Abstract
With the aim of investigating whether interleukin 28B gene (IL28B) rs1297860 polymorphism is associated with different hepatitis C (HCV) infection statuses, we compared IL28B allelic distribution in an Italian case series of 1050 patients with chronic infection and different outcomes, 47 individuals who spontaneously cleared HCV, and 178 blood donors. Furthermore, we compared IL28B variants among 3882 Caucasian patients with chronic infection, 397 with spontaneous clearance, and 1366 blood donors reported in PubMed. Overall data confirmed a relation between IL28B C allele and HCV spontaneous clearance. Furthermore, we found that IL28B T allele had a weak relation with chronic HCV progression to hepatocellular carcinoma. Study findings are in accordance with the hepatocellular carcinogenic model where IL28B TT genotype, by promoting a persistent chronic hepatitis which leads to both hepatocyte injury and chronic inflammation, could facilitate HCC development. Conversely, patients with lymphoproliferative disorders had not any significantly different IL28B rs1297860 allelic distribution than those with chronic HCV, but, like all chronic HCV-related diseases, they showed a lower CC frequency than patients who spontaneously cleared HCV. Study results confirmed the model of persistent HCV infection as a risk factor for the pathogenesis of both liver and lymphoproliferative disorders.
1. Introduction
Hepatitis C virus (HCV), a positive-strand RNA virus widely diffused (about 2-3%) around the world, is a major causative agent of chronic liver diseases, including hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) [1, 2]. HCV infection is also associated with lymphoproliferative disorders, represented by mixed type II cryoglobulinemia (MC) and B-cell non-Hodgkin's lymphomas (NHLs) [3–6]. The reason for such different outcome of HCV infection among patients is still largely unknown.
Traditional therapy for treatment of chronic HCV involves pegylated type I IFN (PEG-IFN) in combination with ribavirin. This treatment eradicates HCV infection in only 40%–50% of patients infected with genotypes 1 or 4 and 75%–90% of those infected with genotypes 2 or 3 [7, 8]. However, adverse effects due to this treatment regimen frequently lead to poor tolerance.
Recent studies have reported that human genetic variants, in particular rs1297860 single nucleotide polymorphism (SNP), within or near the interleukin 28B gene (IL28B) are significantly associated with spontaneous HCV clearance as well as different outcomes of treatment with IFN/ribavirin therapy for chronic HCV infection [9–19]. The relation is more evident for HCV genotype 1 infection. These observations suggest an important role of IL28B against HCV infection and natural history of HCV outcomes.
IL28B is a member, along with IL-29 and IL-28A, of type III interferons, also termed interferon-lambda (IFN-λ). They have in common a strong antiviral function and induction of interferon stimulated genes (ISGs) [20–24].
IFN-λ signals through interferon-lambda receptor1 (IFNLR1) and interleukin-10 receptor2 (IL-10R2) result in STAT phosphorylation, dimer formation, nuclear translocation, and then induction of ISG expression and upregulation of major histocompatibility complex (MHC) class I [25].
IFN-λ responsiveness is restricted to specific cell types [26], partially because IFNLR1 tissue distribution is highly restricted [27]. In contrast to the epithelial-like cells, fibroblasts and endothelial cells were completely unresponsive to IFN-λ. A good IFN-λ-induced response was shown in hepatic cells, with an increased IFNLR1 expression after viral infection [28]. Naïve B and T cells, lymphocytes, and monocytes, although express adequate amounts of IFNLR1 receptor, respond poorly or not at all to IFN-λ [28]. This implies the presence of specific mechanisms on the lymphoid tissues that may inhibit the IFN-λ response. This important functional difference in tissue response to IFN-λ may provide a clinical advantage from IFN-α as a treatment for chronic HCV infection, as IFN-λ is less likely to induce the leucopenia most often associated with IFN-α therapy and may be used in HCV patients who are resistant to IFN-α. Moreover, it was found that IFN-λ upregulates the production of cytokines by natural killer (NK) and T cells [29] and reduces the proliferation of regulatory T cells [30]; thus [31] it is supposed that lymphoid cells respond to IFN-λ by different signals than hepatic cells. Due to the responsiveness of most cell types to IFN-λ and a limited toxicity, IFN-λs are under study for clinical use not only for anti-HCV therapy but also as anticancer and antiviral drug [22].
