Abstract
Adenoviruses are linear double stranded DNA viruses that infect human and rodent cell lines, occasionally transform them and cause tumors in animal models. The host cell challenges the virus in multifaceted ways to restrain viral gene expression and DNA replication, and sometimes even eliminates the infected cells by programmed cell death. To combat these challenges, adenoviruses abrogate the cellular DNA damage response pathway. Tip60 is a lysine acetyltransferase that acetylates histones and other proteins to regulate gene expression, DNA damage response, apoptosis and cell cycle regulation. Tip60 is a bona fide tumor suppressor since mice that are haploid for Tip60 are predisposed to tumors. We have discovered that Tip60 is degraded by adenovirus oncoproteins EIB55K and E4orf6 by a proteasome-mediated pathway. Tip60 binds to the immediate early adenovirus promoter and suppresses adenovirus EIA gene expression, which is a master regulator of adenovirus transcription, at least partly through retention of the virally encoded repressor pVII on this promoter. Thus degradation of Tip60 by the adenoviral early proteins is important for efficient viral early gene transcription and for changes in expression of cellular genes.
Keywords: HAT, Tip60, EIA, Adenovirus
Introduction
Adenoviruses are non-enveloped, linear, double stranded DNA viruses. Discovered in early 1950s, almost 50 human adenoviruses clustered in 6 different species have been identified. Besides lytic infection, adenoviruses are also known to transform host cells through their oncogenes EIA and EIB in cooperation with other cellular proteins.1 Although there is no concrete evidence that adenoviruses are involved in tumor formation in humans they can transform human cells and can induce tumors in newborn rats. Adenovirus has developed different strategies to counter host cell defense and usurp the cellular machinery, both for viral replication and cell transformation. Indeed several paradigms of how viral oncoproteins transform cells by inactivating cellular tumor suppressors were elucidated in cells transformed by adenovirus. For example, adenovirus EIA protein binds and inactivates Rb protein to promote entry of cells into S phase, while EIB55K and E4orf6 oncoproteins bind and direct p53 for degradation by proteasome to prevent cell-cycle checkpoint activation and apoptosis. 2–4 Adenovirus also degrades MRN complex proteins, DNA ligase IV, TOPBP1 and integrin alpha 3 (ref.5–8).
Many studies have shown widespread changes in DNA and histone modifications during tumorigenesis.9 Several viral oncoproteins interact with different histone modifying enzymes of the host cell and deregulate the epigenetic program.10,11 Recently we showed that the Human Papillomavirus (HPV) oncoprotein E6 interacts with and destabilizes the cellular histone acetyltransferase protein Tat interacting protein 60 kDa, Tip60 (ref.12). Intriguingly, the degradation of Tip60 relieved cellular repression of the viral early promoter. Another recent study showed that human cytomegalovirus (HCMV) pUL27 can induce Tip60 degradation by proteasome, though the contribution of this degradation to the viral life cycle was unclear. 13
Tip60 is a lysine acetyltransferase protein of the MYST family. It modifies the chromatin structure by acetylating histones and thus plays an important role in transcriptional regulation.14–16 Tip60 is essential for viability since its disruption causes embryonic lethality.17 Mice that lack one allele of Tip60 show acceleration in tumorigenesis.18 Tip60 has also been implicated in the DNA damage response: it activates the apex kinases in the checkpoint pathways, the ATM/ ATR kinases and it stops the DNA damage response by promoting the dephosphorylation of phosphoH2AX at the DNA damage sites.19–20 Another non-histone protein acetylated by Tip60 is p53. Tip60 acetylates p53 on lysine120 and shifts the balance towards apoptosis and away from cell-cycle arrest. 21–22
Tumor suppressors like p53 and Rb are inactivated by HPV and adenovirus oncoproteins. Since Tip60 is a tumor suppressor that is targeted for degradation through proteasome mediated pathway by the HPV E6 oncoprotein, we investigated whether Tip60 was also degraded by adenovirus oncoproteins. We discovered that the adenovirus oncoproteins EIB55K and E4orf6 also direct Tip60 for degradation in a proteasome-dependent manner and by doing so they relieve the repression imposed by cellular Tip60 on viral early gene expression. The results highlight a possible role for Tip60 during adenovirus replicative cycle, and suggest that downregulation of the tumor suppressor Tip60 may be important for multiple oncogenic viruses.
Results
Adenovirus destabilizes Tip60 protein
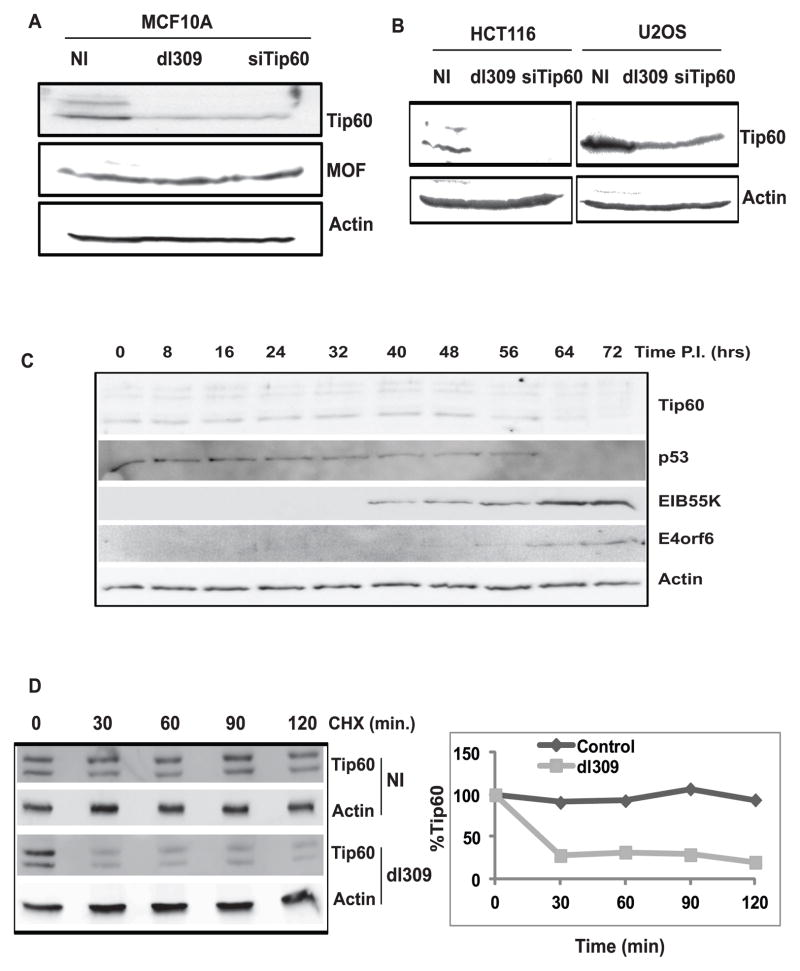
To check the effect of adenovirus on Tip60 levels, MCF10A breast epithelial cells were infected with dl309 virus (phenotypically wild type adenovirus). Note that multiple splice-isoforms of Tip60 are expressed at varying ratios in different cell-lines. Tip60 levels were decreased in the virus (dl309)-infected MCF10A cells compared to mock-infected cells while the levels of another HAT protein MOF was not decreased (Figure 1A). Similar results were obtained in U2OS osteosarcoma cells and HCT116 colon cancer cells (Figure 1B). Evaluating MCF10A lysates harvested at various times post-infection showed that both p53 and Tip60 decreased at about the same time: 64 hr post infection (Figure 1C). The time of decrease corresponded with significant expression of EIB55K and E4orf6 oncoprotein (Figure 1C). The half life of Tip60 was determined by shutting off new protein synthesis by cycloheximide. The half life decreased to <30 min in virus (dl309)-infected cells compared to >120 min in mock-infected cells (Figure 1D). These data suggest that adenovirus infection destabilizes cellular Tip60 protein.
