Abstract
Oncogenic KRAS mutation is the signature genetic event in the progression and growth of pancreatic ductal adenocarcinoma (PDAC), an almost universally fatal disease. Although it has been appreciated for some time that nearly 95% of PDAC harbor mutationally activated KRAS, to date no effective treatments that target this mutant protein have reached the clinic. A number of studies have shown that oncogenic KRAS plays a central role in controlling tumor metabolism by orchestrating multiple metabolic changes including stimulation of glucose uptake, differential channeling of glucose intermediates, reprogrammed glutamine metabolism, increased autophagy, and macropinocytosis. Here we review these recent findings and address how they may be applied to develop new PDAC treatments.
Keywords: autophagy, glutaminolysis, glycolysis, macropinocytosis, metabolism
Mutant KRAS drives PDAC development
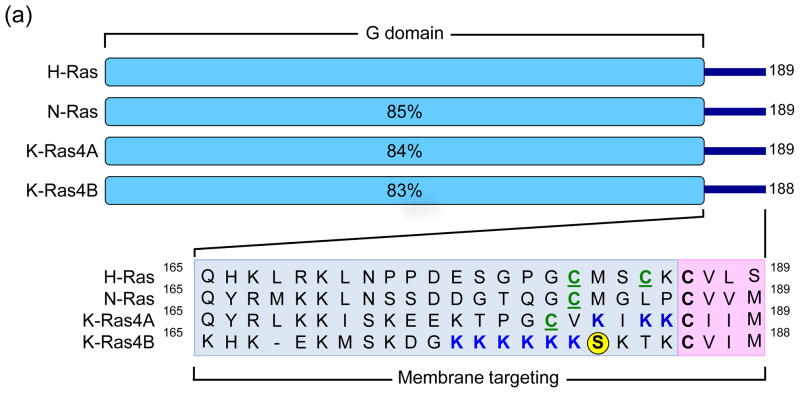
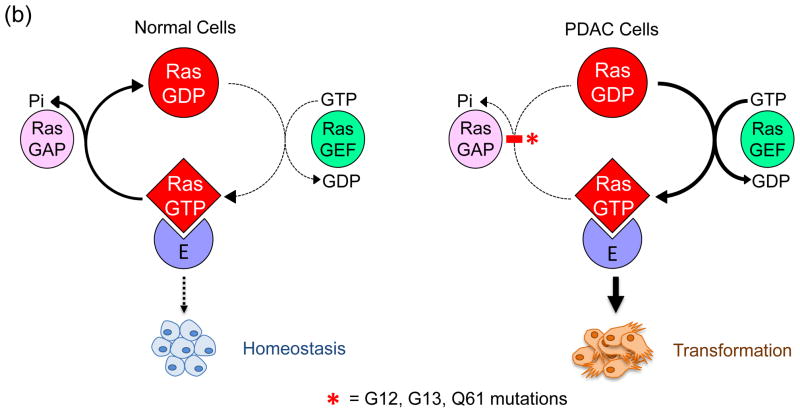
The recognition over thirty years ago that mutationally activated RAS genes are present in human cancer cells led to intense efforts to understand the Ras protein structure, biochemistry, and biology; studies that arguably shaped our understanding of the molecular biology of cancer today [1,2]. The three human RAS genes (HRAS, NRAS, and KRAS) encode four highly homologous (83–90% sequence identity) ~21 kDa small GTPases (Figure 1a), meaning that they cycle between a GTP-bound “on” state and a GDP-bound “off” state (Figure 1b). This cycle is mediated by two other classes of proteins: guanine nucleotide exchange factors (RasGEFs; e.g., Sos1), which promote activation of Ras by aiding in the exchange of GDP for GTP, and GTPase-activating proteins (RasGAPs; e.g., neurofibromin), which drive Ras-mediated GTP hydrolysis and hence inactivation. Aside from a few rare exceptions, mutationally activated Ras proteins found in human cancers result predominantly from one of three single point mutations at residues G12, G13 or Q61. Oncogenic substitution of G12 or G13 leads to constitutive activation of Ras by creating steric hindrance that prevents the formation of van der Waals interactions between Ras and RasGAPS [3], whereas substitutions of Q61 interfere with the coordination of a water molecule necessary for GTP-hydrolysis [4]. Constitutive activation of Ras results in the persistent stimulation of its multitude of downstream signaling partners, which results in many of the phenotypic hallmarks of cancer including increased proliferation, the suppression of apoptosis, altered metabolism, alteration of the tumor microenvironment, evasion of the immune response, and metastasis [5].
Figure 1.
Human Ras proteins are small GTPases. (a) The human RAS genes encode 188 or 189 amino acid proteins that share strong (82–90% overall) amino acid identity; percentages indicate identity with H-Ras. KRAS encodes two related proteins (K-Ras4A or K-Ras4B; 90% identity) due to alternative exon four utilization, with KRAS4B the predominant transcript in pancreatic tissue. Residues 1–164 comprise the G domain that binds and hydrolyses GTP (93–99% sequence identity). The remaining 24/25 C-terminal residues (shown in inset) comprise the membrane targeting sequence (16–40% identity), where the C-terminal four residues comprise the CAAX motif (shaded in pink; C = cysteine, A = aliphatic; X = terminal amino acid) that signals for farnesyltransferase-catalyzed covalent addition of a C15 farnesyl group to the cysteine residue. The 20/21 amino acids upstream of the CAAX motif comprise the hypervariable (HV) region (shaded in blue) where the Ras proteins exhibit the greatest sequence divergence. Within the HV domain are additional membrane targeting sequence elements that include cysteines that are covalently modified by addition of a palmitate fatty acid (green, underlined ‘C’) or polybasic sequences that promote association with the membrane (blue K). K-Ras4B contains a serine (S181) that is reversibly phosphorylated (yellow circle), regulating localization between the plasma and endomembranes. (b) Mutant K-Ras is persistently GTP-bound and active. Wild type K-Ras cycles between an active GTP-bound and an inactive GDP-bound state. In normal quiescent cells, K-Ras is predominantly GDP-bound. Upon growth factor stimulation, RasGEF activation promotes transient formation of K-Ras-GTP, promoting its binding to downstream effectors (E; e.g., Raf, PI3K). The cycle is terminated by the action of RasGAPs, returning K-Ras to the inactive GDP-bound state. Single amino acid substitutions at G12, G13 or Q61 impair the intrinsic GTP binding and hydrolytic activity of K-Ras, and additionally render the protein insensitive to GAP stimulation, favoring accumulation of persistently GTP-bound and active K-Ras in PDAC. Arrow line thickness corresponds to the level of signaling, i.e. in normal cells Ras is predominantly in the GDP-bound state due to relatively higher GAP activity and thus signals less to downstream effectors.