The aim of this paper is to investigate whether a relation between IL28B rs12979860 polymorphism and severity of liver or lymphoproliferative diseases exists.
2. Material and Methods
2.1. Patients
We identified and reviewed papers published from 2009 to 2013 analyzing Caucasian HCV-infected patients for distribution of IL28B genetic variants, using the online database pubMed. Selected studies included in the analysis along with inclusion criteria were reported in Table 1.
Table 1.
Caucasian patients included in IL28B rs12979860 analysis with different HCV-related outcomes reported in the literature (PubMed) between 2009 and 2013.
| Article | Blood donors (HCV neg.) |
Spontaneous clearance of HCV | Chronic HCV | Mild/moderate fibrosis | Cirrhosis | Hepatocellular carcinoma | Mixed cryoglobulinemia S. |
|---|---|---|---|---|---|---|---|
| Mangia et al. (2013) [13] | 178 | 47 | 122 | ||||
| Sharafi et al. (2012) [17] | 142 | 921 | |||||
| Piluso et al. (2013) [15] | 231 | 250 | |||||
| Falleti et al. (2011) [11] | 428 | 429 | 200 | ||||
| Knapp et al. (2011) [12] | 74 | 89 | 234 | ||||
| Fabris et al. (2011) [32] | 344 | 235 | 199 | 35 | |||
| Sarrazin et al. (2011) [16] | 200 | 645 | |||||
| Thomas et al. (2009) [18] | 261 | 381 | |||||
|
| |||||||
| Total | 1366 | 397 | 2534 | 664 | 399 | 35 | 250 |
We also analyzed the IL28B rs12979860 polymorphism in a case series of 1050 HCV-infected patients, 47 individuals who experienced spontaneous HCV clearance, and 178 blood donors, from a cohort of HIV- and HBV-negative Italian patients, referred to the following centers: MASVE Center, University of Florence, Florence; “Santa Maria degli Angeli” General Hospital, Pordenone; National Cancer Institute “Fondazione Pascale”, Naples; “Casa Sollievo della Sofferenza” Hospital, San Giovanni Rotondo; “Azienda Ospedaliero-Universitaria Consorziale Policlinico”, Bari; University Hospital “Santa Maria della Misericordia”, Udine; Ospedale san Gerardo-University of Milano-Bicocca, Monza; and National Cancer Institute “Centro di Riferimento Oncologico”, CRO Aviano National Cancer Institute. Our series of 1050 patients with chronic HCV infection included group of patients with absence of HCC or any sign/symptom of definite MC or NHL (CHC, n = 536), patients with HCC (n = 95), patients with a definite MC syndrome, according to previously described criteria [33] (n = 352), and presence of a definite malignant B-cell NHL (n = 67).
2.2. IL28B Genotyping
Genomic DNA was extracted from peripheral blood which has been previously stored at −80°C.
IL28B genotyping was performed using a specific customized TaqMan SNP-genotyping Assay (rs12979860; Applied Bosystem, Foster City, CA, USA) based on allele-specific labeled probes on a Rotor Gene 6000 (Corbett Research, Sidney, Australia). Amplicon sequencing was used to validate the genotyping techniques.
2.3. Statistical Analysis
Frequencies (f) of IL28B rs12979860 C and T alleles and genotypes polymorphisms were determined individually by direct counting of the positive individuals for a specific genotype/allele polymorphism. The observed and expected genotype frequencies among the study groups were analyzed using the Hardy-Weinberg equilibrium theory. Differences in f between groups of selected patients were evaluated using Fisher's exact test, Pearson's chi-square (χ 2) test (with Yates correction), or Cochran-Armitage z 2 test for trend in frequencies as appropriate [34]. A two-sided P value <0.05 was considered as statistically significant.