Figure 1. Tip60 is destabilized in adenovirus-infected cells.
(A) Immunoblot with indicated antibodies of MCF10A cells (NI- no infection; dl309- infected with dl309 adenovirus; siTip60- transfected with siTip60). Cells were harvested 68 hrs post infection. Knockdown of Tip60 confirms the identity of the Tip60 bands.
(B) Destabilization of Tip60 by adenovirus in other cell lines. HCT116 and U2OS cells were lysed 40 hours after infection or 72 hr after siRNA transfection prior to immunoblotting.
(C) Tip60 destabilization is concurrent with p53 degradation in adenovirus-infected cells. dl309 infected MCF10A cells were harvested at different time points as indicated after infection and lysates were used to probe with indicated antibodies.
(D) Tip60 protein half life decreases in adenovirus-infected cells. dl309-infected and uninfected MCF10A cells were treated with cycloheximide (added at 68hrs post infection) for different time points as indicated and lysates were resolved for Western blot by indicated antibodies. (Graph) Levels of Tip60 were quantitated using Gene Tool Software (Syngene). Tip60 levels in both 0 hours lanes are taken as 100%.
EIB55K and E4orf6 oncoproteins are involved in proteasomal mediated degradation of Tip60
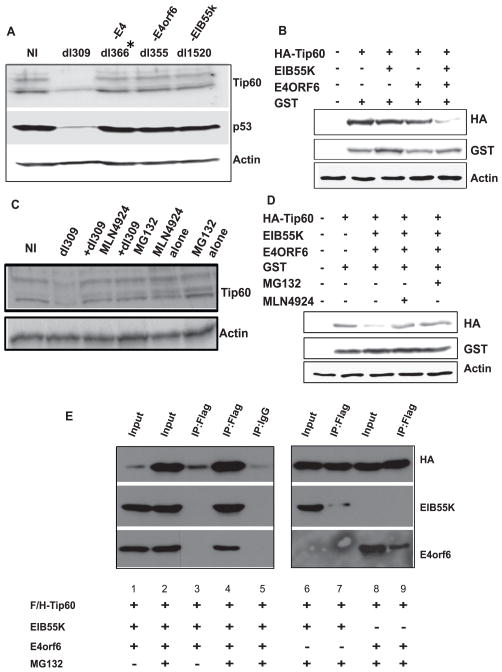
To test whether the oncoproteins of adenovirus are involved in the destabilization of Tip60, we tested different mutants of adenovirus under similar conditions. dl366* virus with a deletion of the entire E4 region did not affect Tip60 levels, similar to dl355 virus with a mutation specifically in the E4orf6 gene. dl1520 virus with a deletion in EIB55K gene also did not decrease Tip60 (Figure 2A). These results suggest the involvement of EIB55K and E4orf6 proteins in the destabilization of Tip60. To confirm this result and to rule out a role of other viral proteins, a plasmid expressing HA/flag-Tip60 was co-transfected with plasmids expressing EIB55K or E4orf6 singly or in combination into U2OS cells. A co-transfected plasmid expressing GST serves as a transfection control. Tip60 levels were reduced in the cells with both EIB55K and E4orf6 but not with either oncoprotein on its own (Figure 2B).
Figure 2. Adenovirus oncoproteins EIB55K and E4orf6 promote proteasome-dependent Tip60 degradation.
(A) MCF10A cells were infected with different strains of adenovirus and lysates prepared 72 hours after infection were probed with indicated antibodies.
(B) U2OS cells were transformed with indicated plasmids. Cells were harvested 48 hours after transfection and lysates immunoblotted with indicated antibodies. GST-expressing plasmid was cotransfected as a transfection control.
(C) Inhibition of the proteasome or Cullins activity stabilizes Tip60 in dl309-infected cells. MCF10A cells were infected by dl309 strain and MG132 or MLN4924 was added to infected cells at 4 or 24 hours before harvesting (72hrs post infection), respectively. Lysates were probed with indicated antibodies.
(D) U2OS cells were transfected with indicated plasmids and addition of either MG132 or MLN4924 stabilizes the Tip60 protein. Rest is as in (B).
(E) Interaction between Flag/HA-Tip60, EIB55K and E4orf6. U2OS cells were transfected with indicated plasmids for 48 hours and treated for 5 hours with or without MG132 before harvest. Anti-flag antibodies and control IgG were used for immunoprecipitation and immunoprecipitated proteins resolved and probed with indicated antibodies. Immunoblot shows co-precipitation of Flag/HA-Tip60, EIB55K, and E4orf6. Input lanes contain 5% of lysates input into the immunoprecipitates.
