Abstract
The ability to determine structure-activity relationships (SAR) and identify cellular targets from cell lysates and tissues is of great utility for kinase inhibitor drug discovery. We describe a streamlined mass spectrometry-based chemoproteomics workflow to examine the SAR and target profiles of a small library of kinase inhibitors that consists of the drug dasatinib and a panel of general type II inhibitors. By combining a simplified affinity enrichment and on-bead protein digestion workflow with quantitative proteomics, we achieved sensitive and specific enrichment of target kinases using our small molecule probes. We applied the affinity matrices in competition experiments with soluble probes in HeLa cell lysates using less than 1 mg of protein per experiment. Each pull-down experiment was analyzed in a single nano LC-MS run. Stringent selection criteria for target identification were applied to deduce 28 protein targets for dasatinib and 31 protein targets for our general type II kinase inhibitor in HeLa cell lysate. Additional kinase and protein targets were identified with the general type II inhibitor analogs, with small structural changes leading to divergent target profiles. We observed surprisingly high sequence coverage on some proteins, enabling further analyses of phosphorylation sites for several target kinases without additional sample processing. Our rapid workflow profiled cellular targets for six small molecules within a week, demonstrating that an unbiased proteomics screen of cellular targets yields valuable SAR information and may be incorporated at an early stage in kinase inhibitor development.
Introduction
Protein phosphorylation cascades regulate numerous important cellular processes in mammalian cells. Intra-cellular protein phosphorylation is mediated by protein kinases and phosphatases, which have opposing effects on this post-translational modification1, 2. Protein kinases are a large enzyme family that have emerged as highly attractive drug targets due to their susceptibility to small molecule (SM) inhibition and the roles that dysregulated kinases play in a number of diseases3. Due to the fact that most potent SM inhibitors target the highly conserved ATP-binding site, achieving selectivity for a desired kinase is often challenging4–6. While strides have been made in predicting which kinases are the likely off-targets of an ATP-competitive inhibitor, true kinome selectivity must be empirically determined. Therefore, techniques that allow rapid and comprehensive profiling of the kinome are integral tools for inhibitor development7, 8. Methods interrogating full-length protein kinases in their cellular environment are particularly attractive9, 10.
Affinity enrichment with immobilized SMs on agarose beads has been in use since the 1970s11 and has been successfully applied to identify important cellular targets of bioactive SMs and drugs12. Classical affinity capture methods use grams of protein and rely on multiple biochemical fractionation steps to isolate a single protein target; optimizing each step of the protein purification requires considerable effort. Because numerous fractionation and sample processing steps may be required to purify a protein target to homogeneity, it is challenging to identify low affinity binding partners or associated protein complexes with traditional approaches. Recently, the combination of quantitative proteomics and affinity pull-downs has provided sensitive and specific detection of protein-bait interactions, where the bait molecule is a protein, peptide, nucleic acid, or SM. We described an approach using stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics and affinity matrices to quantitatively measure even small (~20%) changes in protein enrichment by SM probes over control pull-downs, allowing sensitive and specific identification of direct targets and higher-order interactors of SM probes from cell lysates13, 14. We applied our SILAC target ID approach to identify novel targets of immunophilin ligands13, natural products15, and kinase inhibitors13, 16–18. Our SILAC target ID workflow applies mild non-ionic detergents, yielding bona fide targets with one milligram of input protein and allows the identification of lower affinity interactions (Kd ~ 40 µM)13.
Results and Discussion
Bantscheff et. al and others have used affinity matrices of non-selective kinase inhibitors to broadly enrich the kinome, profiling kinases displaced by individual kinase inhibitors with quantitative proteomics9, 19, 20. One of our long term goals is to achieve targeted enrichment of a sub-kinome of interest for in-depth analyses of kinases. Here we describe an approach utilizing a quantitative proteomics-affinity purification workflow to rapidly profile and compare targets of our newly developed affinity reagents in whole cell lysates (Fig. 1). We evaluated the target profiles of reagents derived from dasatinib 1, the general type II inhibitor 2 and several of its derivatives (4–8; Fig. 2). We found that our novel affinity reagents are highly efficient at enriching tyrosine kinases and other kinases, and that there is substantial overlap in the target profiles of dasatinib and our type II inhibitor probes 4–8. The high sequence coverage obtained for the target kinases facilitated the study of post-translational modifications (PTMs), such as phosphorylation. Further, from the quantitative target profiles of several analogs of 2, valuable structure activity relationship (SAR) information regarding type II inhibitor interactions could be deduced.