3. Results
Reviewed data on IL28B variant distribution in different HCV outcomes and limited to Caucasian subjects were reported in Table 2. The genotype frequencies of the IL28B rs12979860 polymorphism were found in Hardy-Weinberg equilibrium in the blood donors group (Pearson's χ 2 = 0.09; df = 1; P = 0.76). The IL28B rs12979860 C allele was related to a significant increase in spontaneous clearance of the virus in a dominant genetic model (CC versus CT + TT) employed to compare HCV+ patients presenting spontaneous viral clearance versus all other HCV+ subjects (Pearson's χ 2 = 148.9; P < 0.001). Data indicates that C allele had an important role in the control of infection (Fisher's test, P < 0.001, Figure 1).
Table 2.
IL28B rs12979860 variant frequencies in blood donors and patients with HCV infection and different outcomes. Caucasian individuals reported in literature (PubMed), 2009–2013.
|
Blood donors (HCV neg.) |
Spontaneous clearance of HCV | HCV patients | HCV patients total | |||||
|---|---|---|---|---|---|---|---|---|
| Chronic HCV | Mild/moderate fibrosis | Cirrhosis | Hepatocellular carcinoma | Mixed cryoglobulinemia S. | ||||
| Number (%) | Number (%) | Number (%) | Number (%) | Number (%) | Number (%) | Number (%) | Number (%) | |
| IL28B genotype | ||||||||
| CC | 631 (46.2) | 273 (68.8) | 1004 (39.6) | 212 (31.9) | 126 (31.6) | 6 (17.1) | 99 (39.6) | 1447 (37.3) |
| CT | 599 (43.8) | 102 (25.7) | 1225 (48.3) | 373 (56.2) | 204 (51.1) | 21 (60.0) | 113 (45.2) | 1936 (49.9) |
| TT | 136 (10.0) | 22 (5.5) | 305 (12.0) | 79 (11.9) | 69 (17.3) | 8 (22.8) | 38 (15.2) | 499 (12.8) |
| Total | 1366 (100.0) | 397 (100.0) | 2534 (100.0) | 664 (100.0) | 399 (100.0) | 35 (100.0) | 250 (100.0) | 3882 (100.0) |
| χ 2 (P value)a | — | Ref. | 118.8 (<0.001) | 135.9 (<0.001) | 112.4 (<0.001) | 40.5 (<0.001) | 55.7 (<0.001) | 148.9 (<0.001) |
|
| ||||||||
| IL28B dominant model | ||||||||
| CC | 631 (46.2) | 273 (68.8) | 1004 (39.6) | 212 (31.9) | 126 (31.6) | 6 (17.1) | 99 (39.6) | 1447 (37.3) |
| CT + TT | 735 (53.8) | 124 (31.2) | 1530 (60.4) | 452 (68.1) | 273 (68.4) | 29 (82.9) | 151 (60.1) | 2435 (62.7) |
| Total | 1366 (100.0) | 397 (100.0) | 2534 (100.0) | 664 (100.0) | 399 (100.0) | 35 (100.0) | 250 (100.0) | 3882 (100.0) |
| χ 2 (P value)a | — | Ref. | 118.6 (<0.001) | 135.9 (<0.001) | 110.1 (<0.001) | 37.5 (<0.001) | 53.4 (<0.001) | 148.6 (<0.001) |
|
| ||||||||
| IL28B allele | ||||||||
| C | 1861 (68.1) | 648 (81.6) | 3233 (63.8) | 797 (60.0) | 456 (57.1) | 33 (47.1) | 311 (62.2) | 4830 (62.2) |
| T | 871 (31.9) | 146 (18.4) | 1835 (36.2) | 531 (40.0) | 342 (42.9) | 37 (52.9) | 189 (37.8) | 2934 (37.8) |
| Total | 2732 (100.0) | 794 (100.0) | 5068 (100.0) | 1328 (100.0) | 798 (100.0) | 70 (100.0) | 500 (100.0) | 7764 (100.0) |
| χ 2 (P value)a | — | Ref. | 97.4 (<0.001) | 106.7 (<0.001) | 112.1 (<0.001) | 45.8 (<0.001) | 60.3 (<0.001) | 117.7 (<0.001) |
aPearson's chi-square with spontaneous clearance of HCV as reference group.
Figure 1.