To test whether the destabilization of Tip60 is due to degradation by proteasomes, dl309-infected cells were treated with the proteasomal inhibitor MG132. Tip60 decrease was abrogated by MG132 (Figure 2C) implicating proteasomes in the Tip60 degradation. MLN4924 is a global inhibitor of all cullin Ring E3 ligases, because it inhibits the critical neddylation of cullins that is essential for their activity. Treatment with MLN4924 also prevented adenovirus mediated decrease in Tip60 (Figure 2C). Similarly addition of MG132 and MLN4924 prevented the destabilization of transfected HA-Tip60 by co-transfected EIB55K and E4orf6 expression (Figure 2D).
Cullin-5 and the elongins are known to co-operate with E1B55K and E4orf6 to degrade p53 [4]. In order to determine which cullin was involved in the adenovirus-mediated degradation of Tip60, we knocked down individual cullin proteins by siRNA transfection in dl309 virus-infected cells. While p53 was stabilized, as expected, by si-Cullin-5, Tip60 was not stabilized (Figure S1). si-Cullin3 stabilizes Tip60, but since Cullin-3 is known to degrade Tip60 even in uninfected cells,23 we cannot be certain that Cullin-3 is specifically recruited by EIB55K+E4orf6 to degrade Tip60. To ascertain whether the elongins may be involved in the degradation of Tip60, we knocked down elongin B by siRNA in dl309 virus-infected cells. Knockdown of elongin B stabilized p53 but not Tip60 (data not shown) suggesting that elongins may not be involved in the degradation of Tip60 by E4orf6+EIB55K.
The results suggest that as with p53, the E1B55K+E4orf6 viral proteins may interact with Tip60, to recruit Tip60 to the E3 ligase. To test this, plasmids expressing HA/flag-Tip60 were co-transfected into U2OS cells with plasmids expressing EIB55K and E4orf6 (Figure 2E). Flag/HA-Tip60 was increased in cell lysates when it was stabilized by MG132 (lane 2). Immunoprecipitation of Tip60 from the MG132-treated cells co-immunoprecipitated EIB55K and E4orf6 proteins (lane 4). When E4orf6 or EIB55K were transfected singly with Tip60 and the Tip60 was immunoprecipitated, the viral proteins were co-immunoprecipitated, but to a much lower extent than when they were present together (lanes 7, 9). These results suggest that EIB55K and E4orf6 together form a complex that binds to Tip60 and possibly recruits Tip60 to an unknown cullin.
Tip60 suppresses EIA expression
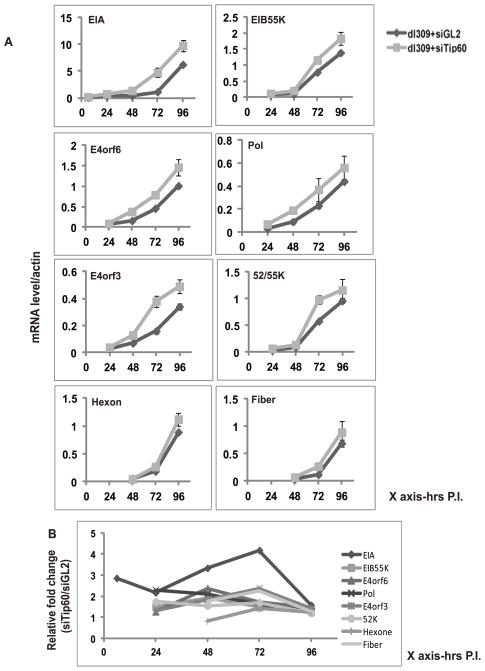
Since Tip60 is a HAT protein and can act as a coregulator for gene transcription we hypothesized that degradation of Tip60 might promote viral gene expression. We therefore checked the expression profiles of different viral genes in cells with wild type Tip60 compared to those with siRNA mediated Tip60-depletion. Tip60 is degraded in dl309 infected cells around 62 hr post-infection, consequently any difference in viral gene expression due to the experimental knockdown of Tip60 is expected to be transient, before the virus naturally directs degradation of Tip60. Expression of all the early genes was increased in siTip60 cells at early times of infection (Figure 3A). Comparing viral gene expression in siTip60 cells to that in siGL2 cells shows that, EIA expression increased by 3–4 fold in Tip60 depleted cells, while the other viral genes showed at most a 2-fold increase in expression (Figure 3B). The E1A protein was also increased in siTip60 cells (Figure S2A). However, expression of all viral genes became comparable in control and siTip60 cells at 96 hr post infection when the virus degrades Tip60, consistent with our expectation. These results show that the presence of Tip60 suppresses the EIA mRNA level early in viral infection.
Figure 3. Tip60 represses expression of viral transcription factor EIA.
(A) Levels of mRNA of different viral genes; EIA (early), EIB55K (early), E4orf6 (early), Pol (early), E4orf3 (early), 52/55K (early/late) Hexone (late) and Fiber (late) were measured at indicated time points after viral infection by RT-qPCR and normalized to β actin. MCF10A cells transfected with siGL2 or siTip60 were infected with dl309. Mean ± SD (n=3).
(B) Relative expression of different viral genes in siTip60 transfected cells relative to siGL2 transfected cells. EIA mRNA shows most enhancement at 72 hr after infection in cells where Tip60 was knocked down.
The increase in EIA expression after knockdown of Tip60 could be secondary to virus replication. To rule this out, we added hydroxyurea (HU) 48 hrs after infection and harvested the cells at 72 hrs post infection and measured EIA transcript levels. HU did not prevent the increase in EIA expression when Tip60 was knocked down showing that the stimulation of E1A expression by siTIP60 is not secondary to an increased number of templates because of viral DNA replication (Figure S2B).
