Abstract
Phosphoinositide-dependent kinase-1 (PDK-1) is a serine/threonine protein kinase that phosphorylates members of the conserved AGC kinase superfamily, including AKT and PKC, and is implicated in important cellular processes including survival, metabolism and tumorigenesis. In large cohorts of nevi and melanoma samples, PDK1 expression was significantly higher in primary melanoma, compared with nevi, and was further increased in metastatic melanoma. PDK1 expression suffices for its activity, due to auto-activation, or elevated phosphorylation by phosphoinositide 3'-OH-kinase (PI 3-K). Selective inactivation of Pdk1 in the melanocytes of BrafV600E::Pten−/− or BrafV600E::Cdkn2a−/−::Pten−/− mice delayed the development of pigmented lesions and melanoma induced by systemic or local administration of 4-HT. Melanoma invasion and metastasis were significantly reduced or completely prevented by Pdk1 deletion. Administration of the PDK1 inhibitor GSK2334470 (PDKi) effectively delayed melanomagenesis and metastasis in BrafV600E::Pten−/− mice. Pdk1−/− melanomas exhibit a marked decrease in the activity of AKT, P70S6K and PKC. Notably, PDKi was as effective in inhibiting AGC kinases and colony forming efficiency of melanoma with Pten WT genotypes. Gene expression analyses identified Pdk1-dependent changes in FOXO3a-regulated genes and inhibition of FOXO3a restored proliferation and colony formation of Pdk1−/− melanoma cells. Our studies provide direct genetic evidence for the importance of PDK1, in part through FOXO3a-dependent pathway, in melanoma development and progression.
Keywords: PDK1, FOXO3a, melanoma, Braf, Pten, GSK2334470
Introduction
PDK1 is a serine/threonine protein kinase that phosphorylates over 20 members of the conserved AGC kinase superfamily, including members of the protein kinases A (PKA), G (PKG, including PKB/AKT) and C (PKC) families.1, 2 PDK1 is activated upon phosphorylation of a threonine or serine residue in the activation loop (T-loop), which is mediated by either auto-phosphorylation or by the phosphoinositide 3'-OH-kinase (PI 3-K).3 Consequently, PDK1 activity can be dependent on phosphatidylinositol-3,4-bisphosphate (PtdIns-3,4-P2) and phosphatidylinositol-3,4-5-trisphosphate (PtdIns-3,4,5-P3), the lipid products of PI 3-K, which requires it to anchor to the plasma membrane.4, 5 It can also function independently of anchoring to the membrane as an active kinase through its auto-activation.4, 5 Accordingly, PDK1 is an important regulator of cellular processes including protein translation, cell survival, metabolism, and tumor development and progression.
In cancer, PDK1 activation has been associated with genomic and epigenetic mechanisms. A gain of the 16p13.3 locus, which spans the PDPK1 region harboring the PDK1 gene, was found in lymph node metastasis and in castration-resistant prostate cancer samples,1 has been associated with poor differentiation of late stage lung cancer2 and with poor prognosis of breast cancer patients.4 Increased PDK1 activity is implicated in enhanced tumor cell proliferation, reduced apoptosis, and angiogenesis.4, 5 PDK1 was shown capable of augmenting tumorigenesis in tissues harboring ERBB2 amplifications,4 PTEN deletions,5 and mutations in the catalytic subunit of phosphoinositide 3-kinase (PIK3CA).6 Inhibition of PDK1 is therefore expected to attenuate tumors associated with deregulated PIK3CA/PTEN signaling. Indeed, hypomorphic mutation of PDK1 in Pten+/− mice delays the onset of tumorigenesis,7 and small molecule inhibitors of PDK1 inhibit tumor xenografts and lung colonization.8, 9 Further, Pdk1 inactivation effectively attenuated the development of Kras oncogene-driven pancreatic cancer, but not NSCLC 10, further supporting the importance of PDK1 in tumor development, albeit, in select cancer types. PDK1 expression in melanoma has not been assessed, nor was the significance of its genetic inactivation for melanoma development and progression evaluated.
Crosstalk between the MAPK and AGC signaling pathways has been implicated in the development and progression of melanoma and for its resistance to therapy.11–13 Our earlier studies showed that crosstalk between PKC and JNK augments the activities of JNK,14 and that crosstalk between ERK and c-Jun increases both the transcription and activity of c-Jun.15 c-Jun is an important transcriptional activator of PDK1.16 Notably, expression of PDK1 is sufficient to restore tumor growth after c-Jun knockdown in melanoma cells,16 suggesting that PDK1 is an important mediator of c-Jun oncogenic activities.
To assess the role of PDK1 in melanoma formation and progression, we used a genetic mouse model driven by melanocyte-specific expression of BrafV600E and inactivation of Pten, achieved using the Tyr::CreER transgene that encodes conditionally active CreERT2 specifically in melanocytes. These mice develop melanoma with 100% penetrance, short latency, and with metastases in lymph nodes, lungs, and spleen.17, 18 Given the relevance of Cdkn2a locus in melanoma, we have also developed a new model in which the Cdkn2a locus has been deleted, on the background of the Braf/Pten mutant animals.