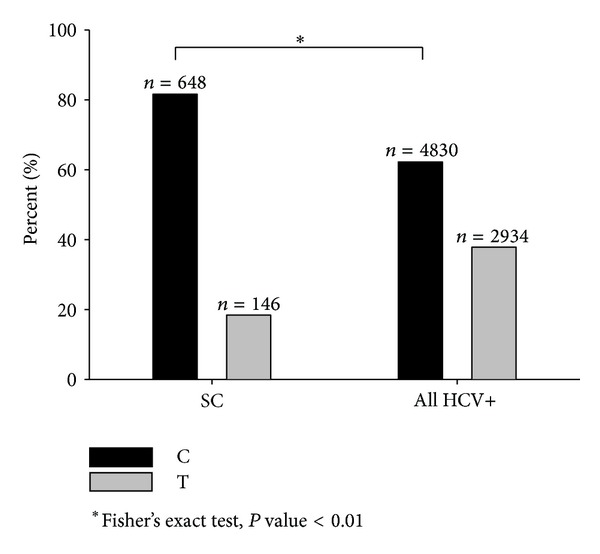
IL28B rs12979860 C/T allele frequencies in patients with spontaneous clearance (SC) of HCV infection versus all HCV+ patients. Caucasian individuals reported in literature (PubMed), 2009–2013.
A significant negative linear trend was detected in the order of the CC genotype frequencies of patients with an increasing severity of liver diseases (from 40% in CHC to 17% in HCC, Figure 2; Cochran-Armitage's z 2 = 22.38, P < 0.001), whereas a significant positive linear trend emerged for TT genotype frequencies (from 12.0% in CHC to 22.8% in HCC, Figure 2; Cochran-Armitage's z 2 = 8.42, P = 0.004), suggesting an important role of CC allele in the control of liver disease progression.
Figure 2.
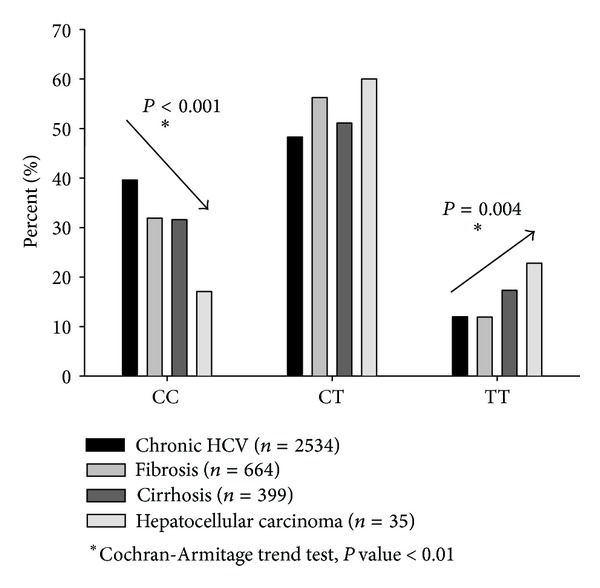
IL28B rs12979860 genotype frequencies in patients with HCV infection and different outcomes. Caucasian individuals reported in literature (PubMed), 2009–2013.
IL28B rs12979860 genotype frequencies found in our case series were similar to those reported in the literature (Table 3). We found a slightly increasing trend in TT genotype frequency in HCC (18.9%) with respect to that of CHC (12.7%), but difference was not statistically significant (Fisher's test, P = 0.106) (Figure 3). Given the modest increase in HCC risk we found and the low number of HCC cases included in this study (35 from the literature and 95 in our case series), replication studied using larger series of HCV+ HCC would need to confirm this finding. However, carriage of TT genotype in blood donors (7.9%) and in patients who spontaneously cleared the virus (2.1%) was lower than that found in our HCC series (Fisher's test, P = 0.009 and 0.003, resp.). Moreover, similar to that found in data from the literature, the frequency of CC genotype was significantly higher among patients who spontaneously cleared the virus compared to patients with CHC (Fisher's test, P < 0.001) (Table 3 and Figure 3).
Table 3.
