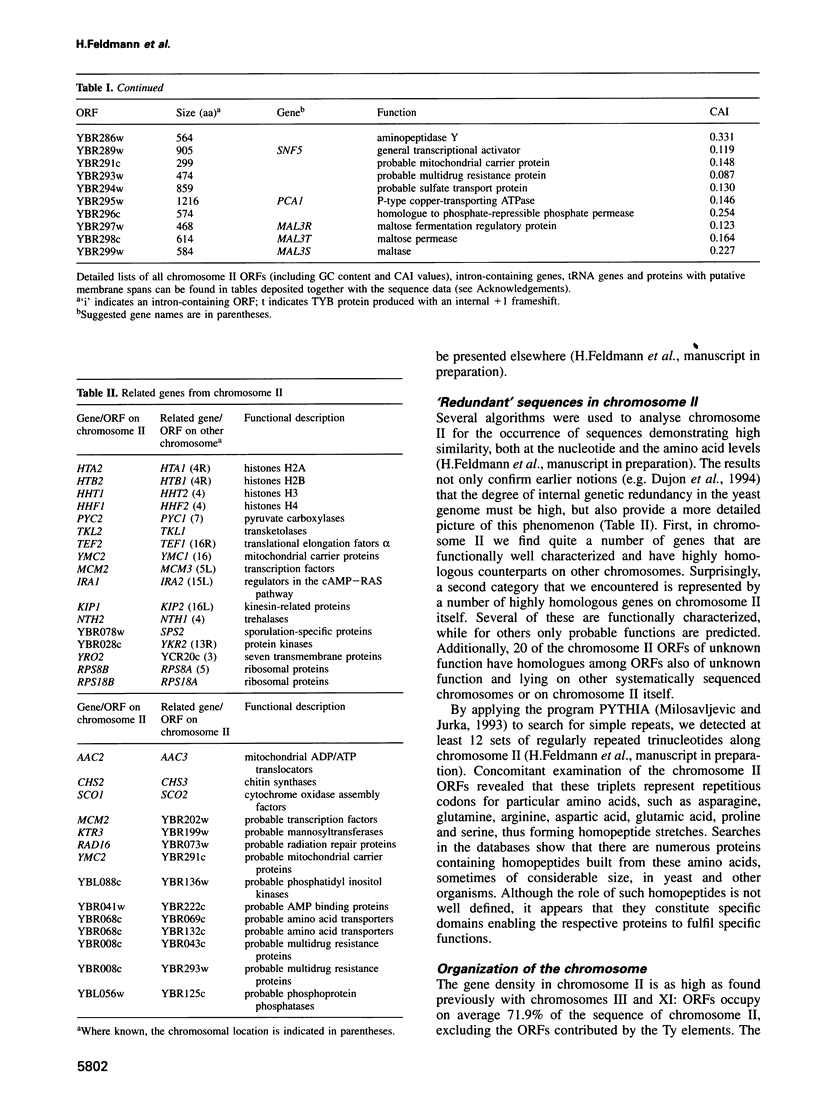
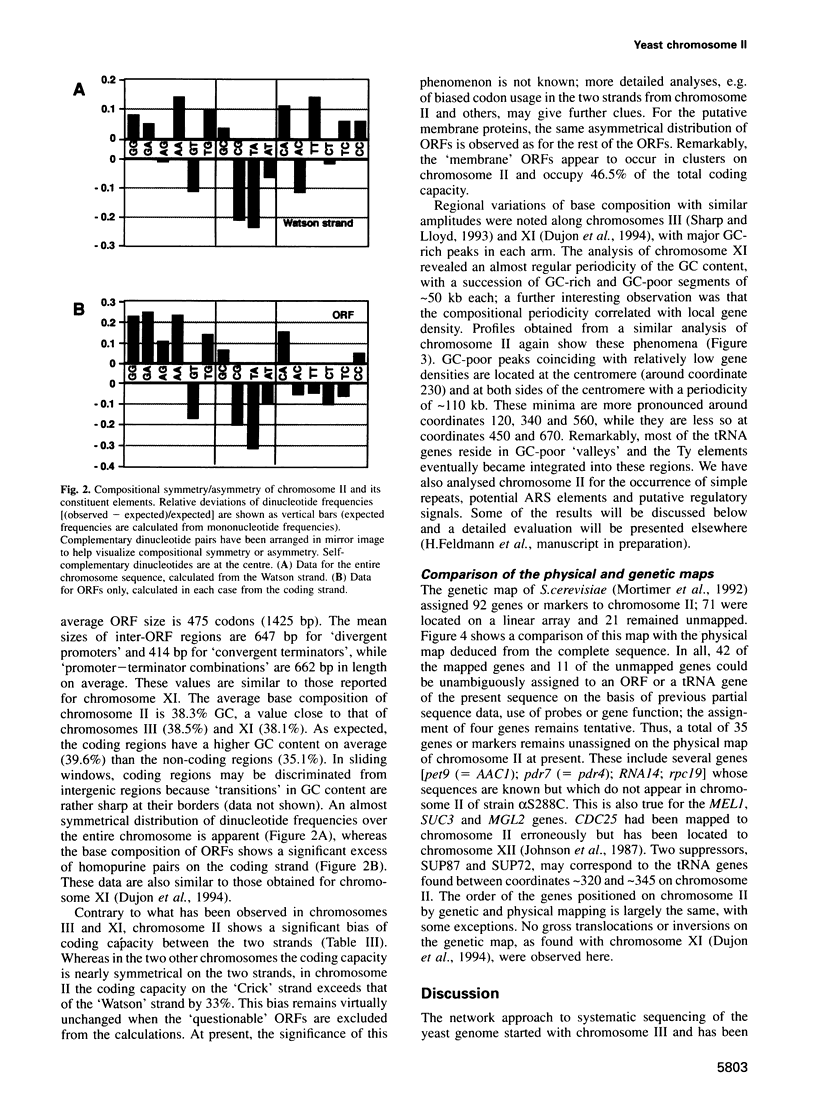
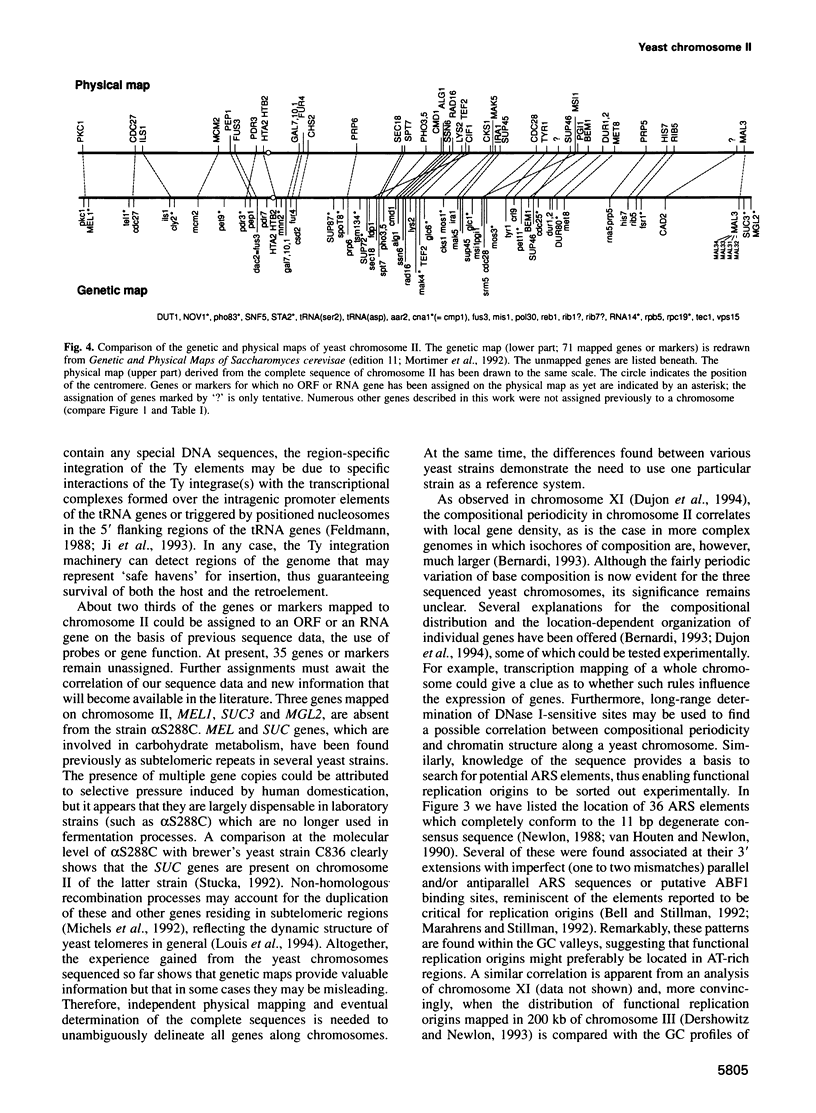
In the framework of the EU genome-sequencing programmes, the complete DNA sequence of the yeast Saccharomyces cerevisiae chromosome II (807 188 bp) has been determined. At present, this is the largest eukaryotic chromosome entirely sequenced. A total of 410 open reading frames (ORFs) were identified, covering 72% of the sequence. Similarity searches revealed that 124 ORFs (30%) correspond to genes of known function, 51 ORFs (12.5%) appear to be homologues of genes whose functions are known, 52 others (12.5%) have homologues the functions of which are not well defined and another 33 of the novel putative genes (8%) exhibit a degree of similarity which is insufficient to confidently assign function. Of the genes on chromosome II, 37-45% are thus of unpredicted function. Among the novel putative genes, we found several that are related to genes that perform differentiated functions in multicellular organisms of are involved in malignancy. In addition to a compact arrangement of potential protein coding sequences, the analysis of this chromosome confirmed general chromosome patterns but also revealed particular novel features of chromosomal organization. Alternating regional variations in average base composition correlate with variations in local gene density along chromosome II, as observed in chromosomes XI and III. We propose that functional ARS elements are preferably located in the AT-rich regions that have a spacing of approximately 110 kb. Similarly, the 13 tRNA genes and the three Ty elements of chromosome II are found in AT-rich regions. In chromosome II, the distribution of coding sequences between the two strands is biased, with a ratio of 1.3:1. An interesting aspect regarding the evolution of the eukaryotic genome is the finding that chromosome II has a high degree of internal genetic redundancy, amounting to 16% of the coding capacity.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amati B. B., Gasser S. M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- Baur A., Schaaff-Gerstenschläger I., Boles E., Miosga T., Rose M., Zimmermann F. K. Sequence of a 4.8 kb fragment of Saccharomyces cerevisiae chromosome II including three essential open reading frames. Yeast. 1993 Mar;9(3):289–293. doi: 10.1002/yea.320090308. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome and its evolutionary history--a review. Gene. 1993 Dec 15;135(1-2):57–66. doi: 10.1016/0378-1119(93)90049-9. [DOI] [PubMed] [Google Scholar]