Results
Inactivation of Pdk1 prolongs latency and reduces size of Braf/Pten melanoma
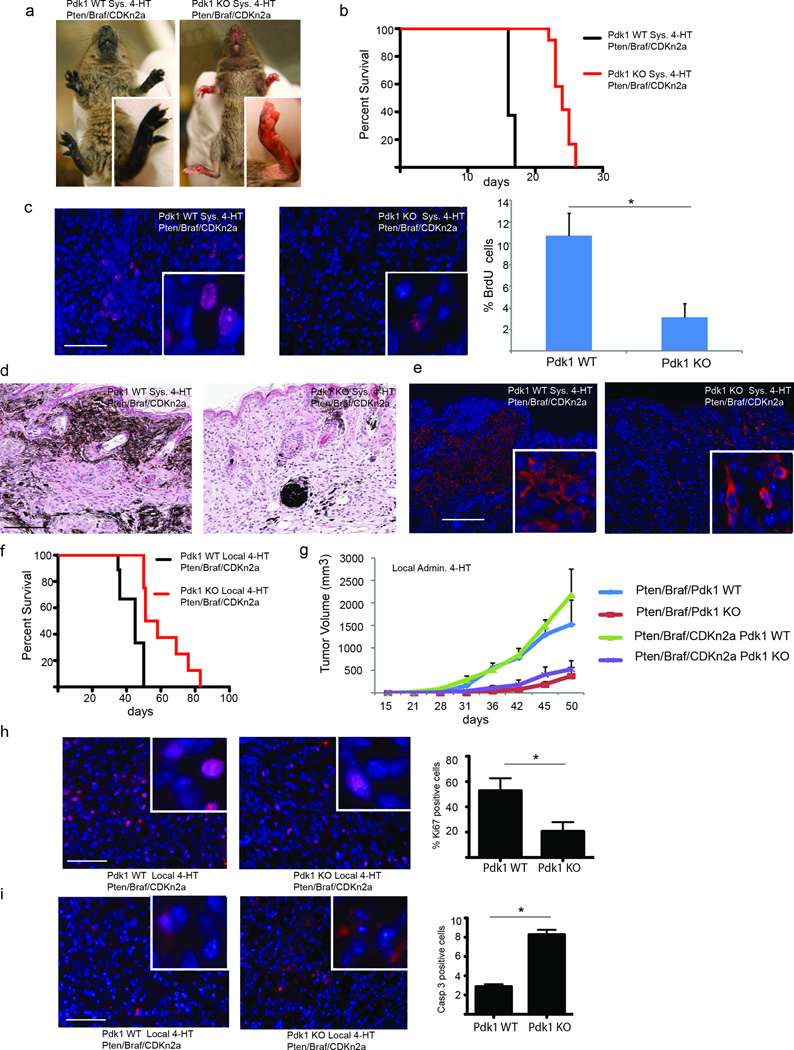
Systemic administration of 4-hydroxytamoxifen (4-HT) to the Tyr::CreER::BrafV600E::Ptenlox/lox and Tyr::CreER::BrafV600E::Cdkn2alox/lox::Ptenlox/lox mice (days 1, 3, 5 following birth) resulted in the appearance of highly pigmented lesions within 7–10 days (Fig. 1a, Fig. S1c, S2e-f). Selective inactivation of Pdk1 in the melanocytes and tumors developed in these animals (see Fig. S1a, S1b, S1d) delayed the development of pigmented lesions increasing the overall survival (17 days in BrafV600E:: Cdkn2a−/−::Pten−/−::Pdk1+/+ and 20 days in BrafV600E::Pten−/−::Pdk1+/+, versus 26 days in BrafV600E:: Cdkn2a−/−::Pten−/−::Pdk1−/−, and 28 days in BrafV600E::Pten−/−::Pdk1−/− animals; Fig. 1b and Fig. S2a). Correspondingly, rate of melanocyte proliferation was markedly attenuated (~80%; Fig. 1c). Histological analysis of primary skin lesions revealed highly pigmented cells with variably shaped enlarged nuclei (Fig. 1d and Fig. S2b), which were confirmed to be of melanocytic origin by immunostaining for tyrosinase (Fig. 1e). The number of pigmented melanoma cells found throughout the dermis and subcutis with pagetoid spread into the epidermis was decreased in the Pdk1−/− genotype (Fig. 1d), providing the initial indication that PDK1 plays a role in melanocyte biology (i.e., pigmentation) and transformation.
Figure 1. Loss of PDK1 delays onset of melanoma development.
(a) Representative pictures of pigmented animals 17 days following perinatal administration of 4-HT to Pdk1 WT (BrafV600E::Pten−/−::Cdkn2a−/−::Pdk1+/+) or Pdk1 KO (BrafV600E::Pten−/−::Cdkn2a−/−::Pdk1−/−) animals. (b) Kaplan-Meier survival curve of mice with the indicated Pdk1 WT (n = 8) and KO (n = 12) genotypes under the genetic background of BrafV600E::Pten−/−::Cdkn2a−/−, following perinatal administration of 4-HT. Log-rank (Mantel-Cox) test of survival plots reveals a statistically significant difference between the Pdk1 WT and KO genotypes (p < 0.0001). (c) Representative immunostaining for proliferation marker (BrdU; red) in skin sections from mice harboring the Pdk1 WT or KO genotypes (on the background of BrafV600E::Pten−/−::Cdkn2a−/−). Quantification shown is of >500 cells in three separate analyses (*P < 0.05 using t-test). (d and e) H&E stain (d) and Tyrp1 immunostaining (e) of skin from BrafV600E::Pten−/−::Cdkn2a−/−::Pdk1+/+ and BrafV600E::Pten−/−::Cdkn2a−/−::Pdk1−/− 17 days following 4-HT administration. (f) Kaplan-Meier survival curve of mice with the indicated Pdk1 WT or KO genotypes (n = 9 for each group) under the genetic background of BrafV600E::Pten−/−::Cdkn2a−/−, following local administration of 4-HT. Log-rank (Mantel-Cox) test of survival plots reveals a statistically significant difference (p = 0.0005) between the Pdk1 WT and KO genotypes. (g) Tumor growth curves for BrafV600E/Pten−/− and BrafV600E/Pten−/−::Cdkn2a−/− genotypes (n = 9 for each group). (h) Immunostaining for proliferation marker (Ki67 stain, panel H) and programed cell death (cleaved caspase 3, panel i) of locally induced melanomas of the indicated Pdk1 WT or KO genotypes (BrafV600E::Pten−/−::Cdkn2a−/−). Quantification of >1000 cells in four analyses is shown. Error bars represent SEM. *P < 0.05. Bar = 50 µm.