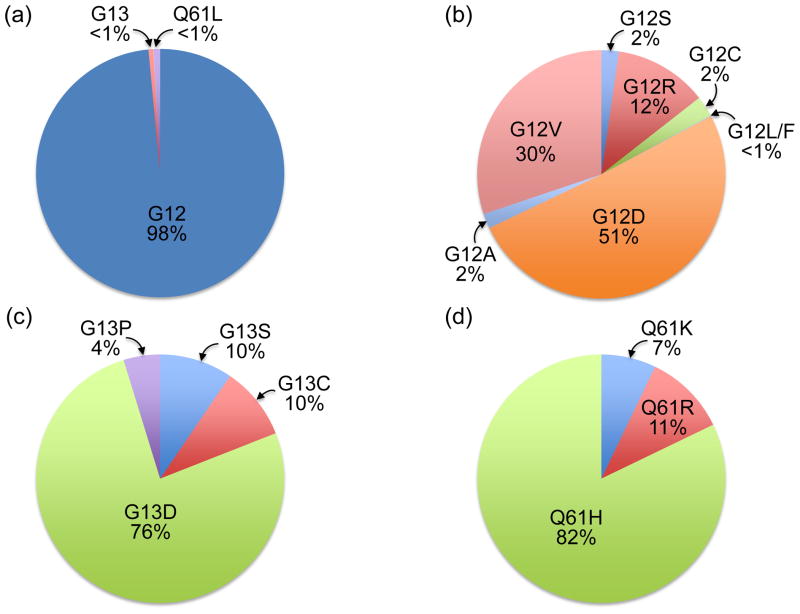
Although it is evident that the different RAS isoforms are unequal in terms of their role in human cancer, the mechanisms governing this observation are not understood. There is nearly exclusive mutation of KRAS in pancreatic, colon, and lung cancer; whereas HRAS mutations are associated with skin (keratinocyte), head and neck, and bladder tumors; and NRAS mutations are present in haematopoietic and skin (melanocyte) malignancies [2]. In terms of the distribution of missense RAS mutations across the entire cancer genomic landscape, mutation of KRAS occurs most frequently, accounting for 85% of RAS mutations, followed by NRAS (12%), with HRAS mutations relatively rare (3%) [1]. Although oncogenic KRAS was first associated with PDAC decades ago, recent exome sequencing established that KRAS is the most frequently mutated gene in PDAC (95%), the predominant form of pancreatic cancer (~90%) [6,9]. The frequency and specific substitutions show cancer type differences, with 98% of KRAS mutations in PDAC occurring at position G12 (Figure 2). Of the eight different substitutions found at this position, the predominant substitution is G12D. In PDAC, KRAS mutation is an early and initiating event as it has been shown that over 90% of low-grade pancreatic intraepithelial neoplasia (PanIN) lesions (Figure 3) harbor oncogenic KRAS mutations [7]. PanINs are graded from stage I to III, and along this continuum they display increasing disorganization and nuclear abnormalities, with high-grade PanINs ultimately transforming into true PDAC. Molecular profiling studies have reinforced the PanIN-to-PDAC progression by demonstrating that KRAS mutation is followed by subsequent inactivation of the tumor suppressor gene CDKN2A (or INK4A/ARF, which encodes p14/Arf and p16/INK4A), followed by inactivation of two other tumor suppressor genes TP53 and SMAD4 (Figure 3) [8]. Comprehensive genetic analysis has revealed that PDAC is an extremely complex and heterogeneous disease [6,9], and throughout the progressive accumulation of mutations, KRAS continues to drive PDAC development by contributing to tumor maintenance [10–12]. Furthermore, gene expression analysis has revealed the presence of at least three different subtypes of PDAC, which exhibit distinct differences in many important aspects of cancer cell biology including cell morphology, gene expression, and in vitro response to therapy [13].
Figure 2.
KRAS mutations in PDAC. (a) Missense mutations result in single amino acid substitutions primarily at G12 (98%), and at lower frequencies at G13 (21%) or Q61 (28%). (b) At G12, eight different amino acid substitutions have been identified, with G12D the predominant mutation (51%). (c) At G13, four different mutations have been described, with the majority G13D. (d) Mutation at Q61 also results in constitutive activation, and among the three mutations described, Q61H occurs most frequently. There is increasing evidence that the different activating mutations may not have the same biochemical and biological consequences. Data were compiled from COSMIC (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/).
Figure 3.