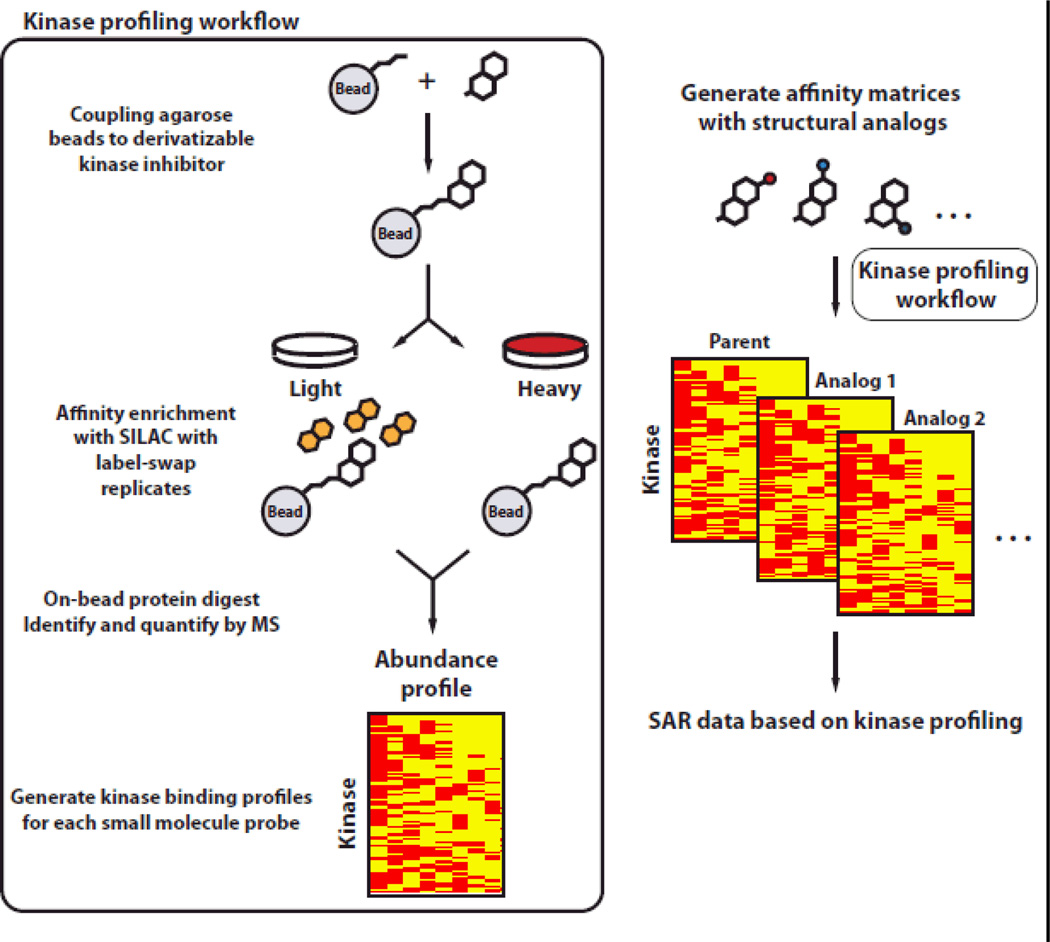
Figure 1.
Schematic of our optimized, rapid, and quantitative chemoproteomic workflow. By evaluating the cellular target profiles of inhibitor analogs with unbiased quantitative proteomics (SILAC), SAR data for the corresponding scaffold can be obtained on a proteome-wide scale.