IL28B rs12979860 variant frequencies in 178 blood donors and 1097 patients with HCV infection and different outcomes. Italian case series.
|
Blood donors (HCV neg.) |
Spontaneous clearance of HCV | HCV patients | HCV patients total | ||||
|---|---|---|---|---|---|---|---|
| Chronic HCV | Hepatocellular carcinoma | Mixed cryoglobulinemia S. | Non-Hodgkin's lymphoma | ||||
| Number (%) | Number (%) | Number (%) | Number (%) | Number (%) | Number (%) | Number (%) | |
| IL28B genotype | |||||||
| CC | 75 (42.1) | 37 (78.7) | 197 (36.7) | 28 (29.5) | 144 (40.9) | 22 (32.9) | 391 (37.2) |
| CT | 89 (50.0) | 9 (19.2) | 271 (50.6) | 49 (51.6) | 155 (44.0) | 37 (55.2) | 512 (48.8) |
| TT | 14 (7.9) | 1 (2.1) | 68 (12.7) | 18 (18.9) | 53 (15.1) | 8 (11.9) | 147 (14.0) |
| Total | 178 (100.0) | 47 (100.0) | 536 (100.0) | 95 (100.0) | 352 (100.0) | 67 (100.0) | 1050 (100.0) |
| χ 2 (P value)a | — | Ref. | 31.9 (<0.001) | 31.4 (<0.001) | 24.4 (<0.001) | 23.5 (<0.001) | 32.8 (<0.001) |
|
| |||||||
| IL28B dominant model | |||||||
| CC | 75 (42.1) | 37 (78.7) | 197 (36.7) | 28 (29.5) | 144 (40.9) | 22 (32.9) | 391 (37.2) |
| CT + TT | 103 (57.9) | 10 (21.3) | 339 (63.3) | 67 (70.5) | 208 (59.1) | 45 (67.1) | 659 (62.8) |
| Total | 178 (100.0) | 47 (100.0) | 536 (100.0) | 95 (100.0) | 352 (100.0) | 67 (100.0) | 1050 (100.0) |
| χ 2 (P value)a | — | Ref. | 31.7 (<0.001) | 30.7 (<0.001) | 23.9 (<0.001) | 23.3 (<0.001) | 32.5 (<0.001) |
|
| |||||||
| IL28B allele | |||||||
| C | 239 (67.1) | 83 (88.3) | 665 (62.0) | 105 (55.3) | 443 (62.9) | 81 (60.4) | 1294 (61.2) |
| T | 117 (32.9) | 11 (11.7) | 407 (48.0) | 85 (44.7) | 261 (37.1) | 53 (39.6) | 806 (38.8) |
| Total | 356 (100.0) | 94 (100.0) | 1072 (100.0) | 190 (100.0) | 704 (100.0) | 134 (100.0) | 2100 (100.0) |
| χ 2 (P value)a | — | Ref. | 25.9 (<0.001) | 30.7 (<0.001) | 23.8 (<0.001) | 21.2 (<0.001) | 27.4 (<0.001) |
aPearson's chi-square with spontaneous clearance of HCV as reference group.
Figure 3.
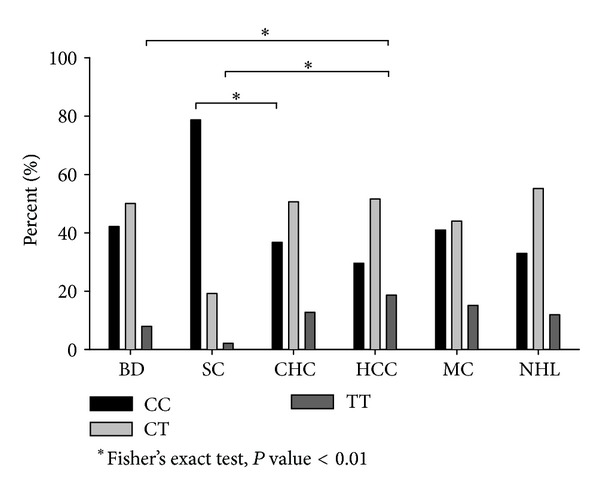
Genotype frequencies of IL28B rs12979860 C/T polymorphism in 178 blood donors (BD), 47 patients with spontaneous clearance (SC) of HCV, 536 patients with chronic HCV (CHC), 95 patients with hepatocellular carcinoma (HCC), 352 patients with mixed cryoglobulinemia syndrome (MC), and 67 patients with non-Hodgkin's lymphoma (NHL). Italian case series.