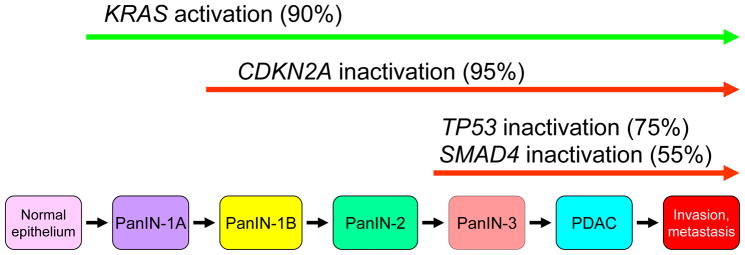
Histologic and genetic progression of pancreatic ductal adenocarcinoma. Pancreatic intraepithelial neoplasias (PanINs), small microscopic abnormal duct structures, are believed to be precursors to invasive pancreatic cancer. While the true precursor cell for PDAC development remains to be fully elucidated, mouse model studies where targeted KRAS activation in acinar cells causes PanIN lesion development suggest that this is the cell of origin for PDAC. The four predominant gene mutations appear to occur in a temporal fashion with PanIN progression. KRAS activating mutations can be found in normal pancreas and PanIN-1. CDKN2A inactivating mutations appear to occur early, in PanINs with low- to intermediate-grade dysplasia, whereas TP53 and SMAD4 inactivating mutations appear to be late events, and occur in PanINs with high-grade dysplasia and in invasive cancer. CDKN2A encodes p14/Arf and p16/Ink4a, an inhibitor of CDK4/6 and G1 cell cycle progression. TP53 encodes the p53 tumor suppressor, a regulator of the cell cycle that is responsible for maintaining cellular and genetic stability. SMAD4 encodes Smad4, a downstream component of the TGF-β signaling network. Oncogenes are indicated with green arrows and tumor suppressors with red arrows.
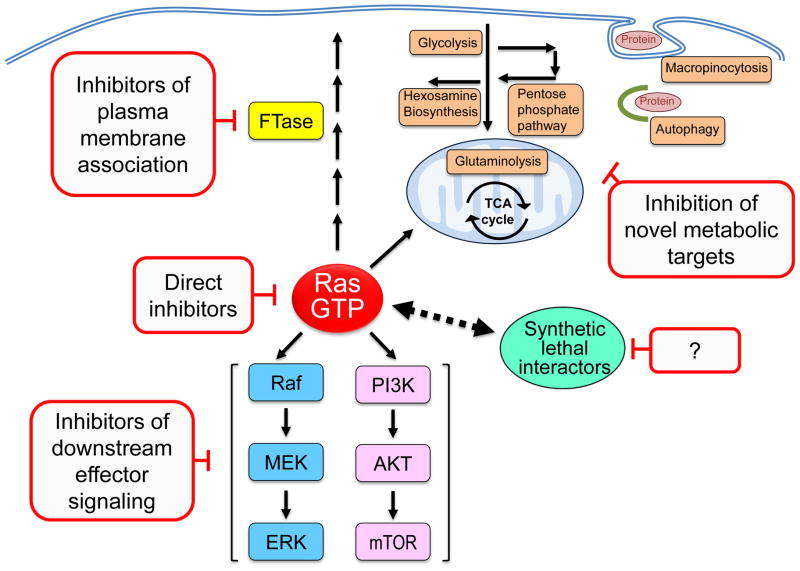
The considerable experimental evidence for the role of mutant KRAS as a driver of cancer development and growth has prompted an intensive effort to identify pharmacologic approaches to block aberrant Ras function for cancer treatment (Figure 4). However, to date, these strategies have not transitioned any effective anti-Ras strategies into the clinic. The development of potent and selective direct inhibitors of Ras have not been successful, leading to a widely held perception that Ras is “undruggable”, although recent studies argue that this approach may still hold promise [14–16]. The intensive efforts in the 1990s to develop inhibitors of Ras posttranslational modifications that promote the membrane association and subcellular localization critical for Ras to drive cancer growth led to the development of farnesyltransferase (FTase) inhibitors (FTIs) (Figure 4). Despite promising preclinical results, clinical trials with FTIs were disappointing as, although FTIs blocked the association of H-Ras with the plasma membrane, they lacked antitumor activity in cancers involving mutant K-Ras and N-Ras due to the unforeseen compensatory activity of the FTase-related enzyme geranylgeranyltransferase-I, which modifies RAS with a geranylgeranyl, rather than a farnesyl group. However, recent approaches to disrupt K-Ras membrane association continue to be pursued. Of particular interest has been the recent development of a small molecule inhibitor of PDEδ, a protein that binds to Ras and regulates its trafficking between membrane compartments [17]. The efficacy of an inhibitor of PDEδ-Ras interaction would not be bypassed by alternative prenylation mechanisms like FTIs; however, it is to soon to anticipate the unexpected consequences of the inhibition of Ras regulation by PDEδ. The identification of synthetic lethal partners of mutant KRAS generated considerable excitement for novel anti-Ras strategies that was then quickly dampened by concerns about the robustness of some of the identified hits [18,19]. Despite these setbacks, there is renewed interest and efforts in the search for anti-Ras drugs.
Figure 4.
Anti-Ras drug discovery. Past and ongoing efforts to develop anti-K-Ras inhibitors have included inhibitors of membrane association and downstream effector signaling. Inhibitors of FTases advanced to Phase III clinical evaluation but did not show significant anti-tumor activity for pancreatic cancer. Currently, there are 22 inhibitors of Raf, MEK and/or ERK under clinical evaluation (clinicaltrials.gov). Similarly, 43 inhibitors of the class I PI3K lipid kinases and their downstream targets, the AKT and mTOR protein kinases, are currently under clinical evaluation. Only recently have small molecules that directly bind to K-Ras and cause perturbations in function been described and characterized in cell culture models. Unbiased genetic functional RNAi screens have identified genes (synthetic lethal interactors) whose functions are essential for the growth of KRAS mutant but not wild type cells. The dashed two-headed arrow indicates a functional relationship between KRAS and the interactor that is typically not mediated through a clear signaling mechanism that connects the two components.