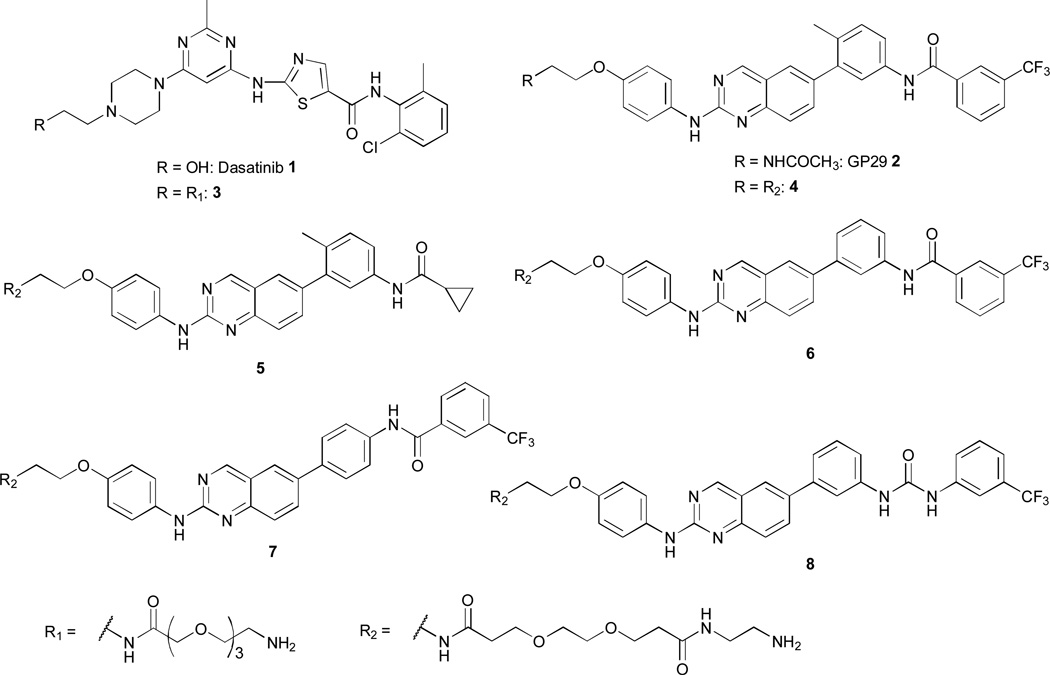
Figure 2.
Inhibitors and affinity reagents used in this study.
To determine the utility of the SILAC target ID workflow for rapidly profiling kinase inhibitor targets, we selected two fairly non-selective ATP-competitive inhibitors, dasatinib 1 and compound 2 (Fig. 2), that have previously been characterized by proteomics methods21–23. Dasatinib 1 is a clinically approved Src/Abl inhibitor that potently inhibits multiple tyrosine kinases. 2 is a type II inhibitor that contains a general pharmacophore for kinases that adopt the DFG-out inactive conformation21, 22, 24. Both inhibitors were derivatized with a flexible amine-containing linker at positions predicted to minimally perturb their interactions with kinase targets to yield affinity reagents 3 and 4 (Fig. 2). These were then coupled to carboxy-functionalized sepharose beads according to our published procedure24, yielding the corresponding affinity resins derived from dasatinib analog 3 and type II probe 4 (for details see ESI).
In order to allow rapid parallel processing of multiple samples, we investigated the digestion of proteins directly on-bead with Lys-C and trypsin in lieu of a SDS-PAGE gel-based separation and in-gel protein digestion procedure. After optimizing affinity capture and wash conditions (see ESI), we found that it was possible to enrich and analyze specific kinases and other proteins with immobilized affinity reagents 3 and 4 using our simplified procedure. Briefly, 250 µl of “light” or “heavy” SILAC-labeled HeLa cell lysate (≤1 mg of protein) was pre-incubated with a competitor-DMSO solution (50 µM competitor final) or DMSO alone (vehicle) for 20 minutes at 4 °C. The treated lysate was then added to 25 µl of a 50% (v/v) affinity bead slurry (~50–80 µM immobilized affinity reagent final, ~1:1 competition) and the mixture was rotated end-over-end for 3 hours at 4 °C. The beads were then washed successively with modified RIPA buffer and Tris-NaCl pH 7.8 to remove non-specific binders and detergent. The samples were re-suspended in 8 M urea, cysteines reduced with tris(2-carboxyethyl)phosphine and capped with 2-chloroacetamide. Peptides were obtained by diluting the sample with 0.1 M TEAB pH 8.5, subsequent treatment with endoproteinase Lys-C and trypsin for 16 hours at 37 °C, followed by acidification and desalting on C18 StageTips25. Peptides were then analyzed by nano-scale LC-MS. A label-swap experiment, where the SILAC state and affinity pull-down condition is reversed, was performed with the same set of lysates. For each affinity probe-competitor pair, we ran two label-swap replicate SILAC experiments, resulting in a total of four replicate pull-down experiments. Because proteins competitively eluted by the soluble SM should have SILAC ratios that invert within a label-swap set, we can easily distinguish contaminants and false positives from true hits. By omitting the gel separation and in-gel digestion processing of samples, this workflow significantly reduces sample processing and the number of LC-MS runs, allowing the rapid and unbiased profiling of kinase inhibitor targets from cell lysates. Thus, using this streamlined procedure, the full process of affinity capture and sample processing (40 pull-downs total) can be completed within a day, by a single operator and LC-MS analysis performed within four days (40 samples).