Notably, inactivation of the cell cycle regulator Cdkn2a in BrafV600E::Pten−/− mice results in more aggressive and faster growing tumors, consistent with the known role of Cdkn2a in melanoma development. Significantly, the contribution of Pdk1 to melanoma development was even more pronounced in the BrafV600E::Pten−/−::Cdkn2a−/− mice, as shown by the increased survival rate upon inactivation of Pdk1 (Fig. 1b).
Local, rather than systemic, administration of 4-HT to adult animals (day 21) allows tumor formation to be monitored at the point of application.17 As was observed upon systemic administration of 4-HT, overall survival was prolonged by Pdk1 deletion (Fig. S1e), from 59 to 88 days for the BrafV600E::Pten−/−::Cdkn2a+/+ mice and from 47 to 82 days for the BrafV600E::Pten−/−::Cdkn2a−/− mice (Fig. 1f and Fig. S2c). The tumor volumes in these mice were also markedly reduced upon Pdk1 inactivation, from 1400 mm3 to 300 mm3 in BrafV600E::Pten−/−::Cdkn2a+/+ and from 2000 mm3 to 400 mm3 in the BrafV600E::Pten−/−::Cdkn2a−/− mice (Fig. 1g). Moreover, rate of melanocyte proliferation was reduced by 50% and cell death was increased 3-fold by Pdk1 inactivation in BrafV600E::Pten−/− mice (Fig. 1h, Fig. 1i). A decreased nuclear to cytoplasmic ratio, often associated with altered PDK1/AGC kinase activity, was noted in the Pdk1 deficient melanomas (see Fig. S2d).
The median survival time of BrafV600E::Pten−/− mice was extended by inactivation of Pdk1 by ~40% (from 20 to 28 days) in animals receiving 4-HT systemically (Fig. S2a) and by ~55% (from 52 to 82 days) in mice with locally administered 4-HT (Fig. S2b). These findings provide genetic evidence for the involvement of Pdk1 in melanoma development, as reflected by the longer disease latency and reduced tumor size in the Pdk1−/− genotypes. Analysis of Pdk1+/− genotypes did not reveal such differences (data not shown), suggesting that partial inhibition of Pdk1 may not suffice to inhibit melanoma development and or progression.
Pdk1 is required for melanoma metastases
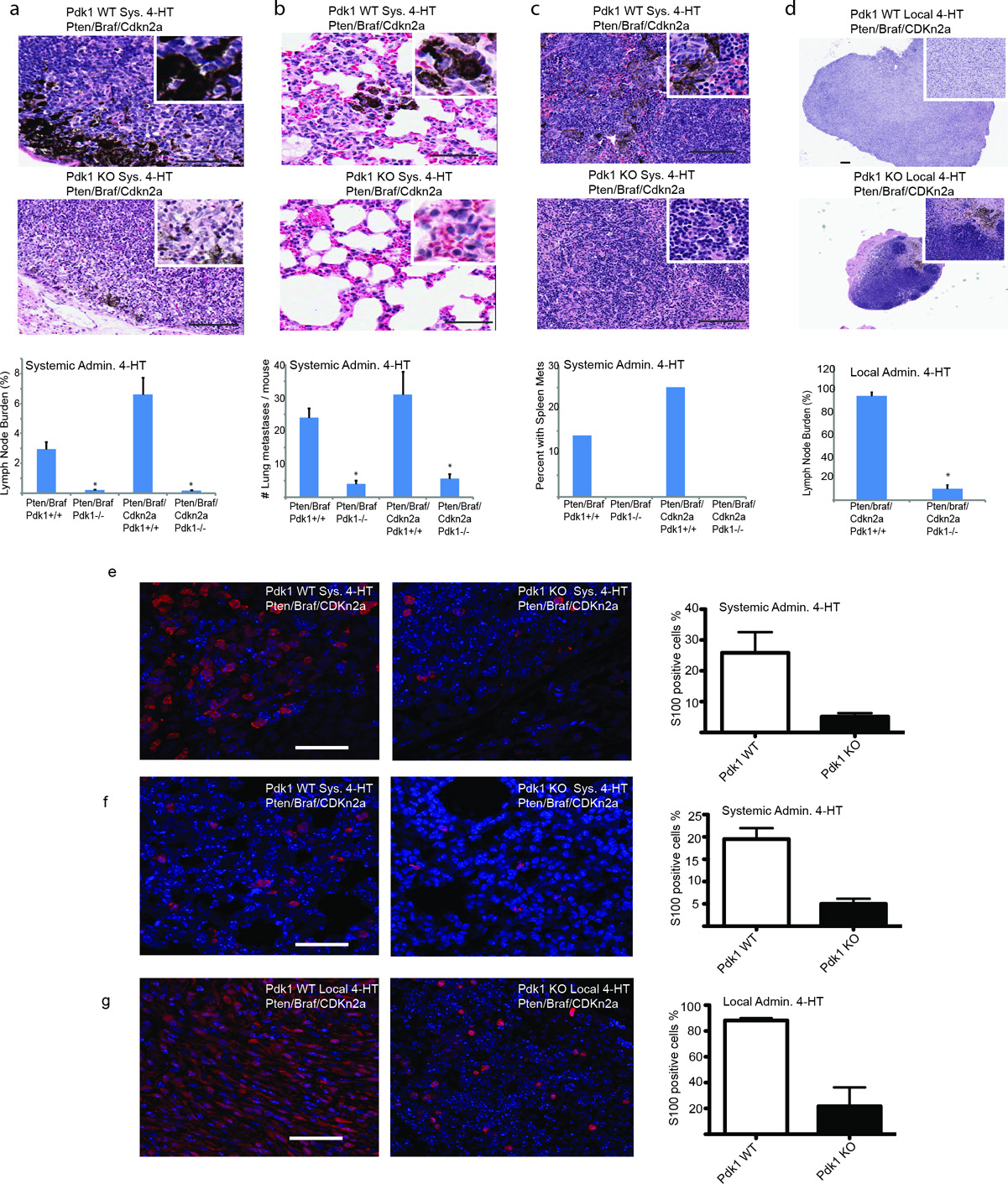
Mice that received 4-HT systemically showed evidence of melanoma spread into the regional lymph nodes and had lung and spleen metastases, consistent with earlier reports.17 Metastasis to lymph nodes, lungs, and spleen was significantly lower in the Pdk1−/− genotypes of both mouse models, with a more pronounced inhibition seen in the BrafV600E::Pten−/−::Cdkn2a−/− animals (Fig. 2a-c, Fig. S2e-f). Independent assessment for the degree of metastases was performed by staining lymph nodes and lungs with S100, a marker with excellent sensitivity for pigmented and amelanotic melanoma (Fig. 2e-f), although within the lymph node compartment it may also stain follicular dendritic cells, which are distinguishable based on the cytologic features and location within the lymph node. Melanin however is remarkably good at absorbing light and therefore S100 stain could not detect those clusters of tumor cells with strong melanin pigmentation that were easily detected by H&E. Notably, quantitation based on both S100 immunofluorescence and H&E staining revealed a pronounced inhibition of metastases in the BrafV600E::Pten−/−::Cdkn2a−/− animals (Fig. 2a-c, 2e, 2f). Similarly, metastatic lesions in lymph nodes of animals subjected to local 4-HT administration were also significantly inhibited by deletion of Pdk1 (~80% reduction) (Fig. 2d and Fig 2g). These findings suggest that Pdk1 plays a critical role in metastasis of the BrafV600E::Pten−/− melanoma. The inhibition of metastasis is likely to be uncoupled from the delay in tumorigenesis, as degree of inhibition seen in lymph nodes and lungs was remarkable, likely to be independent of delayed proliferation.