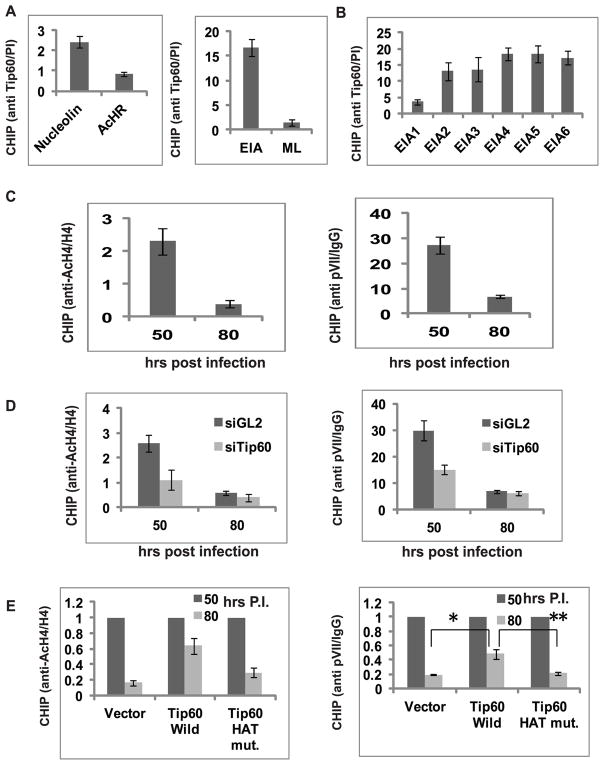
We next used chromatin Immunoprecipitation (ChIP) experiments to determine if Tip60 was preferentially associated with the EIA compared to the Major Late promoter, which remains unchanged after siTip60. Tip60 was detected at the EIA promoter but not at the ML promoter (Figure 4A). A cellular promoter known to be regulated by Tip60, nucleolin, and another unaffected by Tip60, acetylcholine receptor, served as positive and negative controls, respectively. Primer pairs spanning 6 different regions of the E1A promoter (Figure S3) indicated that more Tip60 binds near the transcriptional start site of EIA (Figure 4B). ChIP for histone H4 acetylation measures Tip60 activity at the promoter. pVII is an adenovirus late protein that is known to bind to E1A promoter, represses it and is removed when transcription of viral DNA starts.24 Both acetylated histone H4 (H4Ac) and pVII protein were bound to the EIA promoter at 50 hours after infection when Tip60 is not degraded, and both disappear from the promoter at 80 hours after infection when Tip60 is degraded (Figure 4C). The strong correlation of Tip60 presence at the E1A promoter with H4Ac, and with pVII which acts as an anti-correlative marker for transcription synthesis, supports the hypothesis that by binding to the E1A promoter and through the acetylation of histone H4, Tip60 represses the E1A promoter.
Figure 4. Tip60 binds to EIA promoter, acetylates histone H4.
(A) Tip60 binds to EIA promoter. ChIP assay was performed in MCF10A cells 72 hours after infection with dl309 strain. Ratio of qPCR signal in Tip60 immunoprecipitate relative to that in pre-immune serum immunoprecipitate shows occupancy of Tip60 on EIA promoter. ML- major late promoter. Nucleolin and AcHR were taken as positive and negative control cellular promoters for Tip60 binding. Mean ± SD (n=3).
(B) Primers covering 6 regions of the (Supp Fig. S3) EIA promoter were used in the ChIP assay for Tip60 as in (A). Tip60 binds more near the start site of the EIA transcript. Mean ± SD, n=3.
(C) Acetylated histone H4 and pVII at the EIA promoter decreases after Tip60 degradation at 62 hr post infection. ChIP analysis at EIA promoter at 50 hr and 80 hrs after infection. The rest is as in (A). Mean ± SD, n=3.
(D) Tip60 knockdown accelerates the decrease of H4 acetylation and pVII occupancy at the EIA promoter. ChIP assay was performed in dl309-infected MCF10A cells transfected with control siGL2 or siTip60. Level of acetylated H4 and pVII determined by ChIP assay 50 and 80 hrs after infection. mean ± SD, n=3.
(E) Tip60 HAT activity stimulates H4 acetylation and pVII occupancy at EIA promoter. Relative Q-PCR signal ratio at 80 hrs post infection (50 hrs ratio=1) in cells stably transfected with empty vector or plasmids expressing Tip60 wild type or HAT mutant. ChIP for acetylated H4 and pVII. Mean ± SD, n=3. (*P<0.005and **P<0.005 by Student’s t test).
To test this hypothesis, we knocked down Tip60 by siRNA and measured the amount of H4Ac and pVII at the EIA promoter at 50 and 80 hr post infection. H4Ac and pVII at the EIA promoter decreased prematurely at 50 hr post infection upon knockdown of Tip60 (Figure 4D). To investigate the role of the acetyltransferase activity of Tip60 in retention of H4Ac at the E1A promoter, the ChIP assays were repeated after virus infections in cells with vector alone or overexpressing Tip60 wild type or Tip60 HAT mutant. The decrease in H4Ac and in pVII at the E1A promoter normally seen at 80 hr post-infection was attenuated by wild type Tip60 but not by the catalytically dead Tip60 (Figure 4E). These results indicate that catalytically active Tip60 acetylates H4 and thus represses the EIA promoter.
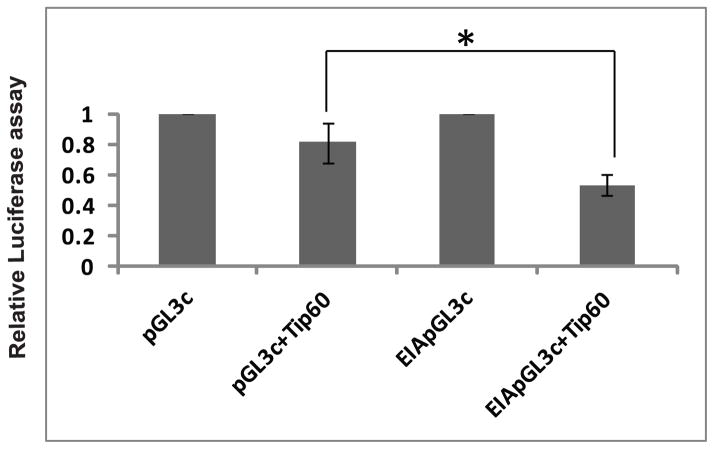
To test whether Tip60 can repress the EIA promoter directly, U2OS cells stably over-expressing Tip60 were transiently transfected with a luciferase reporter driven by the EIA promoter (E1ApGL3c). The over-expressed Tip60 significantly repressed the luciferase activity from E1ApGL3c compared to that driven by the control promoter, pGL3c (Figure 5).