A new direction for anti-Ras efforts is suggested by recent studies linking Ras with altered cellular metabolism. Recently, both mouse genetic and in vitro experiments have implicated KRAS, TP53, MYC, and LKB1 not only in increased PDAC proliferation, but also in the alteration of glucose and glutamine utilization and metabolism [20]. Herein, we will specifically review papers describing oncogenic KRAS-driven metabolic variations that contribute to PDAC progression and indicate the potential ways these pathways could be targeted therapeutically.
The metabolic requirements of proliferation
Aberrant metabolism is now considered one of the hallmarks of cancer [21]. In normal, non-proliferating cells in the presence of oxygen, glucose is metabolized to carbon dioxide by the oxidation of pyruvate (the product of glycolysis) via the tricarboxylic acid (TCA) cycle in mitochondria. The NADH (nicotinamide adenine dinucleotide (NAD+), reduced) produced by the TCA cycle fuels oxidative phosphorylation, which produces a maximal amount of ATP per glucose molecule with minimal lactate production. The study of cancer cell metabolism has a rich history beginning in the 1920’s with the observation by Otto Warburg and colleagues that under aerobic conditions, tumor tissues metabolize approximately tenfold more glucose to lactate than their normal cell counterparts [22]. This observation was originally misinterpreted as evidence that cancer cells have defective mitochondria and impaired aerobic respiration, and are thus dependent on glycolytic metabolism; however, subsequent work has shown that mitochondrial function is not impaired in most cancer cells [23]. Although an explanation of the advantages of increased glycolysis in cancer cells remains controversial [24], a prevalent hypothesis is that while inefficient in terms of net ATP production, aerobic glycolysis supplies rapidly proliferating cells with high levels of glycolytic intermediates that can be incorporated into the biomass required for the generation of new cells [25]. This hypothesis is supported by the observation that several signaling molecules that have traditionally been implicated in cell proliferation, in particular K-Ras, also regulate metabolic pathways.
Oncogenic K-Ras alters glucose uptake and utilization
Constitutive K-Ras-mediated signaling drives the uncontrolled proliferation of PDAC. In order to proliferate, cancer cells must have sufficient energy and biosynthetic building blocks. Previous studies have suggested that the RAS oncogene can promote upregulated glycolysis [26–29]. KRAS signaling promotes glucose uptake by increasing glucose transporter GLUT1 expression, which in turn speeds up glycolytic activity and increases lactate production [30,31]. This metabolic alteration provides a distinct survival advantage as it has been shown that cell lines harboring oncogenic KRAS were able to survive long-term culture in a low glucose environment [31].
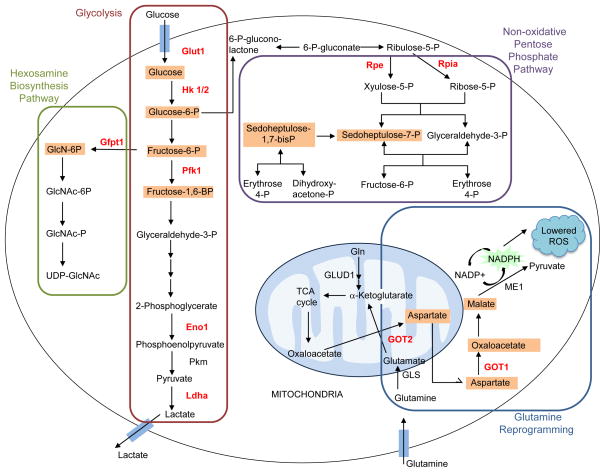
PDAC cells have been shown to have altered metabolism that is consistent with increased aerobic glycolysis [32]. Recently, it was demonstrated that mutant KRAS enhances expression of GLUT1 and several other genes encoding rate-limiting glycolytic enzymes, including hexokinase 1 and 2 (Hk1, Hk2), and phosphofructokinase-1 (Pfkl), as well as lactate dehydrogenase A (Ldha), the enzyme responsible for converting pyruvate to lactate (Figure 5) [11]. Together, this pattern of upregulation leads to an increased glycolytic flux. Additionally, through the upregulation of other rate-limiting enzymes, mutant K-Ras leads to a shunting of glycolytic intermediates into anabolic pathways including the hexosamine biosynthesis pathway (HBP) and the non-oxidative arm of the pentose phosphate pathway (PPP) (Figure 5) [11]. The non-oxidative arm of the PPP can generate the ribose 5-phosphate necessary for nucleic acid biosynthesis thus fueling proliferation. The HBP has been associated with posttranslational protein modification by glycosylation and the synthesis of glycolipids, proteoglycans, and glycosylphosphatidylinositiol anchors. The end product of the HBP, UDP-N-acetylglucosamine (UDP-GlcNAc), is the donor sugar nucleotide used by O-GlcNAc transferase (OGT) in the post-translational modification, O-GlcNAcylation. Hyper-O-GlcNAcylation has been observed in PDAC and this modification appears to aid in the avoidance of apoptosis through NF-κB activation [33]. Likewise, it has been demonstrated in lung cancer lines that Pfk1, a key glycolytic enzyme, was inactivated due to O-GlcNAcylation, resulting in the redirection of glycolytic flux through the PPP [34].
Figure 5.
Glucose and glutamine metabolism is altered by oncogenic K-Ras. Oncogenic K-Ras directs glucose metabolism into biosynthetic pathways in PDAC by upregulating many key enzymes in glycolysis. Oncogenic K-Ras induces nonoxidative pentose phosphate pathway (PPP) flux to fuel increased nucleic acid biosynthesis and activates the hexosamine biosynthesis and glycosylation pathways. PDAC cells also utilize a non-canonical pathway to process glutamine, through which it is used to maintain redox and support growth. Enzymes that show increased activity under the control of oncogenic K-Ras are shown in red, and metabolites that are increased are boxed in orange. Glut1, glucose transporter 1; Hk 1/2, hexokinase 1/2; Pfk1, phosphofructokinase 1; Eno1, enolase 1; Pkm, pyruvate kinase; Ldha, lactate dehydrogenase A; Gfpt1, glucosamine-fructose-6-phosphate aminotransferase-1; GlcN, glucosamine; GlcNAc, N-acetylglucosamine; Rpe, ribulose-5-phosphate-3-epimerase; Rpia, ribulose-5-phosphate isomerase; GLUD1, glutamate dehydrogenase 1; GOT, aspartate transaminase; GLS, glutaminase; ME1, malic enzyme.