Figure 2. Genetic inactivation of Pdk1 inhibits metastases.
(a-c) Representative pictures for &E stain of lymph node (a), lung (b), and spleen (c) 16 days after systemic administration of 4-HT to induce expression of BrafV600E and inactivation of Pten, Cdkn2a and Pdk1. Quantifications shown on the lower panels represent analyses from eight animals for each group. P < 0.0005. The quantification of tumor metastasis was performed using immunohistochemical and H&E stains in a blinded manner by an expert histopathologist (MB). Pigment deposition in lung metastases correlates well with tumor deposits in the lung, but does not in lymph nodes, where melanin pigment-laden macrophages are commonly observed in lymph node sinuses. Lung metastases were uniformly small (less than 0.5 mm) occurred as clusters of tumor cells. These clusters were counted on a full cross section of lung to quantitate metastatic burden. The lymph node deposits in Pdk1 WT (and untreated) melanomas were larger and more irregular, so the percentage of the cross sectional area of a full section of lymph node was used to determine metastatic burden to lymph nodes. (d) Representative H&E stain of lymph node after local induction of melanoma in BrafV600E::Pten−/−::Cdkn2a−/− for Pdk1 WT and Pdk1 KO genotypes. Eight lymph nodes were quantified from eight animals for each group. (e-f) Representative pictures of S100 stain of lymph node (e) and lung (f) 16 days after systemic administration of 4-HT to induce expression of BrafV600E and inactivation of Pten, Cdkn2a and Pdk1. (g) Representative S100 stain of lymph node after local induction of melanoma in BrafV600E::Pten−/−::Cdkn2a−/− for Pdk1 WT and Pdk1 KO genotypes. Quantifications shown on the right panels represent analyses from three animals for each group. Error bars represent SEM. *P < 0.005. Bars in panels a-b, e-g = 50 µm, bar in panel c = 100 µm, bar in panel d = 200 µm.
Key AGC kinases and FOXO3a mediate Pdk1 signaling in melanomagenesis
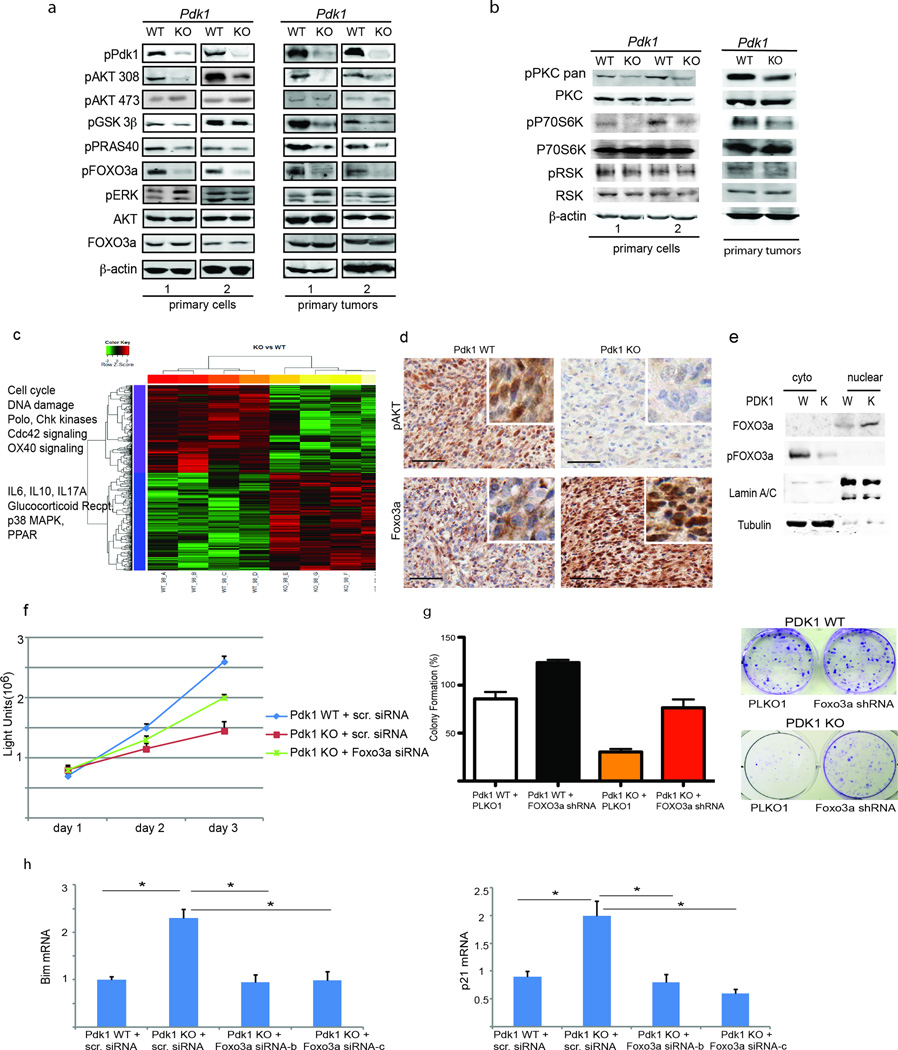
To determine the molecular mechanisms by which PDK1 contributes to melanomagenesis, we analyzed changes in key signaling pathways associated with PDK1. Tumors and tumor-derived primary cultures from BrafV600E::Pten−/−::Cdkn2a−/−::Pdk1−/− mice showed markedly decreased phosphorylation of AKTT308, the primary PDK1 phosphoacceptor site, and of the AKT substrates PRAS40 and GSK3β (Fig. 3a), demonstrating that AKT signaling is significantly reduced in the absence of functional PDK1. AKT analysis was made using pan AKT antibodies, recognizing all three AKT forms. Correspondingly, phosphorylation of S6K, another AGC kinase substrate for PDK1, was markedly attenuated (Fig. 3b). We expect that other AKT substrates were also affected upon PDK1 inactivation. Members of the PKC family are also among the AGC kinases that require phosphorylation by PDK1 for maturation, stabilization, and subsequent activation.19, 20 Consistent with this, both the level and extent of phosphorylation of PKC were notably lower in BrafV600E::Pten−/−::Cdkn2a−/−::Pdk1−/− melanomas, while the decrease observed in RSK phosphorylation was not (Fig. 3b). These findings demonstrate that Pdk1 inactivation in melanocytes impairs key AGC signaling pathways.
Figure 3. Pdk1 inactivation attenuates AKT and PKC signaling.