Figure 5. Tip60 represses the EIA promoter.
Tip60 represses EIA promoter. Firefly plasmid pGL3C or EIA promoter/pGL3C plasmids were transfected in control or Tip60 overexpressing U2OS cells. Renilla Luciferase plasmid was cotransfected as normalizing control. The ratio of Luciferase activity of firefly to renilla was calculated and normalized values are shown. Mean ± SD of two experiments (n=2) done in triplicate. * P<0.05 by Student’s t test.
Collectively these results suggest that Tip60 binds to the EIA promoter and represses the EIA promoter through acetylation of Histone 4.
Tip60 knockdown favors initial establishment of viral DNA synthesis
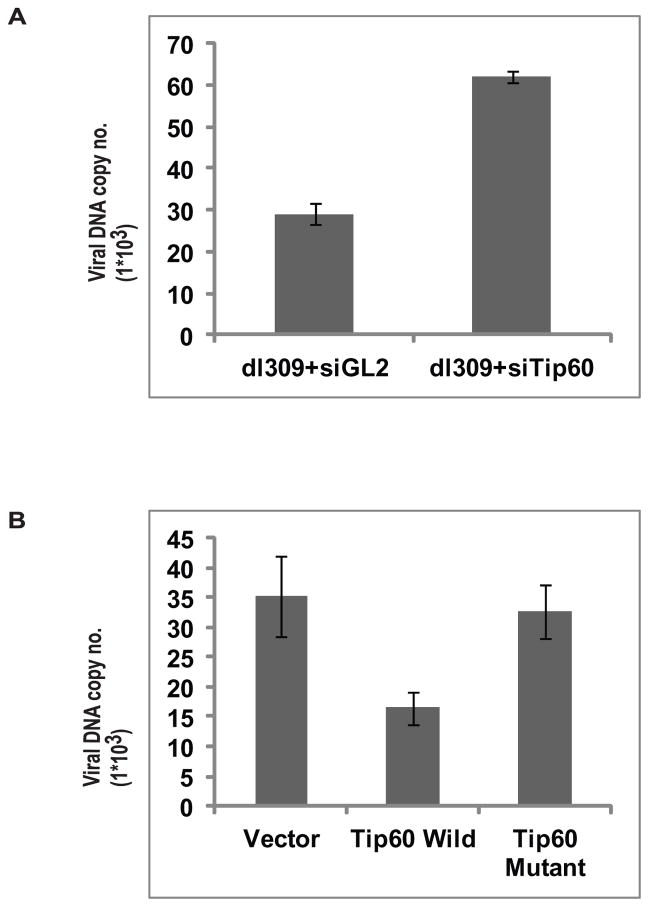
Viral DNA synthesis is dependent on early viral gene expression. Tip60 represses the master regulator of early viral transcription, EIA. Therefore we hypothesized that adenovirus degrades Tip60 because the latter has a negative effect on viral DNA replication. To test this hypothesis, cells transfected with siGL2 or siTip60 were infected with the wild type dl309 virus to determine if diminishing the levels of Tip60 at early times of viral infection leads to greater viral DNA replication. Indeed, Tip60 knockdown resulted in greater viral DNA levels at 72 hr post infection (Figure 6A). This result suggest that Tip60 can suppress virus life cycle.
Figure 6. Tip60 null condition favors viral DNA synthesis.
(A) Tip60 knockdown increases viral DNA production. Viral DNA copy number was measured in MCF10A cells infected with dl309 strain and treated with control siGL2 or siTip60 at 72 hrs post infection. Mean ± SD (n=3).
(B) Tip60 HAT activity is required for suppression of viral DNA titer. MCF10A cell line stably over-expressing either wild Tip60 or catalytically dead Tip60 or stably transfected with vector alone were infected with dl309 strain and viral DNA copy number was measured at 72 hrs post infection as in (A). Mean ± SD (n=3).
To ascertain whether the lysine acetyltransferase activity of Tip60 has any role in the suppression of viral DNA replication, we made cell lines stably transfected with either empty vector or with plasmids expressing wild Tip60 or catalytically dead Tip60 (Figure S4). These cells were then infected with dl309 strain and harvested at 72 hrs post infection. The cells expressing wild type Tip60 had less viral DNA in comparison to cells expressing catalytically dead Tip60 or transfected with empty vector (Figure 6B). These results show that the acetyltransferase activity of Tip60 suppresses viral DNA synthesis.
Tip60 degradation alters expression of cellular genes
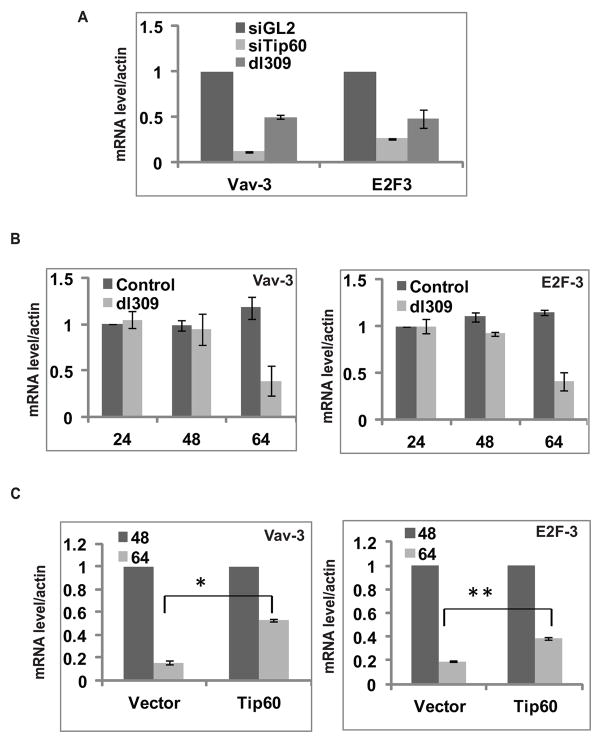
Tip60 acetylates Histones and changes the epigenetic environment of promoters to change the gene expression profile. We hypothesized that Adenovirus-dependent degradation of Tip60 might also affect expression of cellular targets of Tip60. Microarray analysis after Tip60 knockdown in HCT116 cells revealed several genes to be regulated by Tip60 (Dutta and Dutta, unpublished data). Among these genes Vav-3 and E2F-3 were downregulated by Tip60 knockdown or after dl309 infection (Figure 7A). The decrease in expression of both genes coincides with the time of degradation of Tip60 in virus-infected cells: 64 hrs post infection (Figure 7B). These results suggest that the decrease in expression of Vav-3 and E2F-3 in dl309 virus infected cells may be dependent on Tip60 degradation.
