In this issue of Circulation Research, Gregoire and colleagues report their discovery that Yin Yang 1 (YY1) is a critical factor for the transition from cardiac progenitor to differentiated cardiomyocyte1. The authors made clever use of currently available Transcription Factor (TF) binding databases to perform a bioinformatic-based screen for factors regulating cardiac progenitor genes. They compared their dataset of genes enriched in mouse embryonic stem cell (ES)-derived cardiac progenitor cells (CPCs) with freely available TF binding databases. They initially screened 32 candidate transcription factors and surprisingly found that YY1, a known repressor of sarcomeric gene expression, is resident on many CPC genes.
The heart is the first organ to develop in the vertebrate embryo. Precardiogenic mesoderm is initially derived from anterior lateral plate mesoderm. Signals from surrounding tissues promote lateral plate to form cardiogenic mesoderm by inducing a cardiogenic transcriptional network. The cardiogenic mesoderm contains CPCs that differentiate into primitive cardiomyocytes. The transition from CPC to cardiomyocyte is a critical developmental transition point that needs further study and is the focus of the study by Gregoire. Importantly, the cardiogenic developmental process can be mimicked by conversion of ES cells into CPCs and then into cardiomyocytes. ES-derived cardiomyocytes have been used to identify factors regulating heart development and have the added advantage of potentially becoming a source of cells in regenerative cell therapies.
Like the cardiac lineage in embryos, murine ES cell-derived CPC express one of the earliest cardiac genes, the Nkx2.5 transcription factor. Although Nkx2.5 is the mammalian orthologue of tinman, which acts as a master regulator of cardiac development in Drosophila, Nkx2.5 itself is insufficient to promote cardiogenic development. Commitment of early embryonic mesoderm to the cardiac lineage requires signaling molecules such as BMP and Activin to activate expression or function of numerous transcription factors in addition to Nkx2.5 2.
Gregoire and colleagues now report that YY1 is an essential component of the transcriptional loop regulating early heart development. YY1 is a member of the Gli-Kruppel family of zinc finger proteins and as its name implies, shows context dependent activity. YY1 was first cloned as a factor that regulates activity of the adeno-associated virus promoter3. Expressed ubiquitously, YY1 regulates a wide array of biological processes often by modulating transcription in divergent ways. In some contexts, YY1 functions as a repressor while in others it is an activator. The ability of YY1 to recruit both activators and repressors of transcription accounts for its contextual functions.
During transcriptional repression, YY1 physically interacts with histone deacetylaces (HDACs) to repress gene transcription and prevent cardiac hypertrophy 4. In muscle gene regulation YY1 forms a complex with the EZH2 methyltransferase, a component of the polycombe repressor complex, and HDAC1 to repress gene expression 5. Recent work has also shown that YY1 recruits the Xist non-coding RNA to the X inactivation center during X chromosome inactivation indicating that YY1 is also an RNA binding protein 6. In contrast during transcriptional activation, YY1 can directly interact with the histone acetyltransferase, P300 to enhance histone acetylation and thereby enhance gene expression 3. YY1 also recruits PRMT1, a Histone H4 (Arg3) methyltransferase to YY1 regulatory elements and this also promotes transcription (7 and Fig.1).
Fig.1. Conceptual model of YY1 and GATA4 transcriptional activation of Nkx2-5.