(a and b) Western blot analysis was performed using protein lysates from melanoma tumors and their derived primary melanomas cultures under the genetic background BrafV600E::Pten−/−::Cdkn2a−/− with the indicated antibodies. (c) Heat map showing select genes and pathways that were up- or down-regulated in Pdk1–/::BrafV600E::Pten−/−::Cdkn2a−/− melanomas (n = 4). The selected genes have p-values less than or equal to 0.05 and fold changes are >2. The normalized expression signals in the heat map are shown from green (lower) to red (higher signal). Significant genes with similar expression pattern are indicated in bars at the left of the heat map (see Tables S1 to S4 for details). (d) Representative immunostaining of pAKT 308 and Foxo3a in the Pdk1 WT and KO melanomas (BrafV600E::Pten−/−::Cdkn2a−/−). Enlarged areas are shown in insets. (e) Nuclear and cytoplasmic fractionation was performed using protein lysates from primary melanoma cultures under the genetic background BrafV600E::Pten−/−::Cdkn2a−/−, western blot was performed and the membranes were probed with the indicated antibodies. (f) Primary melanoma cultures established from the indicated Pdk1 WT and KO tumors (BrafV600E::Pten−/−::Cdkn2a−/−) were analyzed for cell proliferation (ATP Lite assay) following their transfection with Foxo3a siRNA expression vector. (g) Cultures used in panel (e) were monitored for colony formation assay (representative image of colonies formed in culture is shown on the right panel) following their infection with Foxo3a shRNA expression vector. The graph represents the quantification of the number of colony-forming cells after 7 days in culture. Analysis was performed in triplicates and repeated two times. (h) Cultures used in panel (f) were monitored for changes in gene expression following transfection with two different Foxo3a siRNAs, using QPCR analysis for Bim and p21 mRNA levels. *P < 0.0005.
We next performed a non-biased assessment of changes in gene expression in melanomas from Pdk1-inactivated BrafV600E::Pten−/−::Cdkn2a−/− mice by performing gene expression profiling analysis. Principal component analysis (PCA) for the microarray data confirmed that Pdk1-inactivation appeared to be the most significant effect since the biological replicates in each group were clustered together (Fig. S3a).
To identify the differentially expressed genes, linear modeling and empirical Bayesian statistics approaches were applied.21 Genes with both a p value below 0.05 and a change over ±2-fold were considered statistically different between BrafV600E::Pten−/−::Cdkn2a−/−::Pdk+/+ and BrafV600E::Pten−/−::Cdkn2a−/−::Pdk1−/− groups, and a total of 827 significant differentially expressed genes were identified. As shown in the heat map (Fig. 3c), 431 genes were up-regulated in Pdk1 knockout tumors (Table S1) and 396 were down-regulated (Table S2). Network and functional analyses using Ingenuity Pathways Analysis (IPA, Ingenuity Systems) revealed that these up- and down-regulated genes were classified into a number of functional groups. Among canonical pathways found to exhibit the significant enriched (Fisher test p value ≤ 0.05) in up-regulated genes are IL-6, IL-10, eicosanoid, and glucocorticoid receptor signaling, as well as fibrosis-associated pathways (Table S3). In contrast, pathways associated with cell cycle, DNA replication, mitosis, and the DNA damage response was significantly enriched in the down-regulated gene set in the Pdk1 knockout tumors (Table S4). The changes observed for representative genes from each of the major three clusters were validated by qPCR analysis (Fig. S3c).
