Abstract
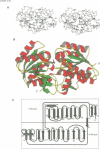
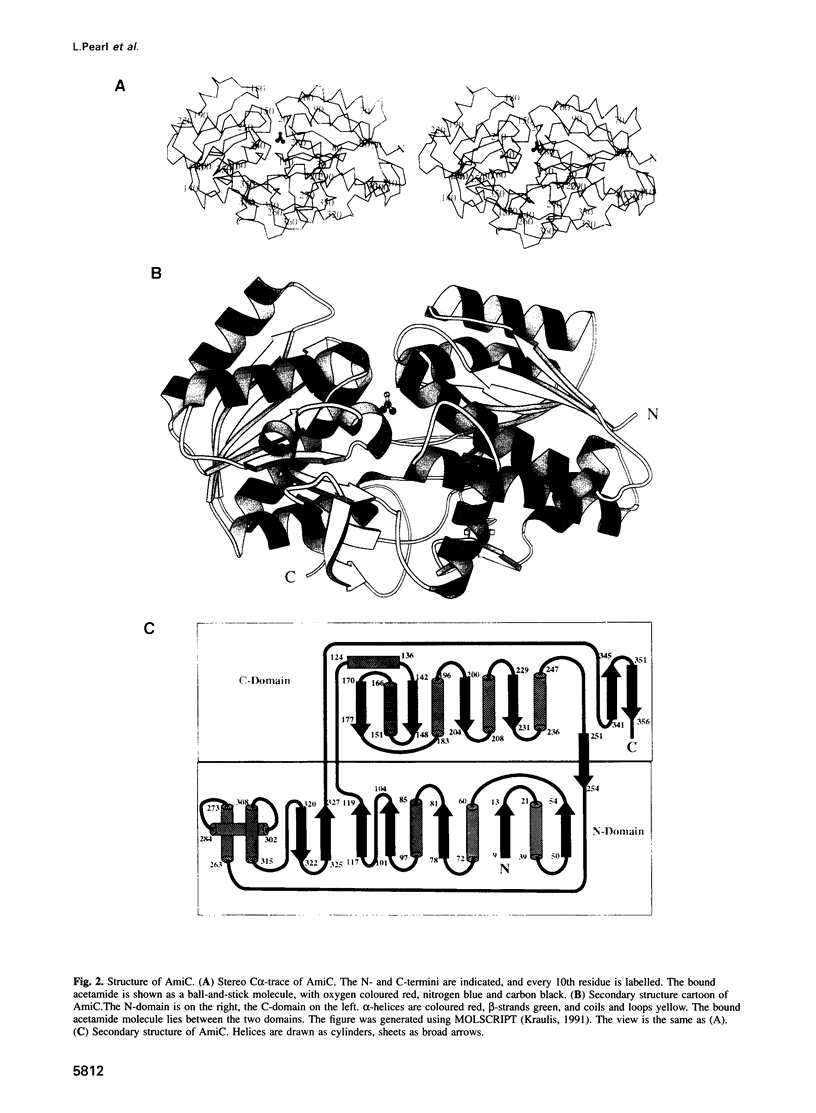
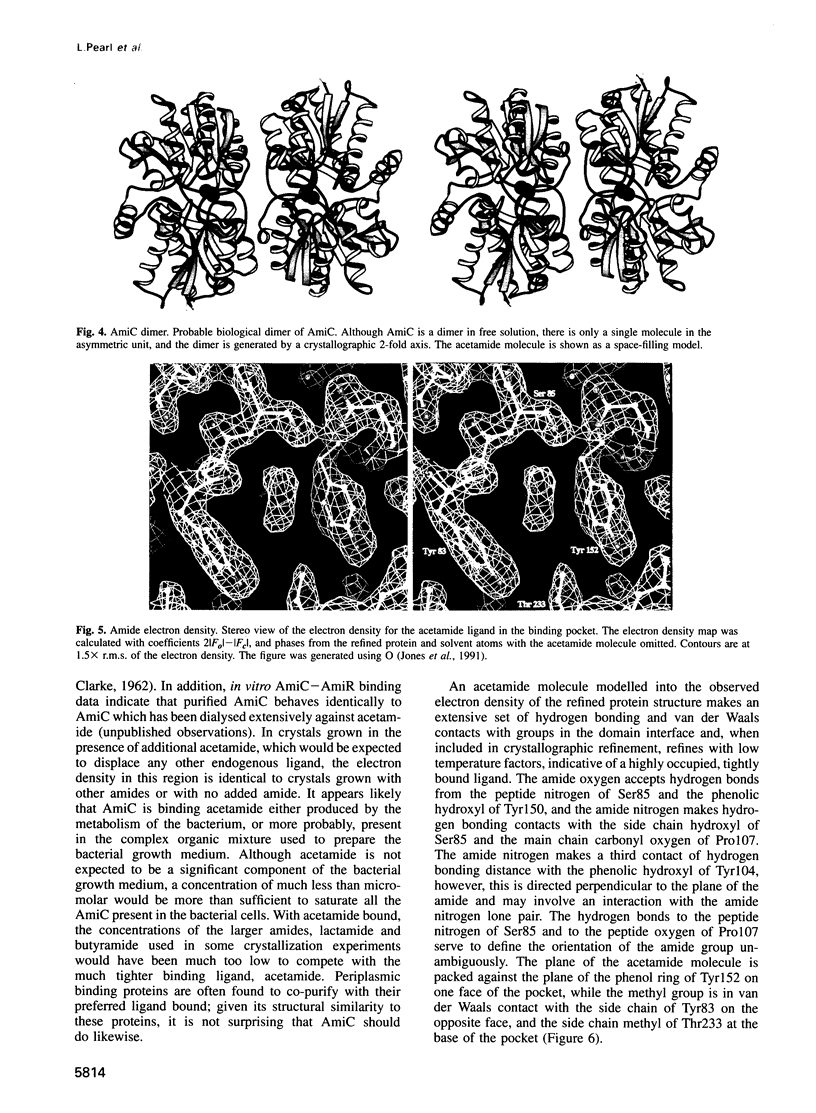
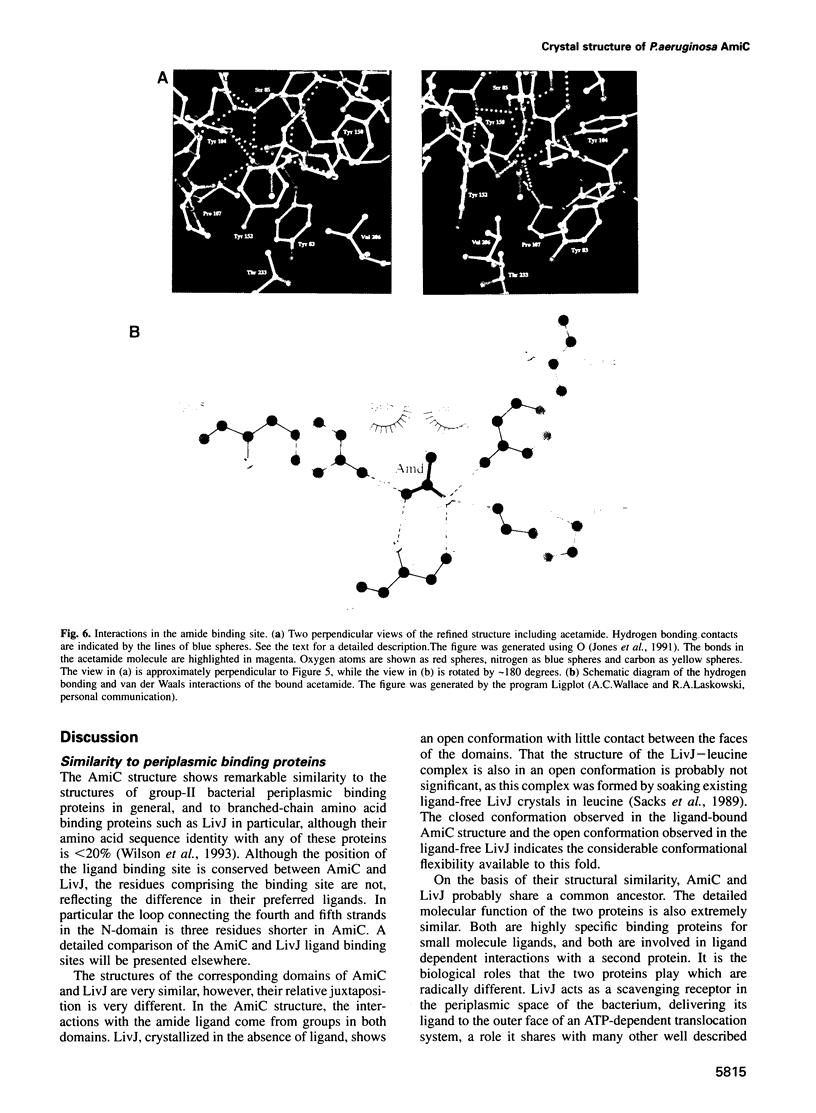
The crystal structure for the negative regulator (AmiC) of the amidase operon from Pseudomonas aeruginosa has been solved at a resolution of 2.1 A. AmiC is the amide sensor protein in the amidase operon and regulates the activity of the transcription antitermination factor AmiR, which in turn regulates amidase expression. The AmiC structure consists of two domains with an alternating beta-alpha-beta topology. The two domains are separated by a central cleft and the amide binding site is positioned in this cleft at the interface of the domains. The overall fold for AmiC is extremely similar to that for the leucine-isoleucine-valine binding protein (LivJ) of Escherichia coli despite only 17% sequence identity, however, the two domains of AmiC are substantially closed compared with LivJ. The closed structure of AmiC is stabilized significantly by the bound acetamide, suggesting a molecular mechanism for the process of amide induction. The amide binding site is extremely specific for acetamide and would not allow a closed conformation in the presence of the anti-inducer molecule butyramide.