Next, we wanted to apply our rapid chemoproteomics profiling workflow to investigate the SAR of the general type II inhibitor 2. To this end, a small panel of four structural analogs, 5–8 (Fig. 2), containing the amine linker moiety for immobilization was synthesized (see ESI). In affinity reagent 5, the characteristic m-trifluoromethyl benzamide moiety extending into the DFG-out pocket of inactive kinases was replaced by the smaller cyclopropane carboxamide moiety. Compound 6 contains the same spatial arrangement of binding moieties as inhibitor 2, but is missing the “flag” methyl group on the 3-anilino ring that bridges the quinazoline core and the m-trifluoromethyl benzamide group. A similar “flag” methyl group in imatinib has been shown to contribute to the overall kinase selectivity of this drug26. Probe 7 contains all of the functional groups of probe 6, but we used a 4-anilino linker instead of the 3-anilino arrangement to obtain a more linear geometry. Compound 8 is identical to probe 6, except that this molecule contains a urea linkage between the 3-trifluoromethylphenyl group and the 3-anilino ring rather than an amide, providing more flexibility and additional functionalities for hydrogen bonding interactions. The corresponding affinity matrices of affinity reagents 5–8 were then used to evaluate their target profile in HeLa lysate as described for the dasatinib probe 3 and type II inhibitor 4 (see above).
From the complete dataset of 40 pull-down experiments, we quantified 1511 of the 2196 total proteins identified. Overall, 161 protein and non-protein kinases in various isoforms were identified, and of these, 145 were quantified (see supplementary Excel file 1). We applied stringent criteria to yield a list of candidate cellular targets. First, only protein hits with significant H/L ratios (MaxQuant Significance B27) in at least one of the replicate soluble competition experiments were considered for downstream data processing. Next, p-values according to the Student’s t-Test with a threshold of p < 0.05 were calculated from all available experiments (see also supplementary Excel file 2). An exception was made if, e.g. the protein was quantified with significant ratios in at least three experiments or when one outlying H/L ratio (out of four) caused the p-value to exceed the <0.05 threshold; these hits are noted in all figures and supplemental tables.
The results from the initial affinity-capture experiments with immobilized dasatinib probe 3 and probe 4 are shown in Fig. 3. In total, 24 kinases and 4 non-kinases were found to bind specifically to dasatinib in our assay. Consistent with previous studies, immobilized dasatinib primarily enriches tyrosine kinases, with members from the ephrin receptor- and Src-family kinases being particularly well represented (Fig. 3)9, 28. The overall yield of targets with immobilized dasatinib using our streamlined workflow is comparable to the 31 identified kinases in Kinobeads/iTRAQ experiments with dasatinib competition,9 and target profiling with an immobilized dasatinib probe28 in K562 leukemia cells and a panel of human cancer cell lines29. We identified 17 kinases that interact specifically with dasatinib in our study and that also were identified in the Kinobeads pulldown in K562 cells. However, 10 kinases were uniquely identified in the Kinobead-dasatinib competition experiment (BTK, HCK, KIT, MAP3K4, MAP4K3, MLK3, SIK2/3, SYK and TAO3). Four kinases (DDR1, LIMK2, PKMYT1 and TESK2) found in the Kinobead dataset also showed a tendency for competition with dasatinib in our experiments with immobilized probe 3, but failed to pass the stringent selection criteria for cellular SM targets (ESI supporting Excel file 1). Seven kinases (DDR2, EPHA2/5/7, PTK6 (TK family), ILK (TKL family), and MAP4K5 (STE family)) were found to be specific binders in our experiments, but were not identified in the Kinobead experiment (Fig. 3). Many of the observed differences in these kinase profiles are likely due to variability in the relative abundance of specific kinases in the two cell lysates tested (K562 versus HeLa), but other factors cannot be ruled out. Gratifyingly, all kinases we identified as specific interactors of dasatinib were also identified as targets in the more recent study by Li et al29. Interestingly, we identified the non-kinases YWHAE, HSPA9, and HSPA1A as specific binders of dasatinib that were previously described by Bantscheff et. al. 9. As an additional non-kinase hit, we identified the ADP-sugar pyrophosphatase (NUDT5). ILK and tyrosine kinase Tec (TEC) were found to uniquely interact with dasatinib probe 3 and not with any of our other probes. Additionally, a set of known interactors of ILK, namely LIMS1, RSU1, and PARVA/B were detected with H/L ratios very similar to this kinase (data not shown)29, 30. This suggests that our rapid profiling method accesses multi-protein complexes interacting with the SM, and may therefore provide additional insights in a SM’s mechanism of action.