Interestingly, a number of these up-regulated and down-regulated genes are commonly regulated by the transcription factor FOXO3a (Fig. S3b), which is negatively regulated by PDK1 substrates AKT or SGK. PDK1-dependent changes in transcription of the FOXO3a-related gene set were confirmed by qPCR, while no changes were observed in pigmented genes expression (Fig. S3c). Furthermore, immunostaining of tumor sections revealed strong nuclear localization of FOXO3a in Pdk1−/− tumors, which correlated with reduced AKT phosphorylation (Fig. 3d), as this kinase has been implicated in the control of FOXO3a phosphorylation-dependent nuclear export.22, 23 These findings suggest that Pdk1 inactivation and concomitant inhibition of AKT activity cause nuclear retention and transcriptional activation of FOXO3a. Nuclear cytoplasmic fractionation of primary melanoma cell lysates confirmed increased nuclear FOXO3a protein in PDK KO cells, thereby corroborating the immunohistochemical data (Fig 3e). To determine the role of a transcriptionally active FOXO3a in melanomagenesis we have inhibited FOXO3a expression (Fig. S3d-e) in BrafV600E::Pten−/−:: Cdkn2a−/−::Pdk1−/− tumor-derived cells. FOXO3a KD increased the rate of proliferation (Fig. 3f) and partially restored the ability of melanoma cells to form colonies (Fig. 3g), while attenuating changes in the expression of FOXO3a-related genes identified in the gene expression microarrays (Fig. 3h). These results establish a regulatory link between PDK1 and FOXO3a and demonstrate its role in melanoma development and progression.
PDK1 inhibitor phenocopies genetic inactivation
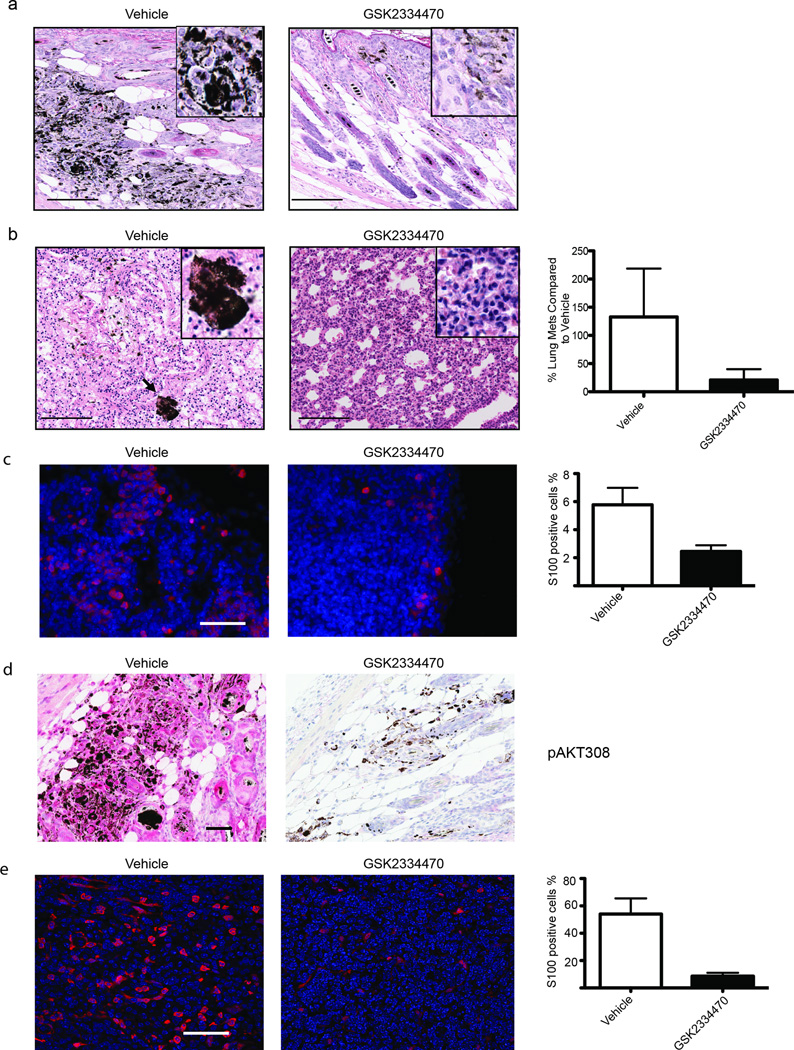
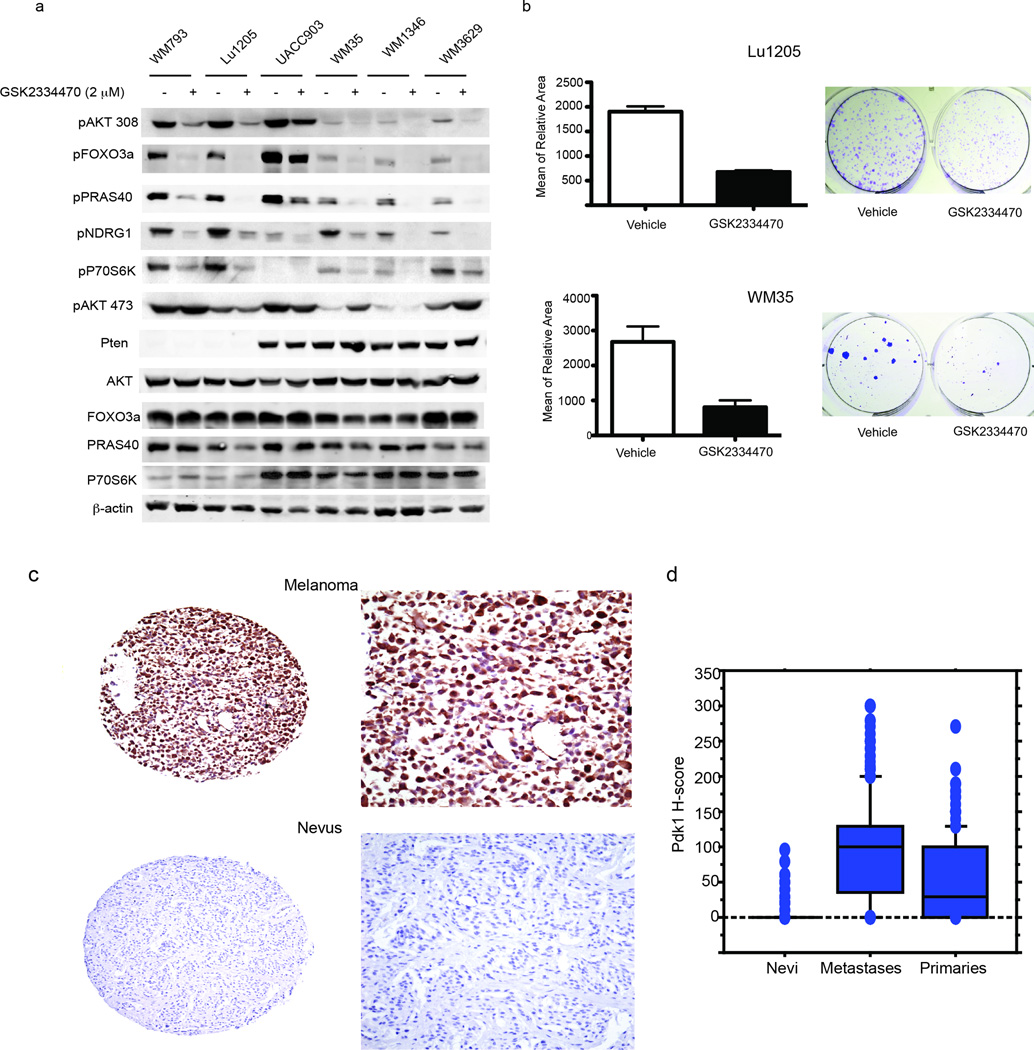
Combinations of PI3K/AKT signaling inhibitors and targeted therapies against BRAF are currently being evaluated in clinical trials for melanoma. Therefore, we asked whether attenuation of PDK1 activity with the available PDK1 inhibitors would phenocopy the genetic inactivation of Pdk1 in the BrafV600E::Pten−/− animals. For this, we tested the efficacy of the PDK1 inhibitor (PDKi) GSK233447024–26 (Fig. S4a) in newborn BrafV600E::Pten−/− mice subjected to systemic administration of 4-HT. Twice weekly administration of PDKi resulted in marked inhibition of pigmented lesions and concomitant melanomagenesis (Fig. 4a), as well as significant inhibition of lung metastases, seen by H&E staining-based quantification (~80%, Fig. 4b), and lymph node metastases as by S100 immunostaining (Fig 4c), similar to the phenotype seen upon genetic ablation of Pdk1 (Fig. 2a, 2b). AKT, but not PKC, signaling was also efficiently inhibited in primary tumor cells (Fig. S4b). Further, immunohistochemistry skin from mice treated with PDKi revealed a marked decrease in pAKT308 (Fig 4d). It is noteworthy that while administration of PDKi did not inhibit development of localized melanomas (local application of 4-HT), it reduced metastasis burden in lymph nodes as quantitated