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey S., Evans R. W., Garratt R. C., Gorinsky B., Hasnain S., Horsburgh C., Jhoti H., Lindley P. F., Mydin A., Sarra R. Molecular structure of serum transferrin at 3.3-A resolution. Biochemistry. 1988 Jul 26;27(15):5804–5812. doi: 10.1021/bi00415a061. [DOI] [PubMed] [Google Scholar]
- Drew R., Lowe N. Positive control of Pseudomonas aeruginosa amidase synthesis is mediated by a transcription anti-termination mechanism. J Gen Microbiol. 1989 Apr;135(4):817–823. doi: 10.1099/00221287-135-4-817. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- KELLY M., CLARKE P. H. An inducible amidase produced by a strain of Pseudomonas aeruginosa. J Gen Microbiol. 1962 Feb;27:305–316. doi: 10.1099/00221287-27-2-305. [DOI] [PubMed] [Google Scholar]
- Louie G. V., Brownlie P. D., Lambert R., Cooper J. B., Blundell T. L., Wood S. P., Warren M. J., Woodcock S. C., Jordan P. M. Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site. Nature. 1992 Sep 3;359(6390):33–39. doi: 10.1038/359033a0. [DOI] [PubMed] [Google Scholar]
- Luck L. A., Falke J. J. Open conformation of a substrate-binding cleft: 19F NMR studies of cleft angle in the D-galactose chemosensory receptor. Biochemistry. 1991 Jul 2;30(26):6484–6490. doi: 10.1021/bi00240a019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer M. E., Lewis B. A., Quiocho F. A. The radius of gyration of L-arabinose-binding protein decreases upon binding of ligand. J Biol Chem. 1981 Dec 25;256(24):13218–13222. [PubMed] [Google Scholar]