- Bussereau F., Mallet L., Gaillon L., Jacquet M. A 12.8 kb segment, on the right arm of chromosome II from Saccharomyces cerevisiae including part of the DUR1,2 gene, contains five putative new genes. Yeast. 1993 Jul;9(7):797–806. doi: 10.1002/yea.320090714. [DOI] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Delaveau T., Delahodde A., Carvajal E., Subik J., Jacq C. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol Gen Genet. 1994 Sep 1;244(5):501–511. doi: 10.1007/BF00583901. [DOI] [PubMed] [Google Scholar]
- Delaveau T., Jacq C., Perea J. Sequence of a 12.7 kb segment of yeast chromosome II identifies a PDR-like gene and several new open reading frames. Yeast. 1992 Sep;8(9):761–768. doi: 10.1002/yea.320080909. [DOI] [PubMed] [Google Scholar]
- Dershowitz A., Newlon C. S. The effect on chromosome stability of deleting replication origins. Mol Cell Biol. 1993 Jan;13(1):391–398. doi: 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doignon F., Biteau N., Aigle M., Crouzet M. The complete sequence of a 6794 bp segment located on the right arm of chromosome II of Saccharomyces cerevisiae. Finding of a putative dUTPase in a yeast. Yeast. 1993 Oct;9(10):1131–1137. doi: 10.1002/yea.320091014. [DOI] [PubMed] [Google Scholar]
- Doignon F., Biteau N., Crouzet M., Aigle M. The complete sequence of a 19,482 bp segment located on the right arm of chromosome II from Saccharomyces cerevisiae. Yeast. 1993 Feb;9(2):189–199. doi: 10.1002/yea.320090210. [DOI] [PubMed] [Google Scholar]
- Dujon B., Alexandraki D., André B., Ansorge W., Baladron V., Ballesta J. P., Banrevi A., Bolle P. A., Bolotin-Fukuhara M., Bossier P. Complete DNA sequence of yeast chromosome XI. Nature. 1994 Jun 2;369(6479):371–378. doi: 10.1038/369371a0. [DOI] [PubMed] [Google Scholar]
- Démolis N., Mallet L., Bussereau F., Jacquet M. RIM2, MSI1 and PGI1 are located within an 8 kb segment of Saccharomyces cerevisiae chromosome II, which also contains the putative ribosomal gene L21 and a new putative essential gene with a leucine zipper motif. Yeast. 1993 Jun;9(6):645–659. doi: 10.1002/yea.320090611. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Civalier C., Tye B. K. Specific interaction between a Saccharomyces cerevisiae protein and a DNA element associated with certain autonomously replicating sequences. Proc Natl Acad Sci U S A. 1988 Feb;85(3):743–746. doi: 10.1073/pnas.85.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes H. G., Robinson B. S., Eisenberg S. At least three distinct proteins are necessary for the reconstitution of a specific multiprotein complex at a eukaryotic chromosomal origin of replication. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11156–11160. doi: 10.1073/pnas.89.23.11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Brewer B. J. A question of time: replication origins of eukaryotic chromosomes. Cell. 1992 Oct 30;71(3):363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- Fondrat C., Kalogeropoulos A. Approaching the function of new genes by detection of their potential upstream activation sequences in Saccharomyces cerevisiae: application to chromosome III. Curr Genet. 1994 May;25(5):396–406. doi: 10.1007/BF00351777. [DOI] [PubMed] [Google Scholar]
- Goffeau A., Nakai K., Slonimski P., Risler J. L., Slominski P [corrected to Slonimski P. ]. The membrane proteins encoded by yeast chromosome III genes. FEBS Lett. 1993 Jun 28;325(1-2):112–117. doi: 10.1016/0014-5793(93)81425-y. [DOI] [PubMed] [Google Scholar]
- Goffeau A., Slonimski P., Nakai K., Risler J. L. How many yeast genes code for membrane-spanning proteins? Yeast. 1993 Jul;9(7):691–702. doi: 10.1002/yea.320090703. [DOI] [PubMed] [Google Scholar]
- Goffeau A. Yeast. Genes in search of functions. Nature. 1994 May 12;369(6476):101–102. doi: 10.1038/369101a0. [DOI] [PubMed] [Google Scholar]
- Hauber J., Stucka R., Krieg R., Feldmann H. Analysis of yeast chromosomal regions carrying members of the glutamate tRNA gene family: various transposable elements are associated with them. Nucleic Acids Res. 1988 Nov 25;16(22):10623–10634. doi: 10.1093/nar/16.22.10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré N., Bergh S., Chanteau S., Doucet-Populaire F., Eiglmeier K., Garnier T., Georges C., Launois P., Limpaiboon T., Newton S. Nucleotide sequence of the first cosmid from the Mycobacterium leprae genome project: structure and function of the Rif-Str regions. Mol Microbiol. 1993 Jan;7(2):207–214. doi: 10.1111/j.1365-2958.1993.tb01112.x. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Moore D. P., Blomberg M. A., Braiterman L. T., Voytas D. F., Natsoulis G., Boeke J. D. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993 Jun 4;73(5):1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- Johnson D. I., Jacobs C. W., Pringle J. R., Robinson L. C., Carle G. F., Olson M. V. Mapping of the Saccharomyces cerevisiae CDC3, CDC25, and CDC42 genes to chromosome XII by chromosome blotting and tetrad analysis. Yeast. 1987 Dec;3(4):243–253. doi: 10.1002/yea.320030405. [DOI] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Kunst F., Devine K. The project of sequencing the entire Bacillus subtilis genome. Res Microbiol. 1991 Sep-Oct;142(7-8):905–912. doi: 10.1016/0923-2508(91)90072-i. [DOI] [PubMed] [Google Scholar]
- Lloyd A. T., Sharp P. M. CODONS: a microcomputer program for codon usage analysis. J Hered. 1992 May-Jun;83(3):239–240. doi: 10.1093/oxfordjournals.jhered.a111205. [DOI] [PubMed] [Google Scholar]
- Logghe M., Molemans F., Fiers W., Contreras R. The two genes encoding yeast ribosomal protein S8 reside on different chromosomes, and are closely linked to the hsp70 stress protein genes SSA3 and SSA4. Yeast. 1994 Aug;10(8):1093–1100. doi: 10.1002/yea.320100811. [DOI] [PubMed] [Google Scholar]
- Louis E. J. Corrected sequence for the right telomere of Saccharomyces cerevisiae chromosome III. Yeast. 1994 Feb;10(2):271–274. doi: 10.1002/yea.320100214. [DOI] [PubMed] [Google Scholar]
- Louis E. J., Naumova E. S., Lee A., Naumov G., Haber J. E. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics. 1994 Mar;136(3):789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Blackburn E. H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993 Apr 23;73(2):347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Mallet L., Bussereau F., Jacquet M. Nucleotide sequence analysis of an 11.7 kb fragment of yeast chromosome II including BEM1, a new gene of the WD-40 repeat family and a new member of the KRE2/MNT1 family. Yeast. 1994 Jun;10(6):819–831. doi: 10.1002/yea.320100612. [DOI] [PubMed] [Google Scholar]
- Mannhaupt G., Stucka R., Ehnle S., Vetter I., Feldmann H. Analysis of a 70 kb region on the right arm of yeast chromosome II. Yeast. 1994 Oct;10(10):1363–1381. doi: 10.1002/yea.320101014. [DOI] [PubMed] [Google Scholar]