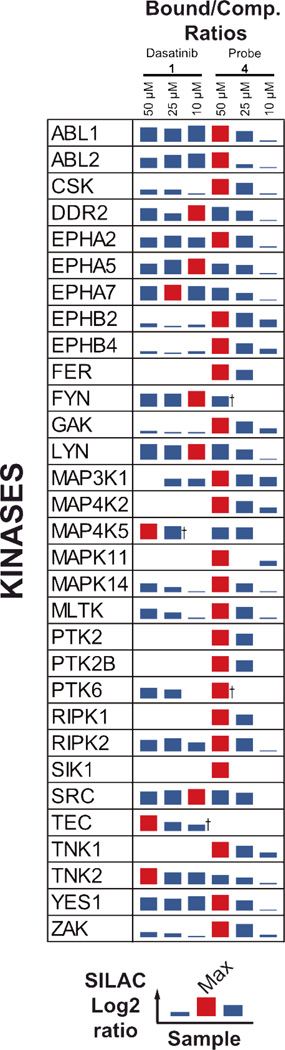
Figure 3.
Target profile of the affinity probes tested. Left panel: Bars represent the average log2 bound/competed ratio of protein hits found in one of the competition experiments. Compounds tested are shown in bold numbers (1, 4–8). Proteins listed were quantified in our experiments with significant H/L-ratios and p-value <0.05 (Student´s t-Test). Right panel: Heat map of the sequence coverage obtained for specific hits found in this study. †: not quantified in one of the four experiments. ‡: Single experimental outlier raises H/L-ratio p-value above the <0.05 threshold.
For probe 4, 29 specific kinase targets and two non-kinase targets were identified, which is in good agreement with our previous proteomics data21, 22. We identified (fps/fes related) tyrosine kinase (FER), MEK kinase 1 (MAP3K1), focal adhesion kinase 1 (FAK/PTK2), and protein tyrosine kinase 2-beta (PTK2B) as targets that bound uniquely to probe 4 (Fig. 3). As mentioned earlier, type II probe 4 shows a bias for specifically binding kinases of the TK and TKL family. Exceptions are p38α/β (CMGC family), MAP3K1, MAP4K2 (STE family), SIK (CAMK family) and GAK. Overall, 17 kinases specifically enriched with probe 4 were detected that were not found in our proteomics profiling studies with closely related probes in A431 cells22 and in Hela cells21 (see ESI Fig. 1 and ESI Table 1). Further, we identified epimerase family protein SDR39U1 (SDR39U1) as a putative non-kinase hit of probe 4 (shared with m-anilino probes 6 and 8). Strikingly, the target profiles of dasatinib probe 3 and 4 are remarkably similar, with 20 kinases, belonging mainly to the TK family, shared between the two compounds (Fig. 3, Fig. 4a). Among the kinases found to bind specifically to the type I inhibitor dasatinib but not type II probe 4 are MAP4K5, MAP2K5 (STE family) and ILK (TKL family). Kinases found to bind specifically to probe 4 but not to dasatinib were MAP3K1, MAP4K2 (STE family), SIK (CAMK family) and RIPK1 (TKL family). These results highlight that it is possible for inhibitors to have very similar selectivity profiles, even if they are based on very different pharmacophores (type I versus type II). We noted that we obtained very high sequence coverage for some kinases in the panel, identifying up to 85% of the protein sequence (e.g. CSK; also see ESI). Other kinases showing high sequence coverage (>70%) for both probe 3 and 4, were LYN and MAPK14. This suggested that the proteins could be efficiently analyzed for PTMs, and indeed a database search for possible phosphorylation sites on serine, threonine, and tyrosine residues revealed that a considerable number of these sites could be identified without further processing of the samples. The 55 phosphosites we identified on protein kinases from our probe-target panel can be found in the supporting information (ESI table 2).