- Sack J. S., Saper M. A., Quiocho F. A. Periplasmic binding protein structure and function. Refined X-ray structures of the leucine/isoleucine/valine-binding protein and its complex with leucine. J Mol Biol. 1989 Mar 5;206(1):171–191. doi: 10.1016/0022-2836(89)90531-7. [DOI] [PubMed] [Google Scholar]
- Vermersch P. S., Tesmer J. J., Lemon D. D., Quiocho F. A. A Pro to Gly mutation in the hinge of the arabinose-binding protein enhances binding and alters specificity. Sugar-binding and crystallographic studies. J Biol Chem. 1990 Sep 25;265(27):16592–16603. [PubMed] [Google Scholar]
- Wilson S. A., Chayen N. E., Hemmings A. M., Drew R. E., Pearl L. H. Crystallization of and preliminary X-ray data for the negative regulator (AmiC) of the amidase operon of Pseudomonas aeruginosa. J Mol Biol. 1991 Dec 20;222(4):869–871. doi: 10.1016/0022-2836(91)90579-u. [DOI] [PubMed] [Google Scholar]
- Wilson S. A., Wachira S. J., Drew R. E., Jones D., Pearl L. H. Antitermination of amidase expression in Pseudomonas aeruginosa is controlled by a novel cytoplasmic amide-binding protein. EMBO J. 1993 Sep;12(9):3637–3642. doi: 10.1002/j.1460-2075.1993.tb06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]