As regards the lymphoproliferative disorders associated with HCV infection, the literature reported only one study including patients with autoimmune MC syndrome [15] and no study on NHL. Data obtained in our case series were reported in Table 3.
In MC, IL28B CC genotype frequency was found similar to that in blood donors (Figure 3); nonetheless, the carriage of TT genotype in MC (15.1%) was weakly higher than that among blood donors (7.9%, Fisher's test, P = 0.019) but not significantly higher than that among CHC patients (12.9, Fisher's test, P = 0.319). HCV positive patients with NHL (11.9%) were very similar to CHC patients; in this case the carriage of TT genotype was not significantly different.
4. Discussion
Several studies have indicated that IL28B rs12979860 polymorphism has a role in the response of HCV infection and liver disease risk, but the knowledge of its overall effect on the natural history of hepatitis viral infection remains partial and limited to certain aspect in individual studies. In addition, data regarding the IL-28B rs12979860 risk towards a HCV-related lymphoproliferative disorder (MC or NHL) is even less known. Thus we conducted this study to evaluate relations between IL-28B rs12979860 CT polymorphism and natural history of HCV infection by using the following inclusion criteria: Caucasian individuals affected by HCV infection (excluding HBV or HIV co-infection) and different liver or lymphoproliferative diseases related to HCV progression. The analysis of aggregated data from the literature clearly indicated that IL-28B rs12979860 allelic frequencies were significantly different in patients who spontaneously cleared HCV infection in comparison to patients with chronic HCV infection. HCV+ patients who spontaneously cleared the infection carried the CC genotype in 68.8% of cases, while patients with chronic HCV infection and liver diseases carried this genotype in a range from 39.6% to 17.1%. The IL-28B C allele was inversely related to the risk of chronic and liver diseases with respect to both blood donors and patients who spontaneously eliminated the virus when the dominant genetic model (CC versus CT + TT) was employed, in agreement with previous idea that C allele is implicated in the control of HCV infection. In addition, although resulting from a limited number of cases analyzed (n = 35 from literature [32], n = 95 in our series), the carriage of TT genotype and the presence of T allele were slightly prevalent in the HCC group of patients. Due to the weak effect of IL28B T allele found on the progression to HCC, replication of this analysis in a large number of cases is required to confirm our study finding. Anyhow, this finding is in accordance with the HCC carcinogenic model where IL28B T allele carriage was much more favorable to persistence of a chronic HCV infection, which leads to both hepatocyte injury and chronic inflammation in the liver, thus facilitating the HCC development [35–37].
We presented for the first time a comparative analysis of IL28B rs12979860 allelic distribution in patients with chronic HCV infection and lymphoproliferative diseases (Table 3 and Figure 3). No statistically significant differences in IL28B rs12979860 allelic distribution emerged between patients with lymphoproliferative disorders and other groups of patients or blood donors. It should be noted, however, that CC genotype was more frequent in patients with MC (144/352, 40.9%) than in those with malignant tumors taken together (HCC + NHL, 50/162, 30.9%, Fisher's test P = 0.031, data not shown in Tables). Moreover, TT genotype was slightly more frequent, without reaching statistical significance maybe because of low numbers, in patients with an HCC (18.9%) than in those with a NHL (11.9%, Figure 3). These findings could suggest that patients with MC syndrome carrying IL28B rs12979860 CC, a genotype more favorable to resolve the infection, should contrast both liver and lymphoproliferative malignant diseases and that patients with TT genotype could be more predisposed to evolve towards an HCC than a malignant lymphoma.
In conclusion, study results on overall data from the literature and our case series confirmed that all chronic HCV-related disorders presented a lower frequency of C allele as compared to patients who spontaneously eliminated HCV, thus supporting the model of persistent HCV infection as an important risk factor for the pathogenesis of both liver and lymphoproliferative disorders [3, 6]. Moreover, an additional risk factor related to TT homozygosis and liver disease progression emerged from this study. Of note, since recently a polymorphism (ss469415590) of a new interferon gene, IFNL4, was found strongly associated with HCV clearance and was in high linkage disequilibrium with IL28B [38]; further studies are needed regarding IFNL4 polymorphism and its possible involvement in the HCV-related malignancies.