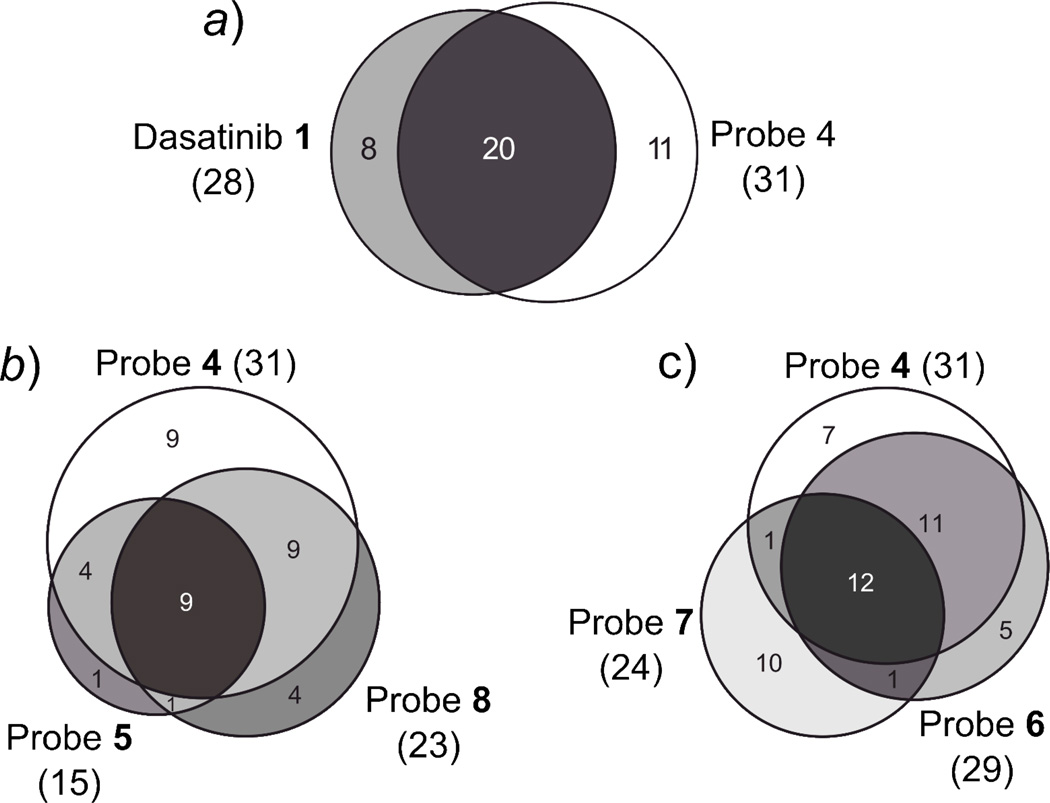
Figure 4.
Venn diagrams showing the overlap of target profiles of the compounds tested. a) Protein target overlap of dasatinib 1 and probe 4. b) Protein target overlap of probe 4, the corresponding m-urea analog 8, and cyclopropane carboxamide 5. c) Protein target overlap of probe 4, the p-amide analog 7, and m-amide 6.
To further prove that the significant H/L ratios observed for the competition experiments with dasatinib 1 and type II probe 4 correspond to a specific interaction with a kinase target, a series of titration experiments were conducted (Fig. 5). Pull-downs with probe matrices 3 and 4 were performed with soluble competitor (1 and 4, respectively) at concentrations of 50 µM, 25 µM and 10 µM (quadruplicate experiments each). For probe 4, a clear dose response of the H/L ratios could be observed for most of the kinase targets, with values falling continuously with lower competitor concentrations (Fig. 5). When titrating probe matrix 3 with dasatinib 1 as the competitor, a dose response could be observed only for TEC, MAPK14 and MLTK. This was most likely due to the higher affinity of dasatinib 1 for its targets (pM for some kinases) as compared to the conjugated probe 3, likely leading to effectively complete competition of the majority of kinase targets at the concentrations tested.
Figure 5.
Titration of the log2 H/L-ratios of kinases specifically targeted by dasatinib 1 and general type II probe 4. Competitor concentration are varied from 10, 25 to 50 µM. Log2 H/L ratios of the probe 4 targets mostly show a graded dose response, whereas only a few dasatinib targets show a graded dose response. †: not quantified in one of the quadruplicate experiments.