Figure 7. Tip60 degradation by adenovirus decreases Tip60 dependent cellular gene expression.
(A) Relative expression of cellular genes in MCF10A cells transfected with siGL2, siTip60 or infected with dl309. Cells were harvested 64 hrs post infection or transfection. Relative expression of genes by RT-qPCR compared to control siGL2. Mean ± SD (n=3).
(B) Decrease in cellular gene expression coincides with Tip60 degradation in dl309- infected cells. MCF10A cells were infected with mock or dl309 virus and cells were harvested at different time points as indicated. Relative mRNA expression of Vav-3 and E2F-3 was measured at indicated times by RT-qPCR taking value of Control 24 hrs sample as 1. Mean ± SD (n=3).
(C) Tip60 overexpression can rescue decrease in cellular genes expression. Relative mRNA expression of Vav-3 and E2F-3 at 48 and 64 hrs post infection in cells stably transfected with empty vector or Tip60 expressing plasmid. Level of expression at 48 hrs in vector or Tip60 overexpressing cells is taken as 1. Mean ± SD, n=3. (*P<0.05and **P<0.05 by Student’s t test).
To verify this hypothesis, we measured the expression of both genes in dl309 virus-infected cells that were stably infected with empty vector or with vector overexpressing Tip60. Overexpression of Tip60 partially alleviated the repression of Vav-3 and E2F-3 seen in adenovirus-infected cells (Figure 7C). These results suggest that the degradation of Tip60 by adenovirus results in repression of two cellular genes that are normally up-regulated by Tip60.
Discussion
We suggest that Tip60-dependent acetylation of H4 at the EIA promoter represses the immediate early viral promoter. Since EIA is critically important for expression of other viral genes, the degradation of Tip60 may promote efficient EIA-dependent viral transcription and regulate expression of cellular genes (Figure 8). While this model offers a novel perspective on how the degradation of Tip60 by the adenovirus E1B55K and E4orf6 affects the course of a productive infection, Tip60 degradation may also have consequences for cell transformation by the adenovirus. Different investigative groups have demonstrated that the EIA protein alters the epigenetic program of the cell. 25–28 EIA binds to the promoters of genes involved in the cell cycle and growth, cause hypoacetylation at H3K18 and thus stimulates the cell cycle. EIA also binds to the promoters of antiviral genes and differentiation genes and causes their transcriptional repression. Thus the degradation of Tip60 is expected to affect the expression of several cellular genes through the induction of E1A. Our results suggest that in addition, the degradation of Tip60 directly affect the expression of cellular genes normally regulated by Tip60 (Figure 8).
Figure 8. Model for relief of Tip60 suppression of the EIA promoter by early viral genes and resultant viral production.
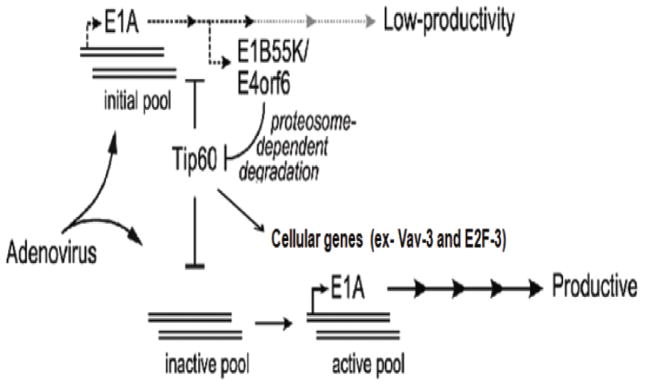
The EIA promoter in a fraction of the input viral genomes is suppressed by Tip60 (inactive pool). Early gene expression from a limited initial pool of active viral genomes leads to expression of the EIB and E4 genes with concomitant degradation of Tip60 through the proteasome. The loss of Tip60 de-represses the EIA promoter which leads to high levels of transcription, viral DNA replication and productive infection.
Adenoviruses can cause tumors in rodents and Syrian hamsters and can transform rodent cells in vitro. In fact, this latter property has been successfully utilized to understand how viral oncoproteins like E1A and E1B55K inactivate a variety of cellular tumor suppressors such as Rb and p53. Since Tip60 is a bona fide tumor suppressor,18 and since Tip60 is known to be degraded by at least one other viral oncogene, HPV E6, we suspect that the degradation of Tip60 by adenovirus may also be important for cell transformation.
Although there is little evidence of the presence of adenovirus in human cancer there are reports that suggest a transient requirement of the virus for cell-transformation, following which the cells lose the virus: the so-called Hit-and-run oncogenesis.29 Increasing evidence suggests that viral oncoproteins can target epigenetic regulators to alter epigenetic regulation of the cells.11 Tip60 is an important epigenetic factor. By acetylating histone H4, Tip60 alters the expression of many cellular genes. Tip60 is also very important for a cell’s response to DNA damage: it activates the checkpoint pathways by activating ATM, 30,31 directs p53 to apoptotic pathways 21,22 and turns off the alarm signal by dephosphorylating phosphoH2AX at the end of the damage response.20, 32–36 Thus it is conceivable that the degradation of Tip60 by an adenovirus infection predisposes the host cell to deleterious mutations caused by DNA damage (Figure 8). Destruction of Tip60 by an adenovirus infection may thus be part of the “Hit-and-run” method of oncogenesis. Once additional mutations convert a cell to the frankly transformed state, the viral oncoprotein may no longer needed.