by S100 stain (Fig. 4e). The reduced effectiveness of PDKi in locally induced melanomas suggests that either the route/frequency of administration and/or dose of PDKi may be insufficient, or that alterations in signaling in melanoma developed in the adult animals (e.g., increased local PI3K signaling24, 27) may have limited PDKi efficacy. It is also possible that enzymes not affected by the PDKi (i.e., PKC 27) may play more pronounced roles in locally induced melanomas.25, 26 We also compared the effect of PDKi on human melanoma cell lines with wild type (WT) versus inactivated Pten (IC50 estimated to be <10 µM for these cell lines). Notably, melanoma cells with inactivated Pten exhibited increased phosphorylation of the PDK1 substrates AKT, when compared to melanoma cells carrying WT Pten. Yet, PDKi effectively attenuated AKTT308 and p70S6K phosphorylation, as well as the phosphorylation of the AKT downstream substrate PRAS40, the phosphorylation of the SGK downstream substrate NDRG1, and phosphorylation of the AKT/SGK substrate FOXO3a, independent of the Pten mutation (Fig. 5a). Notably, inhibition of phosphorylated PRAS40 was seen upon PDKi treatment despite increased AKTS473 phosphorylation; the latter was previously reported and attributed to mTORC2.24 These findings suggest that PDKi effectively attenuates number of key AGC kinases and respective substrates, independent of Pten status and AKT activity. Correspondingly, colony formation assay was used to assess clonal growth of the human melanoma cell lines Lu1205 (Pten inactive) and WM35 (Pten WT) that were either treated with vehicle or PDKi. Notably, PDKi decreased the size of colonies in both Pten WT and mutant melanomas, compared to the vehicle treated cells (Fig. 5b). These data suggest that pharmacological inhibition of PDK1 is as effective in attenuating AGC kinases activated in both Pten WT and mutant melanoma cells.
Figure 4. PDK1 inhibitor GSK2334470 delays melanoma formation and inhibits metastases.
(a and b) representative H&E stain of skin (a) and lungs (b) in vehicle and GSK2334470 treated mice (BrafV600E::Pten−/−::PDK1−/−) following perinatal administration of 4-HT. Quantification shown for number of lung metastases is based on six sections 100 µm apart that were analyzed from three mice. Bar = 100 µm. (c) Representative S100 stain of lymph node in vehicle and GSK2334470 treated mice (BrafV600E::Pten−/−::PDK1−/−) following perinatal administration of 4-HT. Bar = 50 µm (d) Representative immunostaining of pAKT308 in the skin of vehicle and GSK2334470 treated mice following perinatal administration of 4-HT (BrafV600E::Pten−/−::PDK1−/−). Bar = 50 µm. (e) Representative S100 stain of lymph node in vehicle and GSK2334470 treated mice (BrafV600E::Pten−/−::PDK1−/−) following local administration of 4-HT. Bar = 50 µm. Quantifications shown on the right panels of panels c and e represent analyses from three animals for each group. Error bars represent SEM.