PDAC exhibits characteristically low vascular density and a dense stromal surrounding that, in addition to hindering the delivery of therapeutics and contributing to overall PDAC aggressiveness [35], aids in the formation of numerous hypoxic regions [35,36]. Hypoxia has been shown to alter glycolytic flux in PDAC by increasing the transcription of HBP pathway genes [37]. The presence of a transcriptional link between the hypoxic microenvironment and PDAC metabolic signaling remains unclear. The hypoxia-dependent regulation of metabolism is a principal and elemental function of the hypoxia-inducible factor 1 alpha (HIF-1α) transcriptional activator [38]. Although HIF-1α has been shown to mediate GLUT1 expression in HRAS-transformed fibroblasts [39] and Ras-dependent glycolytic upregulation in glioblastoma [40], recent studies contend that KRAS-mediated glucose metabolic reprogramming is not dependent on HIF-1α [11,31]. In PDAC, it appears that downstream of oncogenic K-Ras the ERK mitogen-activated protein kinase (MAPK) pathway is a major driver of glucose metabolism adjustments, likely mediated transcriptionally by the Myc transcription factor [11].
Glutamine metabolism is reprogrammed by oncogenic K-Ras to maintain redox balance
In addition to altered levels of glycolysis, cancer cells also display an increased dependence on glutamine to power anabolic processes [41]. This was first appreciated in 1955, when it was observed that proliferating cells in vitro consumed 10-fold more glutamine than any other amino acid [42]. Glutamine contributes to macromolecular synthesis in cancer cells by providing carbon to fuel the TCA cycle and nitrogen for nucleotide, nonessential amino acid (NEAA), and hexosamine biosynthesis [43,44]. Glutamine can also provide a proliferating cell with a significant amount of NADPH through the activity of malic enzyme (ME1) which oxidatively decarboxylates malic acid, producing carbon dioxide, NADPH and pyruvate [45]. The generation and maintenance of constant intracellular NADPH levels is essentially important as NADPH reduces glutathione to maintain the redox state of the cell.
Processing of the glutamine-derived carbon skeleton in the form of glutamate begins in the mitochondria either through glutamate dehydrogenase (GLUD1) or transaminases. The classical GLUD1-mediated pathway has recently been shown to stimulate cancer proliferation downstream of mTOR [46], and mediate glutaminolysis in oncogenic KRAS-transformed fibroblasts [30]. However, it appears that in the context of PDAC, oncogenic K-Ras expression directs the metabolism of glutamine through a different pathway. A recent report showed that KRAS mutant PDAC uses a unique form of glutamine metabolism to regulate redox balance [47]. They demonstrated that in PDAC, glutamine-derived aspartate is transported into the cytoplasm where aspartate transaminase (GOT1) converts it to oxaloacetate, which is then converted to malate, and then to pyruvate. The conversion of malate to pyruvate by ME1 results in an increased NADPH/NADP+ ratio in the cell, which maintains the cellular redox state (Figure 5). Proteomic analysis supports this unique pattern of protein upregulation in PDAC [35], and mutant K-Ras drives this metabolic reprogramming via increasing GOT1 and decreasing GLUD1 gene expression [47], resulting in increased flux through the GOT1-dependent pathway.
In addition to NADPH production via glutamine metabolism, there are a number of other mechanisms whereby PDAC can maintain redox homeostasis [45,48]. Indeed, a number of studies have suggested that PDAC cells prefer low intracellular ROS levels [49]. K-Ras(G12D) expression was shown to increase the transcription of NRF2, which through stable elevation of the basal Nrf2 antioxidant program, activates a ROS-detoxification program to confer a more reduced intracellular environment [50]. The observation that oncogenic KRAS-driven PDAC requires low ROS for optimal growth is surprising, as increased ROS stress in cancer cells has long been viewed as an event associated with progression [51,52]. This apparent discrepancy could be rationalized based on the dual nature of ROS, where low levels of ROS may be growth promoting but high levels are cytotoxic [53].
K-Ras-driven PDAC growth requires autophagy
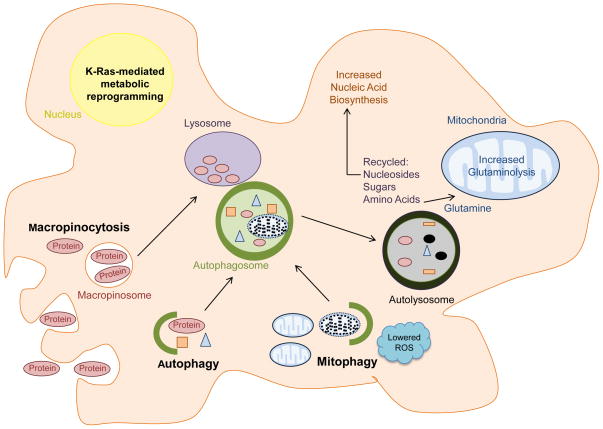
Autophagy is a highly conserved mechanism to degrade intracellular components and promote programmed survival of cells under metabolic stress by providing energy in the form of ATP and building blocks such as amino acids, lipids, sugars and nucleosides [54]. The molecular machinery that controls autophagy is complex with over 30 different autophagy-related genes (ATGs) identified in yeast, of which many are functionally conserved in higher eukaryotes [55]. Briefly, autophagy is controlled by multiple signaling components including those that interpret the cellular energy level (AMPK) and nutrients/amino acid levels (mTOR), as well as growth factors. These pathways converge on a conserved autophagy machinery including the ULK1 kinase complex, a complex between class III phosphoinositide 3-kinase (PI3KC3) and Beclin-1 [56] which nucleates phagophore formation, and a ubiquitin-like conjugation system (including ATG5) that is necessary for autophagosome maturation [57]. Upon autophagy activation, an incipient phagophore engulfs cytoplasmic components, fuses to form a double-membrane autophagosome, and subsequently fuses with the lysosome leading to content degradation within an autolysosomal organelle (Figure 6). The products generated by lysosomal degradation are then released into the cytoplasm where they can be reused.