Acknowledgment
The work was supported by the Italian Association for Cancer Research (AIRC no. 10266).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Zaltron S, Spinetti A, Biasi L, Baiguera C, Castelli F. Chronic HCV infection: epidemiological and clinical relevance. BMC Infectious Diseases. 2012;12(2):S2 pages. doi: 10.1186/1471-2334-12-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberti A, Chemello L, Benvegnù L. Natural history of hepatitis C. Journal of Hepatology, Supplement. 1999;31(1):17–24. doi: 10.1016/s0168-8278(99)80369-9. [DOI] [PubMed] [Google Scholar]
- 3.De Re V, Caggiari L, Garziera M, De Zorzi M, Repetto O. Molecular signature in HCV-positive lymphomas. Clinical and Developmental Immunology. 2012;2012:9 pages. doi: 10.1155/2012/623465.623465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansonno D, Carbone A, De Re V, Dammacco F. Hepatitis C virus infection, cryoglobulinaemia, and beyond. Rheumatology. 2007;46(4):572–578. doi: 10.1093/rheumatology/kel425. [DOI] [PubMed] [Google Scholar]
- 5.Zignego AL, Giannini C, Gragnani L. HCV and lymphoproliferation. Clinical and Developmental Immunology. 2012;2012:8 pages. doi: 10.1155/2012/980942.980942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dammacco F, editor. HCV Infection and Cryoglobulinemia. Springer; 2012. (ISBN-13:978-8847017047). [Google Scholar]
- 7.Zeuzem S, Berg T, Moeller B, et al. Expert opinion on the treatment of patients with chronic hepatitis C. Journal of Viral Hepatitis. 2009;16(2):75–90. doi: 10.1111/j.1365-2893.2008.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangia A, Thompson AJ, Santoro R, et al. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139(3):821.e1–827.e1. doi: 10.1053/j.gastro.2010.05.079. [DOI] [PubMed] [Google Scholar]
- 9.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139(6):1865–1876. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibert S, Roger T, Calandra T, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. The Journal of Experimental Medicine. 2013;210(6):1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falleti E, Bitetto D, Fabris C, et al. Role of interleukin 28B rs12979860 C/T polymorphism on the histological outcome of chronic hepatitis C: relationship with gender and viral genotype. Journal of Clinical Immunology. 2011;31(5):891–899. doi: 10.1007/s10875-011-9547-1. [DOI] [PubMed] [Google Scholar]
- 12.Knapp S, Warshow U, Ho KMA, et al. A polymorphism in IL28B distinguishes exposed, uninfected individuals from spontaneous resolvers of HCV infection. Gastroenterology. 2011;141(1):320.e2–325.e2. doi: 10.1053/j.gastro.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangia A, Santoro R, Copetti M, et al. Treatment optimization and prediction of HCV clearance in patients with acute HCV infection. Journal of Hepatology. 2013;59(2):221–228. doi: 10.1016/j.jhep.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy JJ, Li JH, Thompson A, et al. Replicated relation between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138(7):2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piluso A, Giannini C, Fognani E, et al. Value of IL28B genotyping in patients with HCV-related mixed cryoglobulinemia: results of a large, prospective study. Journal of Viral Hepatitis. 2013;20(4):e107–e114. doi: 10.1111/jvh.12017. [DOI] [PubMed] [Google Scholar]
- 16.Sarrazin C, Susser S, Doehring A, et al. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. Journal of Hepatology. 2011;54(3):415–421. doi: 10.1016/j.jhep.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 17.Sharafi H, Pouryasin A, Alavian SM, et al. Distribution of IL28B genotypes in iranian patients with chronic hepatitis C and healthy individuals. Hepatitis Monthly. 2012;12(12):p. e8387. doi: 10.5812/hepatmon.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng MH, Li Y, Xiao DD, et al. Interleukin-28B rs12979860C/T and rs8099917T/G contribute to spontaneous clearance of hepatitis C virus in Caucasians. Gene. 2013;518(2):479–482. doi: 10.1016/j.gene.2012.12.067. [DOI] [PubMed] [Google Scholar]