The target profiles for our newly developed type II inhibitor analogs 5–8 (Fig. 3) were obtained by the same competition experiment used for profiling probe 4 and dasatinib 1, i.e. by competitively eluting targets from the probe-matrix with a soluble probe. The Venn diagrams shown in Fig. 4b and 4c indicate a high degree of overlap of targets between the parent probe 4 and its analogs, as well as among themselves (5–8, Fig. 2), although we observed that small structural differences led to distinct interactions with subsets of the kinome. Somewhat surprisingly, the parent compound, probe 4, interacts with the largest number (29) of kinases. In contrast to imatinib, removing the “flag” methyl group from this scaffold26, as in 6, did not lead to a more promiscuous compound. Probe 6 specifically interacts with 25 kinase targets, 21 of which are also targets of probe 4. Only two kinases, discoidin domain-containing receptor 1 (DDR1) and Janus kinase 1 (JAK1) are unique for probe 6. Removal of the “flag” methyl in 6 abolished interactions with tyrosine-protein kinases CSK and FER, PTK2, PTK2B, serine/threonine-protein kinase SIK1, activated CDC42 kinase 1 (TNK2), MAP3K1 and MAP4K2, but facilitated interactions with MAPK9 (JNK2) and MAP2K5. With probe 6, we observed high sequence coverage (>70%) for Receptor-interacting serine/threonine-protein kinase 2 (RIPK2) and tyrosine-protein kinase Yes (YES1), suggesting a highly specific enrichment of these kinases. Replacing the amide linkage of probe 6 with a more flexible urea (probe 8) has only a modest effect on kinase selectivity. 22 kinases are specific targets of type II inhibitor analog 8, with only one of these, LIM domain kinase 1 (LIMK1), being unique to this affinity reagent. Transition to the urea linkage abrogates interaction with the tyrosine kinases FYN, SRC, YES1, EPHA2, DDR1, PTK6 and JAK1. Thus, the nature of the hydrogen bond donor-acceptor in the linker connecting the adenine mimic and the moiety occupying the DFG-out pocket has a small effect on kinase selectivity. Also, the sequence coverage profile of probe 8 did not differ significantly from that of probes 4 and 6. For probe 5, in which the m-trifluoromethyl benzamide moiety found in probe 4, 6, and 7 was replaced with a cyclopropane carboxamide moiety, a significant drop in the number of specific interactors (15) and a reduction in average log2 H/L ratios was observed (Fig. 3). 13 kinases that specifically interacted with parent compound 4 were also targets of probe 5. However, introduction of the smaller cyclopropane carboxamide moiety seems to abrogate interaction with 17 kinases, mainly TKs, which specifically interact with probe 4 and not 5 (Fig. 3, see also ESI table 3). Whether this effect stems from the inability of probe 5 to interact with the DFG-out pocket is difficult to judge from our proteomic dataset alone, since binding kinetics might influence the outcome of our experiments. Interestingly, probe 7, in which the m-trifluoromethyl benzamide moiety was moved to the p-position of the anilino linker (Fig. 2), shows relatively low overlap with the target panel of the parent compound 4 (13 proteins; Fig. 3). A total of 10 kinases and non-kinases were found to bind uniquely to probe 7. These kinases are aurora kinases A and B (AURKA/B), calcium/calmodulin-dependent protein kinase kinase 1 (CAMKK1), protein-tyrosine kinase HCK (HCK), MAP4K3, hepatocyte growth factor receptor (MET), serine/threonine-protein kinase PAK2 (PAK2) and the lipid kinase phosphatidylinositol 5-phosphate 4-kinase type-2 gamma (PIP4K2C). Overall, shifting the trifluoromethyl benzamide moiety from the m- to the p-position eliminates the interaction with 17 kinases found to bind to parent compound 4 (see ESI table 3). Especially high sequence coverage (>70%) was obtained for AURKA. A unique non-kinase target, and the protein with the highest average log2 H/L ratio, of probe 7 is all-trans-retinol 13,14-reductase (RETSAT). Thus, small structural changes can affect interactions with non-kinase targets in addition to kinase targets.
In summary, we developed an improved, rapid, and very simple quantitative proteomics workflow to profile targets of immobilized SM kinase inhibitors in native proteomes. The methodology relies on the well-established SILAC approach for quantitative proteomics and an optimized on-bead digest protocol for sample processing. Our pull-down experimental protocol requires less than 1 mg of protein and is analyzed in a single nanoLC-MS run. The identification of targets from label-swap SILAC data was straightforward and false positives were easily eliminated. We profiled affinity probes derived from the clinically approved kinase inhibitor dasatinib 1, as well as the promiscuous type II inhibitor 2 and four of its analogs (5–8; Fig. 2), identifying both previously described and novel targets of these inhibitors. Profiling of six different affinity matrices in 40 distinct experiments was achievable in four days by a single researcher. The quick turnaround and high sensitivity and specificity of our data demonstrate that our rapid chemoproteomics profiling approach to study SAR of SM probes is practical and easily achievable. Incorporating target profiling early in kinase inhibitor development, therefore, has the potential to provide both valuable insight and new leads.