Figure 6.
KRAS-driven transcriptional reprogramming contributes to the utilization of autophagy and macropinocytosis to meet the metabolic needs of PDAC cells. Macropinocytosis is utilized to transport extracellular protein into the cell. Following degradation, the internalized protein yields amino acids such as glutamine, which can enter the mitochondria to fuel central carbon metabolism. Autophagy recycles cellular components to basic building blocks such as nucleosides, sugars, and amino acids that can be exploited to fuel nucleic acid biosynthesis and glutaminolysis. Through mitophagy, Ras-driven tumor cells can remove damaged mitochondria, which would otherwise increase ROS stress.
The role of autophagy in cancer is extremely complex as demonstrated by a growing literature describing situations where autophagy can either promote or inhibit tumorigenesis [58,59]. The majority of evidence suggests autophagy functions in PDAC, as well as more generally in Ras-dependent tumors, to sustain tumor growth. A clinicopathological study of samples from 71 different PDAC patients revealed that PDAC exhibited activated autophagy and that high LC3 protein (a component of autophagosome formation) expression correlated with poor patient outcome [60]. Furthermore, it was demonstrated that pancreatic primary tumors and cell lines showed elevated autophagy, as determined by increased expression of LC3 in PanIN-3, PDAC, and cell lines showing high basal autophagic flux. Correspondingly, genetic depletion of ATG5 or pharmacological inhibition of autophagy with chloroquine suppressed PDAC cell line growth in vitro and tumorigencity in vivo[61]. Chloroquine treatment also increased survival in a K-Ras(G12D)-driven mouse model of PDAC. Suppression of autophagy was associated with increased ROS, elevated DNA damage, and decreased mitochondrial oxidative phosphorylation [61]. Similar analyses in the pancreatic cancer cell line PANC-1 and other RAS mutant human tumor cell lines showed that high basal autophagy is necessary for the growth and survival of these cell lines [62].
Two studies examined the interplay of autophagy dependence in KRAS mutant PDAC tumor cells and mitochondrial function and metabolism. Yang et al. showed in PDAC cell lines that inhibition of autophagy caused a defect in mitochondrial respiration that can be rescued by supplying substrates for the TCA cycle [61]. They concluded that PDAC relies on autophagy to supply bioenergetic intermediates for the TCA cycle. Guo et al. similarly showed a defect in mitochondrial respiration upon autophagy inhibition in immortal, nontumorigenic baby mouse kidney epithelial (iBMK) cells ectopically expressing oncogenic HRAS or KRAS. Similarly to Yang et al., they demonstrated that autophagy inhibition leads to a decrease in TCA cycle metabolites and attenuated mitochondrial respiration; however, this defect in respiration was also accompanied by an accumulation of grossly abnormal mitochondria [62]. They showed that a distinct type of autophagy, mitophagy, appears to play an important role in improving the survival of these cells by removing damaged mitochondria, which otherwise would contribute to increased intracellular ROS levels [63]. While the details diverge in these studies, the unifying reliance upon autophagy in PDAC and other RAS mutant tumors to meet metabolic demand is striking.
Two studies in other cell types further illustrate the complex role of autophagy in tumorigenesis and the importance of cellular context as well as experimental system. Acute ectopic induction of oncogenic H-Ras(V12) expression in immortalized human ovarian surface epithelial cells promoted autophagic cell death rather than growth promotion [64]. A likely basis for the different biological consequence of Ras-induced autophagy is that, unlike the cell types used in Guo et al., the ovarian cells studied did not undergo transformation by oncogenic RAS. Therefore, while oncogenic RAS activated autophagy in this cell line, the lack of growth transformation may impact what role autophagy plays in cellular fate. Likewise, in H-Ras(V12)- transformed fibroblasts it was demonstrated that autophagy mediated RAS-induced senescence [65]. These studies suggest that the biological outcome of the cross talk between RAS and autophagy will vary with cell type and genetic context. Additionally, the levels of autophagic activity may also be critical in dictating tumor cell response to RAS-induced senescence [66].
In addition to the degradation of intracellular components through autophagy, to obtain needed metabolitesoncogenic RAS expressing cells also endocytose extracellular fluid through the process of macropinocytosis [67–69]. A recent report demonstrated that PDAC cells expressing oncogenic KRAS utilized macropinocytosis to transport extracellular protein into the cell and that this endocytosed protein served as a source of glutamine [70]. This scavenging phenotype may be a general property of Ras-driven cancers as Ras transformed cells were recently shown to scavenge extracellular lipids as a critical aspect of their metabolism[71]. Thus, scavenging for various metabolic substrates serves as yet another mechanism by which PDAC cells, as well as Kras-driven cancers in general, can support their unique metabolic needs.