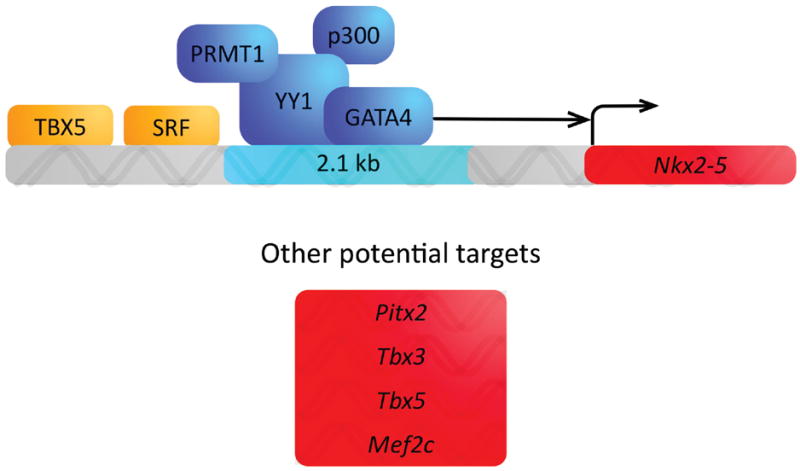
The YY1 and GATA4 binding domain has been mapped to a 2.1kb region upstream of the Nkx2-5 promotor. Additionally, TBX5 and SRF binding motifs are located near the YY1 and GATA4 binding domain. PRMT1 and p300 are known transcription co-activators of YY1. Other potential targets of YY1 transcriptional activation include Pitx2, Tbx3, Tbx5, Mef2c.
In this manuscript, the authors show that YY1 acts as a transcriptional activator to promote both Nkx2.5 expression and CPC commitment. Using a combination of in vitro luciferase-based assays, in vivo chromatin immunoprecipitation (ChIP) and genome-wide sequencing analysis, the authors delineated a 2.1 kb cardiac-specific enhancer element that recruits YY1 to enhance Nkx2.5 transcription. In addition, they show that YY1 function during CPC commitment is through a partnership with Gata4, a transcription factor essential for early heart development. Furthermore, the authors show that the overexpression of YY1 enhanced the cardiogenic differentiation of ES cells into CPCs in vitro using an ES cell-based assay. Lastly, cardiac mesoderm-specific YY1 loss-of-function resulted in early embryonic lethality.
One of the experiments utilized by Gregoire is ChIP-sequencing (ChIP-seq), an approach that allows analysis of chromatin binding sites on a genome-wide scale. ChIP-seq requires massively parallel sequencing that is becoming cheaper to perform as new technologies continue to be developed. Because of its unbiased nature, ChIP-seq has the potential to provide powerful new information about transcriptional regulation. ChIP-seq has already been used to provide insight into transcription factor binding sites throughout the genome in multiple contexts 8.
There are limitations to the data presented by Gregoire. Although CPCs derived from ES cells are useful for working out mechanisms, it is nonetheless an artificial system that requires in vivo validation. Although Gregoire performed considerable in vivo validation, further work in the embryo will be necessary to completely validate their model. A related limitation of the genome-wide approaches, such as ChIP-seq, is that they fail to provide three dimension, spatial information about protein chromatin interactions. Nonetheless, the new findings reported by Gregoire are exciting and will open up new areas of investigation into early cardiac development.
Acknowledgments
Sources of Funding
Supported by NIH grants (JFM),5T32HL007676-23 (JL), AHA NCRP SDG 0930240N (YM).
Footnotes
Disclosures
None
References
- 1.Gregoire S, Karra R, Passer D, Deutsch MA, Krane M, Feistritzer R, Sturzu A, Domian I, Saga Y, Wu SM. Essential and unexpected role of yy1 to promote mesodermal cardiac differentiation. Circ Res. 2013;112:xxx–xxx. doi: 10.1161/CIRCRESAHA.113.259259. in this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noseda M, Peterkin T, Simoes FC, Patient R, Schneider MD. Cardiopoietic factors: Extracellular signals for cardiac lineage commitment. Circulation research. 2011;108:129–152. doi: 10.1161/CIRCRESAHA.110.223792. [DOI] [PubMed] [Google Scholar]
- 3.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor yy1: Are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 4.Sucharov CC, Dockstader K, McKinsey TA. Yy1 protects cardiac myocytes from pathologic hypertrophy by interacting with hdac5. Molecular biology of the cell. 2008;19:4141–4153. doi: 10.1091/mbc.E07-12-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The polycomb ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes & development. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeon Y, Lee JT. Yy1 tethers xist rna to the inactive x nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E. Targeted recruitment of a histone h4-specific methyltransferase by the transcription factor yy1. Genes & development. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]