- Marahrens Y., Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992 Feb 14;255(5046):817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- McCusker J. H., Haber J. E. Cycloheximide-resistant temperature-sensitive lethal mutations of Saccharomyces cerevisiae. Genetics. 1988 Jun;119(2):303–315. doi: 10.1093/genetics/119.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz E. M., Pruitt R. E. Arabidopsis thaliana and Plant Molecular Genetics. Science. 1985 Sep 20;229(4719):1214–1218. doi: 10.1126/science.229.4719.1214. [DOI] [PubMed] [Google Scholar]
- Michels C. A., Read E., Nat K., Charron M. J. The telomere-associated MAL3 locus of Saccharomyces is a tandem array of repeated genes. Yeast. 1992 Aug;8(8):655–665. doi: 10.1002/yea.320080809. [DOI] [PubMed] [Google Scholar]
- Milosavljević A., Jurka J. Discovering simple DNA sequences by the algorithmic significance method. Comput Appl Biosci. 1993 Aug;9(4):407–411. doi: 10.1093/bioinformatics/9.4.407. [DOI] [PubMed] [Google Scholar]
- Miosga T., Zimmermann F. K. Sequence and function analysis of a 2.73 kb fragment of Saccharomyces cerevisiae chromosome II. Yeast. 1993 Nov;9(11):1273–1277. doi: 10.1002/yea.320091115. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Contopoulou C. R., King J. S. Genetic and physical maps of Saccharomyces cerevisiae, Edition 11. Yeast. 1992 Oct;8(10):817–902. doi: 10.1002/yea.320081002. [DOI] [PubMed] [Google Scholar]
- Nasr F., Bécam A. M., Grzybowska E., Zagulski M., Slonimski P. P., Herbert C. J. An analysis of the sequence of part of the right arm of chromosome II of S. cerevisiae reveals new genes encoding an amino-acid permease and a carboxypeptidase. Curr Genet. 1994 Jul;26(1):1–7. doi: 10.1007/BF00326297. [DOI] [PubMed] [Google Scholar]
- Nasr F., Bécam A. M., Slonimski P. P., Herbert C. J. YBR1012 an essential gene from S. cerevisiae: construction of an RNA antisense conditional allele and isolation of a multicopy suppressor. C R Acad Sci III. 1994 Jul;317(7):607–613. [PubMed] [Google Scholar]
- Newlon C. S. Yeast chromosome replication and segregation. Microbiol Rev. 1988 Dec;52(4):568–601. doi: 10.1128/mr.52.4.568-601.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Palladino F., Gasser S. M. Telomere maintenance and gene repression: a common end? Curr Opin Cell Biol. 1994 Jun;6(3):373–379. doi: 10.1016/0955-0674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad M. R., Kirchrath L., Hollenberg C. P. A putative P-type Cu(2+)-transporting ATPase gene on chromosome II of Saccharomyces cerevisiae. Yeast. 1994 Sep;10(9):1217–1225. doi: 10.1002/yea.320100910. [DOI] [PubMed] [Google Scholar]