Targeting K-Ras-mediated metabolic pathways
Although the currently pursued anti-K-Ras therapies for PDAC remain promising, the newly appreciated connections between oncogenic KRAS and dysregulated metabolic signaling in PDAC may provide attractive novel directions for these efforts (Figure 4). Lactate efflux pathways, for example, represent one potential target. To sustain the high glycolytic rates associated with PDAC, cells must secrete large amounts of lactate, as high intracellular lactate inhibits glycolysis by negatively regulating Ldha activity. Oncogenic KRAS contributes to this pathway by transcriptionally upregulating LDHA expression [11]. RNAi silencing of CD147 expression, required for the function and expression of the MCT1 and MCT4 lactate transporters that remove intracellular lactate, inhibited pancreatic cell proliferation [72] and suppressed PDAC cell line xenograft growth in mice [73]. Le et al. used a small molecule (FX-11) to inhibit Ldha and observed that treatment with this inhibitor caused increased ROS production due to its inhibition of the regeneration of NAD+ and cell death. Aside from upregulating LDHA expression, K-Ras(G12D) expression in PDAC also preferentially induces flux through the nonoxidative PPP and if this pathway is blocked, tumorigenic activity is suppressed [11]. Thus, it is possible that PDAC cells could be particularly susceptible to pharmacologic inhibition of the early steps of nucleotide biosynthesis such as ribose-5-phosphate production. Furthermore, Hk2 has been identified as an attractive target for KRAS-driven lung cancers, as systemic whole-body deletion of HK2 in the mouse selectively targets cancer cells [74]. Based on the finding that mutant KRAS enhances expression of HK2 [11], it is possible that it may be an attractive target for pancreatic cancers as well. In addition, both increased glycolytic flux through the HBP [11] and hyper-O-GlcNAcylation [33] have been observed in PDAC, suggesting that targeting enzymes such as O-GlcNAc transferase (OGT) could be a therapeutic strategy. A recently developed OGT inhibitor, (Ac-5SGlcNAc) [75], has been shown to inhibit cell proliferation and soft agar colony formation of MIA PaCa-2 cells via suppression of hyper-O-GlcNAcylation [33].
Historically, following the realization that cancer cells were sensitive to glutamine starvation, a number of glutamine analogs were developed including 6-diazo-5-oxo-L-norleucine, azaserine, and acivicin, all of which showed promising preclinical chemotherapeutic activity; however, they were not successful clinically due to dose-limiting neurotoxicity, gastrointestinal toxicity, and myelosuppression [76]. The current strategy for targeting glutamine utilization by cancer cells is to focus on those processes that glutamine addicted cancer cells rely on while preserving the glutamine metabolism of normal cells. Because reliance on GOT1 for the maintenance of redox homeostasis is unique to PDAC, and GOT1 is dispensable in normal cells, this KRAS controlled enzyme could be an exciting therapeutic target [47]. Furthermore, Son et al. demonstrated that two previously characterized GLS inhibitors, Compound 968 [77] and BPTES [78], have a growth suppressive effect on PDAC cells that is enhanced when GLS inhibition is combined with hydrogen peroxide treatment. Hydrogen peroxide treatment increases ROS, similar to radiation in vivo, revealing that the combination of current PDAC treatments with GLS or GOT1 inhibition could have significant therapeutic implications. However, there are currently no clinically safe, highly specific, yet potent drugs available to target glutaminolysis or glycolysis.
Autophagy and macropinocytosis are both biological mechanisms that provide the cell with amino acids, including glutamine, which aid in driving tumorigenic metabolism. Consequently, by targeting these pathways, glutamine starvation can be achieved. The finding that RAS-transformed cells depend on autophagy for survival [62], suggests that cancers with high levels of mutant Ras proteins will be particularly susceptible to autophagy inhibition. The autophagy inhibitor hydroxychloroquine (HCQ) is already approved by the FDA for the treatment of malaria and various rheumatological diseases. HCQ has been used in combination with chemotherapeutic drugs, radiation, and immunotherapy in cancer treatment clinical trials [79]. There are multiple early phase studies exploring the use of HCQ in pancreatic cancer in various therapeutic combinations and in various stages of the disease (NCT01128296, NCT01506973, NCT01494155, NCT01273805). It should be noted that HCQ is actually an inhibitor of lysosome acidification, and hence is not specific to the autophagy pathway. In fact, this may prove beneficial, as it would inhibit other pathways that use the lysosome for degradation, such as macropinocytosis. However, further elucidation of the specific mediators of autophagy in PDAC could yield surprising new targets with a favorable therapeutic index.
Concluding Remarks
We have summarized and highlighted the recently defined role of oncogenic KRAS in the regulation of altered metabolic signaling pathways in PDAC, a clinically challenging disease in dire need of new treatments. The original, simplistic view that tumor cells exhibit enhanced glycolysis due to impaired mitochondria has been replaced with the understanding that tumor cells rewire metabolic pathways to serve their specific proliferative needs. Furthermore, it is becoming clear that this rewiring is orchestrated by well-known oncogenes, such as KRAS. Oncogenic KRAS increases glucose uptake and redirects the flux of carbon through pathways such as the PPP to fuel biosynthesis, and by increasing the transcription of lactate transporters, KRAS furthers the enhanced glycolytic state of PDAC. To maintain redox balance, mutant KRAS alters glutamine utilization by increasing transcription of transaminases, which send the glutamine carbon skeleton down a metabolic pathway that fuels NADPH production. To supply cells with as much glutamine and other amino acids as possible, oncogenic KRAS also drives macropinocytosis of extracellular fluids in PDAC cells. Additionally, autophagy is upregulated by KRAS in PDAC to support cell survival, potentially by restraining oxidative damage and maintaining energy homeostasis. The elucidation of these links between oncogenic KRAS and metabolic signaling has opened up new lines of investigation, the pursuit of which could lead to the design of new PDAC therapeutics.
Highlights.
KRAS is the most frequently mutated gene in PDAC and drives cancer development and growth.
Oncogenic KRAS alters cellular glucose uptake, glycolytic flux, and glutamine usage.
Autophagy and macropinocytosis are upregulated by mutant KRAS to assist PDAC survival.