The adenovirus oncoproteins EIB55K and E4orf6 are required for the destabilization of Tip60 protein. Chronologically the degradation of Tip60 in adenovirus-infected cells is nearly concurrent to the degradation of p53 protein. E1B55K and E4orf6 are also important for the degradation of p53, and they do so by interacting with p53 and recruiting it to a Cullin-5 complex containing elongins B and C.4 Our experiments suggest that Tip60 is recruited similarly by E1B55K and E4orf6, perhaps to Cullin-3, but also perhaps to an unknown E3 ligase complex that is inhibited by the neddylation inhibitor MLN4924.
Two studies showed that pVII protein remains associated with viral DNA during the early stages of infection. The start of viral transcription releases the pVII protein from viral genome. 24, 37 Our results extend it to say that Tip60 and H4 acetylation at the adenovirus EIA promoter may be correlated with pVII retention at the promoter. We do not know whether the acetylation of H4 at the promoter or of some other protein at or away from the promoter is responsible for the pVII retention and repression of the promoter. The minimum hypothesis is that the acetylation of H4 by Tip60 at the E1A promoter recruits a cellular repressor complex, similar to the recruitment of the Brd4 complex at the HPV major early promoter,12 and that this repressor inhibits EIA transcription.
Since Tip60 suppresses EIA expression, which is the major viral transcription factor, we were also interested to see the effect of Tip60 knockdown on viral DNA synthesis. The difference in viral DNA copy number suggests that Tip60 limits the number of viral genomes recruited for DNA replication.
p53 and Rb are major tumor suppressors that are inactivated by diverse oncogenic viruses. Thus it is quite exciting that oncoproteins from at least two different viruses (HPV and adenovirus) inactivate Tip60, another known tumor suppressor. Whether the inactivation of Tip60 by these multiple viruses contributes to viral tumorigenesis or to the viral life cycle are important questions that need to be addressed. It is worth noting that besides HPV E6 and adenovirus E1B55K+E4orf6, some other viral proteins are known to interact with Tip60: pUL27 of CMV, Tat protein of HIV, Herpesvirus conserved kinases and KSHV LANA protein. 13, 38–40 Therefore degradation of or interaction with Tip60 seems to be important for several types of viruses.
Materials and Methods
Cell culture
MCF10A (non-tumorigenic epithelial cell line) cells were cultured in DMEM- F12 medium containing donor calf serum, while U2OS cells (human osteosarcoma cell line) were maintained in DMEM media containing 10% donor calf serum. HCT116 cells (human colorectal carcinoma cell line) were grown in McCoy media containing 10% fetal bovine serum. Cells were maintained in sub-confluent cultures in a 5% CO2 atmosphere at 37°C.
Antibodies and Western blotting
For western blotting cells were lysed in IPH buffer (50mM Tris-HCl pH-8, 150mM NaCl, 5mM EDTA, 0.5%NP40, 1mM dithiothreitol, 20mM NaF and protease inhibitor mix (Sigma)) and then subsequently sonicated. Lysates were immunobloted with indicated antibodies. Tip60 antibodies were used as described elsewhere.12 Other antibodies are as follows: anti-p53 (Santacruz), anti-Flag (Sigma), anti-pVII, 41 anti-EIB55K, 42 E4orf6, 43 anti-GST (Santacruz), anti-β actin (Sigma).
Viruses
The dl309 strain is phenotypically wild type Ad5 which lacks a portion of E3 region. The dl355 mutant virus contains deletion in E4orf6 region while dl366* mutant virus has deletion in entire E4 region. Mutant dl1520 contains a deletion of 827 bp in the 55kDa protein coding region. 44
Cells were passaged 16–20 hr before infection to density of 3×106 cells. Cells were washed with phosphate buffer saline (PBS) and then the medium was replaced with medium without serum with virus (MOI 5) and 100U/ml penicillin and 100ug/ml of streptomycin. The cells were gently rocked intermittently and incubated for 120 min at 37°C. The virus suspension was then removed and cells were grown in normal medium at 37°C.
Plasmid construction, Transfection and Stable cell lines
Construction of human Tip60-expressing plasmids is described elsewhere. 20 Wild Tip60 and HAT mutant Tip60 expressing cell line were made in MCF10A cells.
For siRNA Transfection, 30nM of annealed siRNA duplex (Invitrogen) with Lipofectamine 2000 RNAi max reagent (Invitrogen) was used as per manufacturer’s instructions. Target sequences of oligonucleotides are provided in Supplemental information.
Real Time PCR and Chromatin Immunoprecipitation assay and Luciferase assay
For measuring viral gene and cellular gene expression RNA was isolated from virus-infected cells by TRIzol reagent (Invitrogen) according to the manufacturer’s instructions (Invitrogen). These cDNAs were used as template for real time PCR using SYBR green PCR master mix (Applied Biosystems). Sequences of primers used are in Supplemental information.
For viral DNA quantitation total DNA was isolated from infected cells and viral DNA was quantitated by qPCR using specific primers from AdenoX qPCR titration kit (Clontech, cat- 632253). ChIP assay was performed as described elsewhere.12 Primers used for ChIP PCR are given in Supplemental information.
Luciferase assay was performed by transfecting the firefly Luciferase gene driven by control (GL3) or EIA promoter (EIAGL3), along with Renilla Luciferase expressing plasmid as control. Cells were harvested 48 hr after Transfection. Luciferase assay was done following manufacturer’s instructions (Promega). The firefly Luciferase activity is normalized to renilla Luciferase. The ratio in pGL3c or EIApGL3c (without Tip60) is held as 1.