- 20.Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. Journal of Interferon and Cytokine Research. 2010;30(8):555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon-δ is functionally an interferon but structurally related to the interleukin-10 family. The Journal of Biological Chemistry. 2009;284(31):20869–20875. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Liu X, Zhou Y, Shao BS. Interferon-λs: the modulators of antivirus, antitumor, and immune responses. Journal of Leukocyte Biology. 2009;86(1):23–32. doi: 10.1189/jlb.1208761. [DOI] [PubMed] [Google Scholar]
- 23.Steen HC, Gamero AM. Interferon-lambda as a potential therapeutic agent in cancer treatment. Journal of Interferon and Cytokine Research. 2010;30(8):597–602. doi: 10.1089/jir.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witte K, Witte E, Sabat R, Wolk K. IL-28A, IL-28B, and IL-29: promising cytokines with type I interferon-like properties. Cytokine and Growth Factor Reviews. 2010;21(4):237–251. doi: 10.1016/j.cytogfr.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko SV, Renauld J-C. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-λ1. Similarities with type 1 interferon signaling. The Journal of Biological Chemistry. 2004;279(31):32269–32274. doi: 10.1074/jbc.M404789200. [DOI] [PubMed] [Google Scholar]
- 26.Witte K, Gruetz G, Volk H-D, et al. Despite IFN- receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes and Immunity. 2009;10(8):702–714. doi: 10.1038/gene.2009.72. [DOI] [PubMed] [Google Scholar]
- 27.Borden EC, Sen GC, Uze G, et al. Interferons at age 50: past, current and future impact on biomedicine. Nature Reviews Drug Discovery. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickensheets H, Sheikh F, Park O, Gao B, Donnelly RP. Interferon-lambda (IFN-lambda) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. Journal of Leukocyte Biology. 2013;93:377–385. doi: 10.1189/jlb.0812395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan WJ, Eskdale J, Srinivas S, et al. Human interferon lambda-1 (IFN-λ1/IL-29) modulates the Th1/Th2 response. Genes and Immunity. 2007;8(3):254–261. doi: 10.1038/sj.gene.6364382. [DOI] [PubMed] [Google Scholar]
- 30.Mennechet FJD, Uzé G. Interferon-λ-treated dendritic cells specifically induce proliferation of FOXP3-expressing suppressor T cells. Blood. 2006;107(11):4417–4423. doi: 10.1182/blood-2005-10-4129. [DOI] [PubMed] [Google Scholar]
- 31.Butchar JP, Mehta P, Justiniano SE, et al. Reciprocal regulation of activating and inhibitory Fcγ receptors by TLR7/8 activation: implications for tumor immunotherapy. Clinical Cancer Research. 2010;16(7):2065–2075. doi: 10.1158/1078-0432.CCR-09-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabris C, Falleti E, Cussigh A, et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. Journal of Hepatology. 2011;54(4):716–722. doi: 10.1016/j.jhep.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Pietrogrande M, De Vita S, Zignego AL, et al. Recommendations for the management of mixed cryoglobulinemia syndrome in hepatitis C virus-infected patients. Autoimmunity Reviews. 2011;10(8):444–454. doi: 10.1016/j.autrev.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Agresti A. Categorical Data Analysis. 3rd edition. New York, NY, USA: John Wiley & Sons; 2013. [Google Scholar]
- 35.Benvegnu L, Alberti A. Risk factors and prevention of hepatocellular carcinoma in HCV infection. Digestive Diseases and Sciences. 1996;41(12):49S–55S. doi: 10.1007/BF02087876. [DOI] [PubMed] [Google Scholar]
- 36.Dring MM, Morrison MH, McSharry BP, et al. Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(14):5736–5741. doi: 10.1073/pnas.1016358108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castello G, Scala S, Palmieri G, Curley SA, Izzo F. HCV-related hepatocellular carcinoma: from chronic inflammation to cancer. Clinical Immunology. 2010;134(3):237–250. doi: 10.1016/j.clim.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Prokunina-Olsson L, Muchmore B, Tang W, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nature Genetics. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]