Mechanisms of oncogenic KRAS-driven metabolism may yield new anti-KRAS therapeutics.
Acknowledgments
We apologize to colleagues whose studies we did not cite due to space limitations. CJD is supported by grants from the NIH, Pancreatic Cancer Action Network-AACR and the Lustgarten Pancreatic Cancer Foundation. KLB is supported by a T32 CA71341 training grant. ACK is supported by grants from the NIH, ACS, DOD, and the Lustgarten Pancreatic Cancer Foundation. JDM is supported by an American Society of Radiation Oncology (ASTRO) Junior Faculty Career Research Training Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheffzek K, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 4.Scheidig AJ, et al. The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7:1311–1324. doi: 10.1016/s0969-2126(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 5.Pylayeva-Gupta Y, et al. Ras oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanda M, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142:730–733. e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hezel AF, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes & Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 9.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins MA, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim KH, Counter CM. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell. 2005;8:381–392. doi: 10.1016/j.ccr.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Collisson EA, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurer T, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Q, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shima F, et al. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc Natl Acad Sci USA. 2013;110:8182–8187. doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmermann G, et al. Small molecule inhibition of the KRAS–PDEdinteraction impairs oncogenic KRAS signaling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 18.Babij C, et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 2011;71:5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- 19.Luo T, et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc Natl Acad Sci USA. 2012;109:2860–2865. doi: 10.1073/pnas.1120589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regel I, et al. Energy metabolism and proliferation in pancreatic carcinogenesis. Langenbecks Arch Surg. 2012;397:507–512. doi: 10.1007/s00423-012-0933-9. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 23.Moreno-Sanchez R, et al. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 24.Lunt SY, Vander Heiden MG. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 25.Vander Heiden MG, et al. Understanding the Warburg Effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Racker E, et al. Glycolysis and methylaminoisobutyrate updake in rat-1 cells transfected with ras or myc oncogenes. Proc Natl Acad Sci USA. 1985;82:3535–3538. doi: 10.1073/pnas.82.11.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiaradonna F, et al. Ras-dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene. 2006;25:5391–5404. doi: 10.1038/sj.onc.1209528. [DOI] [PubMed] [Google Scholar]
- 28.Vizan P, et al. K-ras codon-specific mutations produce distinctive metabolic phenotypes in human fibroblasts. Cancer Res. 2005;65:5512–5515. doi: 10.1158/0008-5472.CAN-05-0074. [DOI] [PubMed] [Google Scholar]
- 29.Chiaradonna F, et al. Acquired glucose sensitivity of k-ras transformed fibroblasts. Biochem Soc Trans. 2005;33:297–299. doi: 10.1042/BST0330297. [DOI] [PubMed] [Google Scholar]
- 30.Gaglio D, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011 doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun J, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou W, et al. Proteomic analysis reveals Warburg effect and anomalous metabolism of glutamine in pancreatic cancer cells. J Proteome Res. 2012;11:554–563. doi: 10.1021/pr2009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Z, et al. Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J Biol Chem. 2013;288:15121–15130. doi: 10.1074/jbc.M113.470047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi W, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neesse A, et al. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60:861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 36.Koong AC, et al. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 2000;48:919–922. doi: 10.1016/s0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- 37.Guillaumond F, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2013;110:3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C, et al. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 40.Blum R, et al. Ras inhibition in glioblastoma down-regulates hypoxia-inducible factor-1alpha, causing glycolysis shutdown and cell death. Cancer Res. 2005;65:999–1006. [PubMed] [Google Scholar]
- 41.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122:501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 43.Hensley CT, et al. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang CV. Links between metabolism and cancer. Genes & Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deberardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Csibi A, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trachootham D, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Kong B, et al. Overview on how oncogenic Kras promotes pancreatic carcinogenesis by inducing low intracellular ROS levels. Front Physiol. 2013;4:1–5. doi: 10.3389/fphys.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matés JM, et al. Oxidative stress in apoptosis and cancer: an update. Arch Toxicol. 2012;86:1649–1665. doi: 10.1007/s00204-012-0906-3. [DOI] [PubMed] [Google Scholar]
- 53.Matés JM, et al. Glutamine homeostasis and mitochondrial dynamics. Int J Biochem Cell Biol. 2009;41:2051–2061. doi: 10.1016/j.biocel.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 54.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang R, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes & Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hippert MM, et al. Autophagy in cancer: good, bad, or both? Cancer Res. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 60.Fujii S, et al. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008 doi: 10.1111/j.1349-7006.2008.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes & Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes & Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moiseeva O, et al. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol Cell Biol. 2009;29:4495–4507. doi: 10.1128/MCB.01868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elgendy M, et al. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 65.Young ARJ, et al. Autophagy mediates the mitotic senescence transition. Genes & Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, et al. Autophagic activity dictates the cellular response to oncogenic RAS. Proc Natl Acad Sci USA. 2012;109:13325–13330. doi: 10.1073/pnas.1120193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walsh AB, Bar-Sagi D. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J Biol Chem. 2001;276:15609–15615. doi: 10.1074/jbc.M010573200. [DOI] [PubMed] [Google Scholar]
- 68.Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233:1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 69.Porat-Shliom N, et al. A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol Biol Cell. 2008;19:765–775. doi: 10.1091/mbc.E07-08-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kamphorst JJ, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci USA. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneiderhan W, et al. CD147 silencing inhibits lactate transport and reduces malignant potential of pancreatic cancer cells in in vivo and in vitro models. Gut. 2009;58:1391–1398. doi: 10.1136/gut.2009.181412. [DOI] [PubMed] [Google Scholar]
- 73.Le A, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patra KC, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24:213–228. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gloster TM, et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahluwalia GS, et al. Metabolism and action of amino acid analog anti-cancer agents. Pharmacol Ther. 1990;46:243–271. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]
- 77.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson MM, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]