Supplementary Material
Acknowledgments
We thank the members of Dutta laboratory for lively discussions and helpful comments. This work was supported by RO1 GM084465 to A.D. A.G. was supported by US army Postdoctoral fellowship for breast cancer research DOD-W81XWH1110687.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Moran E. Interaction of adenoviral proteins with pRB and p53. FASEB J. 1993;7:880–885. doi: 10.1096/fasebj.7.10.8344487. [DOI] [PubMed] [Google Scholar]
- 2.Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, et al. Association between an oncogenes and an anti-oncogene: the adenovirus EIA protein binds to retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 3.DeCapri JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, et al. (1988) SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 4.Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, et al. Degradation of p53 by adenovirus E4orf6 and EIB55k proteins occurs via a novel mechanism involving a cullin containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 6.Baker A, Rohleder KJ, Hanakahi LA, Ketner G. Adenovirus E4 34K and E1b55K oncoproteins target host DNA ligase IV for proteasomal degradation. J Virology. 2007;81:7034–7040. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dallaire F, Blanchette P, Groitl P, Dobner T, Branton PE. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B55- kilodalton E3 ubiquitin ligase complex. J Virology. 2009;83:5329–5338. doi: 10.1128/JVI.00089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackford AN, Patel RN, Forrester NA, Theil K, Groitl P, Stewart GS, et al. Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc Natl Acad Sci USA. 2010;107:12251–12256. doi: 10.1073/pnas.0914605107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seligson DB, McBrian MA, Mah V, Yu H, Tze S, Wang Q, et al. Global levels of histone modifications predict prognosis in different cancers. The American journal of Pathology. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari R, Berk AJ, Kurdistani SK. Viral manipulation of the host epigenome for oncogenic transformation. Nature reviews. 2009;10:290–294. doi: 10.1038/nrg2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niller HH, Wolf H, Minarovits J. Viral hit and run oncogenesis: genetic and epigenetic scenarios. Cancer letters. 2011;305:200–217. doi: 10.1016/j.canlet.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Jha S, Pol SV, Banerjee NM, Dutta AB, Chow LT, Dutta A. Destabilization of Tip60 by Human Papillomavirus E6 results in Attenuation of Tip60 Dependent Transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38:700–711. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reitsma JM, Savaryn JP, Faust K, Sato H, Halligan BD, Scott ST. Antiviral inhibition targeting the HCMV kinase pUL97 requires pUL27 dependent degradation of Tip60 acetyltransferase and cell cycle arrest. Cell Host Microbe. 2011;9:103–114. doi: 10.1016/j.chom.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Sterner DE, Berger SL. Acetylation of histones and transcription related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Gentet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Bio. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozyous disruption of Tip60 gene causes early embryonic lethality. Developmental Dynamics. 2009;238:2912–2921. doi: 10.1002/dvdy.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, et al. Tip60 is a haplo- insufficient tumour suppressor required for an oncogenes induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 19.Sancar A, Lindsey Boltz LA, Unsal-Kacmaz K, Linn S. Molecualr mechanism of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 20.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone Acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–51. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y, Luo J, Zhang W, Gu W. Tip60 dependent acetylation of p53 modulates the decision between cell cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Bhoumik A, Singha N, O’Connell MJ, Ronai ZA. Regulation of Tip60 by ATF2 modulates ATM activation. J Biol Chem. 2008;283:17605–17614. doi: 10.1074/jbc.M802030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Morral N, Engel DA. Transcription releases protein VII from adenovirus chromatin. Virology. 2007;369:411–422. doi: 10.1016/j.virol.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. Epigenetic reprogramming by Adenovirus eIa. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus small eia alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh MK, Harter ML. A viral mechanism for remodeling chromatin structure in G0 cells. Mol Cell. 2003;12:255–260. doi: 10.1016/s1097-2765(03)00225-9. [DOI] [PubMed] [Google Scholar]
- 28.Sha J, Ghosh MK, Zhang K, Harter ML. E1A interacts with two opposing transcriptional pathways to induce quiescent cells into S phase. J Virol. 2010;84:4050–4059. doi: 10.1128/JVI.02131-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nevels M, Tauber B, Spruss T, Wolf H, Dobner T. Hit and Run transformation by adenovirus oncogenes. J Virol. 2001;75:3089–3094. doi: 10.1128/JVI.75.7.3089-3094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone Acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Xu Y, Roy K, Price BD. DNA damage induced acetylation of lysine 3016 of ATM kinase activity. Mol Cell Biol. 2007;27:8502–09. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–87. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 33.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 34.Sapountzi V, Logan IR, Robson CN. Cellular functions of Tip60. Int J Biochem Cell Bio. 2006;38:1496–1436. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, et al. DNA damage dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karen KA, Hearing P. Adenovirus core protein VII protects viral genome from a DNA damage response at early times after infection. J Virology. 2011;85:4135–4142. doi: 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Col E, Caron C, Chable-Bessia C, Legube G, Gazzeri S, Komatsu Y, et al. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 2005;24:2634–2645. doi: 10.1038/sj.emboj.7600734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li R, Zhu J, Xie Z, Liao G, Liu J, Chen MR, et al. Conserved Herpesvirus Kinases Target the DNA damage pathway and Tip60 Histone Acetyltransferase to promote virus replication. Cell Host & Microbe. 2011;10:390400. doi: 10.1016/j.chom.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shamay M, Liu J, Li R, Liao G, Shen L, Greenway M, et al. A protein array screen for Kaposi’s Sarcoma associated Hepresvirus LANA interactors link LANA to Tip60, PP2A activity, and telomere shortening. J Virology. 2012;86:5179–5191. doi: 10.1128/JVI.00169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JS, Osheim YN, Xue Y, Emanuel MR, Lewis PW, Bankovich A, et al. Adenovirus protein VII condenses DNA, represses transcription and associate with transcriptional activator EIA. J Virology. 2004;78:6459–68. doi: 10.1128/JVI.78.12.6459-6468.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zantema A, Fransen JA, Davis-olivier A, Ramaekers FC, Vooijs GP, Deleys B, et al. Localization of the EIB proteins of adenovirus 5 in transformed cells as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
- 43.Marton MJ, Bain SB, Ornelles DA, Shenk T. The adenovirus E4 17kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA binding properties and stimulating EIA independent accumulation of E2 mRNA. J Virology. 1990;64:2345–59. doi: 10.1128/jvi.64.5.2345-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barker DD, Berk AJ. Adenovirus proteins from both E1B reading frames are required for the transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.