- Schaaff-Gerstenschläger I., Baur A., Boles E., Zimmermann F. K. Sequence and function analysis of a 4.3 kb fragment of Saccharomyces cerevisiae chromosome II including three open reading frames. Yeast. 1993 Aug;9(8):915–921. doi: 10.1002/yea.320090811. [DOI] [PubMed] [Google Scholar]
- Schaaff-Gerstenschläger I., Mannhaupt G., Vetter I., Zimmermann F. K., Feldmann H. TKL2, a second transketolase gene of Saccharomyces cerevisiae. Cloning, sequence and deletion analysis of the gene. Eur J Biochem. 1993 Oct 1;217(1):487–492. doi: 10.1111/j.1432-1033.1993.tb18268.x. [DOI] [PubMed] [Google Scholar]
- Scherens B., el Bakkoury M., Vierendeels F., Dubois E., Messenguy F. Sequencing and functional analysis of a 32,560 bp segment on the left arm of yeast chromosome II. Identification of 26 open reading frames, including the KIP1 and SEC17 genes. Yeast. 1993 Dec;9(12):1355–1371. doi: 10.1002/yea.320091210. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Lloyd A. T. Regional base composition variation along yeast chromosome III: evolution of chromosome primary structure. Nucleic Acids Res. 1993 Jan 25;21(2):179–183. doi: 10.1093/nar/21.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skala J., Van Dyck L., Purnelle B., Goffeau A. The sequence of an 8 kb segment on the left arm of chromosome II from Saccharomyces cerevisiae identifies five new open reading frames of unknown functions, two tRNA genes and two transposable elements. Yeast. 1992 Sep;8(9):777–785. doi: 10.1002/yea.320080911. [DOI] [PubMed] [Google Scholar]
- Skala J., Van Dyck L., Purnelle B., Goffeau A. The sequence of an 8.8 kb segment on the left arm of chromosome II from Saccharomyces cerevisiae reveals four new open reading frames including homologs of animal DNA polymerase alpha-primases and bacterial GTP cyclohydrolase II. Yeast. 1994 Apr;10 (Suppl A):S13–S24. doi: 10.1002/yea.320100003. [DOI] [PubMed] [Google Scholar]
- Smits P. H., De Haan M., Maat C., Grivell L. A. The complete sequence of a 33 kb fragment on the right arm of chromosome II from Saccharomyces cerevisiae reveals 16 open reading frames, including ten new open reading frames, five previously identified genes and a homologue of the SCO1 gene. Yeast. 1994 Apr;10 (Suppl A):S75–S80. doi: 10.1002/yea.320100010. [DOI] [PubMed] [Google Scholar]
- Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993 Jul 1;21(13):3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A., Dujon B. Nested chromosomal fragmentation in yeast using the meganuclease I-Sce I: a new method for physical mapping of eukaryotic genomes. Nucleic Acids Res. 1992 Nov 11;20(21):5625–5631. doi: 10.1093/nar/20.21.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A., Fairhead C., Dujon B. The complete sequence of the 8.2 kb segment left of MAT on chromosome III reveals five ORFs, including a gene for a yeast ribokinase. Yeast. 1990 Nov-Dec;6(6):521–534. doi: 10.1002/yea.320060609. [DOI] [PubMed] [Google Scholar]
- Van Dyck L., Pearce D. A., Sherman F. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1994 Jan 7;269(1):238–242. [PubMed] [Google Scholar]
- Van Dyck L., Purnelle B., Skala J., Goffeau A. An 11.4 kb DNA segment on the left arm of yeast chromosome II carries the carboxypeptidase Y sorting gene PEP1, as well as ACH1, FUS3 and a putative ARS. Yeast. 1992 Sep;8(9):769–776. doi: 10.1002/yea.320080910. [DOI] [PubMed] [Google Scholar]
- Van Houten J. V., Newlon C. S. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol Cell Biol. 1990 Aug;10(8):3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Aart Q. J., Barthe C., Doignon F., Aigle M., Crouzet M., Steensma H. Y. Sequence analysis of a 31 kb DNA fragment from the right arm of Saccharomyces cerevisiae chromosome II. Yeast. 1994 Jul;10(7):959–964. doi: 10.1002/yea.320100711. [DOI] [PubMed] [Google Scholar]
- Vassarotti A., Goffeau A. Sequencing the yeast genome: the European effort. Trends Biotechnol. 1992 Jan-Feb;10(1-2):15–18. doi: 10.1016/0167-7799(92)90160-w. [DOI] [PubMed] [Google Scholar]
- Wilson R., Ainscough R., Anderson K., Baynes C., Berks M., Bonfield J., Burton J., Connell M., Copsey T., Cooper J. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994 Mar 3;368(6466):32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Lohan A. J. Sequence around the centromere of Saccharomyces cerevisiae chromosome II: similarity of CEN2 to CEN4. Yeast. 1994 Apr;10 (Suppl A):S41–S46. doi: 10.1002/yea.320100006. [DOI] [PubMed] [Google Scholar]
- Zagulski M., Bécam A. M., Grzybowska E., Lacroute F., Migdalski A., Slonimski P. P., Sokolowska B., Herbert C. J. The sequence of 12.5 kb from the right arm of chromosome II predicts a new N-terminal sequence for the IRA1 protein and reveals two new genes, one of which is a DEAD-box helicase. Yeast. 1994 Sep;10(9):1227–1234. doi: 10.1002/yea.320100911. [DOI] [PubMed] [Google Scholar]