Supplementary Material
Acknowledgements
We wish to thank Dr. Ho-Tak Lau and Danny Suh for contributions to the manuscript and helpful discussions. M. G. was supported by a research fellowship of the Deutsche Forschungsgemeinschaft (DFG). Research reported in this publication was supported in part by the National Institute of General Medical Sciences, National Cancer Institute, and National Institute on Drug Abuse under award numbers R01GM086858 (D.J.M), R21CA177402 (S.E.O and D.J.M), and 5P30DA028846 (S.E.O). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Blume-Jensen P, Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. Nat. Rev. Drug. Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Yang PL, Gray NS. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 5.Noble ME, Endicott JA, Johnson LN. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurty R, Maly DJ. Comb. Chem. High. Throughput. Screen. 2007;10:652–666. doi: 10.2174/138620707782507368. [DOI] [PubMed] [Google Scholar]
- 7.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 8.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Nat. Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 9.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 10.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, Nordin B, Weissig H, Yang Q, Lee JD, Gray NS, Kozarich JW. Chem. Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuatrecasas P, Wilchek M, Anfinsen CB. Proc. Natl. Acad. Sci. U. S. A. 1968;61:636–643. doi: 10.1073/pnas.61.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harding MW, Galat A, Uehling DE, Schreiber SL. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 13.Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, Koehler AN, Marcaurelle LA, Golub TR, Gould RJ, Schreiber SL, Carr SA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margolin AA, Ong SE, Schenone M, Gould R, Schreiber SL, Carr SA, Golub TR. PLoS One. 2009;4:e7454. doi: 10.1371/journal.pone.0007454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, Stern AM, Mandinova A, Schreiber SL, Lee SW. Nature. 2011;475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Kuai L, Ong SE, Madison JM, Wang X, Duvall JR, Lewis TA, Luce CJ, Conner SD, Pearlman DA, Wood JL, Schreiber SL, Carr SA, Scolnick EM, Haggarty SJ. Chem. Biol. 2011;18:891–906. doi: 10.1016/j.chembiol.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong SE, Li X, Schenone M, Schreiber SL, Carr SA. Methods Mol. Biol. 2012;803:129–140. doi: 10.1007/978-1-61779-364-6_9. [DOI] [PubMed] [Google Scholar]
- 18.Wen Q, Goldenson B, Silver SJ, Schenone M, Dancik V, Huang Z, Wang LZ, Lewis TA, An WF, Li X, Bray MA, Thiollier C, Diebold L, Gilles L, Vokes MS, Moore CB, Bliss-Moreau M, Verplank L, Tolliday NJ, Mishra R, Vemula S, Shi J, Wei L, Kapur R, Lopez CK, Gerby B, Ballerini P, Pflumio F, Gilliland DG, Goldberg L, Birger Y, Izraeli S, Gamis AS, Smith FO, Woods WG, Taub J, Scherer CA, Bradner JE, Goh BC, Mercher T, Carpenter AE, Gould RJ, Clemons PA, Carr SA, Root DE, Schreiber SL, Stern AM, Crispino JD. Cell. 2012;150:575–589. doi: 10.1016/j.cell.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Mol. Cell. Proteomics. 2009;8:1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brigham JL, Perera BG, Maly DJ. ACS Chem Biol. 2013;8:691–699. doi: 10.1021/cb300623a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranjitkar P, Perera BG, Swaney DL, Hari SB, Larson ET, Krishnamurty R, Merritt EA, Villen J, Maly DJ. J. Am. Chem. Soc. 2012;134:19017–19025. doi: 10.1021/ja306035v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnamurty R, Brigham JL, Leonard SE, Ranjitkar P, Larson ET, Dale EJ, Merritt EA, Maly DJ. Nat. Chem. Biol. 2013;9:43–50. doi: 10.1038/nchembio.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranjitkar P, Brock AM, Maly DJ. Chem. Biol. 2010;17:195–206. doi: 10.1016/j.chembiol.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rappsilber J, Mann M, Ishihama Y. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann J, Buchdunger E, Mett H, Meyer T, Lydon NB, Traxler P. Bioorg. Med. Chem. Lett. 1996;6:1221–1226. [Google Scholar]
- 27.Cox J, Mann M. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 28.Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Kocher T, Superti-Furga G. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, Bennett KL, Gao J, Song L, Eschrich S, Superti-Furga G, Koomen J, Haura EB. Nat. Chem. Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legate KR, Montanez E, Kudlacek O, Fassler R. Nat. Rev. Mol. Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.