Figure 5. PDK1 inhibition attenuates AGC kinases in both Pten WT and mutant melanomas.
(a) Western blot analysis was performed using protein lysates from human melanoma cells with Pten mutation (793, Lu1205, UACC903) or Pten WT (WM35, WM1346, and WM3629) with the indicated antibodies. (b) Human melanoma cell lines Lu1205 and WM35 were treated with either vehicle or GSK2334470 (2.5 µM, changing the media every other day) and monitored for colony formation assay. The graph represents the quantification of the mean relative area after 7 days (Lu1205) and 12 days (WM35) in culture (representative images of colonies grown in culture are shown on the right panel). Analysis was performed in triplicates and repeated two times. Error bars represent SEM. (c) Expression of PDK1 was studied in a large cohort of primary and metastatic melanomas and in nevi. Examples of strong and weak staining are shown. (d) Box plots demonstrate differences in expression in the three categories of samples, with the PDK1 H-score (scale 0–300) on the Y-axis. Expression was significantly lower in nevi and primary lesions than metastases (P = 0.0001).
Increased PDK1 expression in metastatic human melanoma
We used tumor microarrays to determine whether PDK1 expression levels correlate with melanocytic neoplastic progression and clinical outcome. Using a standard H score system we found that over 90% of the positive tissue samples expressed PDK1 in both cytoplasmic and nuclear compartments (Fig. 5c). This analysis of more than 700 tumors revealed significantly higher expression of PDK1 in melanomas compared with nevi (Fig. 5d; p < 0.001). Similarly, expression was higher in metastatic relative to primary melanoma specimens and higher in melanomas than in nevi (Fig. 5c; p < 0.001 for both comparisons). No association was found between PDK1 expression and melanoma specific survival within the primary or metastatic subsets. PDK1 expression was higher in lesions thicker than 1 mm (p = 0.01) and in ulcerated lesions (p = 0.004). These findings substantiate the importance of PDK1 in melanoma development and progression and point to its possible use as a marker in melanoma.
Discussion
Growing evidence points to the activation of PDK1 in cancer, through genomic or epigenetic changes, often associated with more aggressive tumors and poorer prognosis.1, 2, 4 Consistent with these reports, genetic inactivation of Pdk1 resulted in effective inhibition of Kras-driven pancreatic cancer, but not in non-small cell lung cancer.10 The present study provides the first genetic evidence for the importance of PDK1 in melanoma. Genetic inactivation of Pdk1 delayed the onset of melanomas and almost completely abolished metastases in both the systemic and local BrafV600E::Pten−/− mouse melanoma models. Gene expression profiling has pointed to the possible role of FOXO3a in BrafV600E::Pten−/−:: Cdkn2a−/−::Pdk1−/− melanomas, a finding that was confirmed upon inhibition of FOXO3a expression, which restored the tumor cells’ ability to form colonies in culture. As both AKT and SGK1 are capable to phosphorylate and cause nuclear exclusion of FOXO3a, further studies will determine the possible role of SGK1 in FOXO3a regulation in melanoma cells. Further, PDK1 inhibition was as effective on the AGC kinase AKT, SGK, P70S6K, and downstream substrates NDRG1 and FOXO3a, in both Pten WT and mutant melanomas, indicating that the core components of the PDK1 pathway are equally inhibited, independent of AKT phosphorylation. Future studies could elaborate on the role of distinct AGC kinases in melanoma harboring different genetic backgrounds (i.e., Pten WT, Cdkn2a WT, or Nras mutant). The genetic data supporting an important role for PDK1 in melanoma development and metastasis is supported by the use of PDKi. The administration of PDKi to systemically activated BrafV600E::Pten−/−:: Cdkn2a–/– animals, effectively reduced melanomagenesis and metastatic load, phenocopying the genetic inactivation. Notably, PDKi was not effective in inhibition of tumors initiated in the local model, suggesting that either improved efficacy or combination with other inhibitors would be warranted. The possibility that exposure of PDKi may result in mTORC-2-driven AKT activation and AKT-driven resistance was recently proposed.25 Although this possibility was not evaluated in the present study, we also observed increased AKTS473 phosphorylation, upon PDKi treatment, albeit, with effective inhibition of PRAS40 phosphorylation, a primary AKT substrate. Partial inhibition of Pdk1, as seen in the heterozygous BrafV600E::Pten−/−::Pdk1+/− animals, was not sufficient to inhibit metastases and delay tumor development, indicating that efficient inhibition of PDK1 and its downstream targets will be required for achieving an effective therapeutic index, similar to targeting other therapeutic targets where over 50% of inhibition needs to be achieved. It is important to note that neither the tissue specific genetic inactivation nor the general pharmacological inhibition of PDK1 result in toxicity, implying that higher doses and more potent PDK1 inhibitors can be tolerated. These findings suggest that PDK1 inhibitors could be tested in clinical trials in combination with BRAFi or MEKi. Accordingly, an intervention study with mice that have established large tumors showing the utility of PDKi alone with BRAFi would substantiate the clinical utility of this approach. It should be noted that some studies targeting PDK1 with small molecule inhibitors or RNAi failed to show efficacy in cancer models.28, 29 This difference suggests that PDK1 inhibition may affect tumor growth and metastases via mechanisms that vary with cancer type and involve varying microenvironmental factors (e.g., stroma or innate immunity), which are lacking in commonly used xenograft models. Taken together, our demonstration that PDK1 and its downstream effectors play a critical role in melanoma development and metastases offers new opportunities for evaluation of therapeutic modalities for melanoma and other tumors expressing high levels of PDK1.
Materials and Methods
Primary melanoma cells
Murine melanoma cells from BrafV600E::Pten−/−::Cdkn2a−/− tumors were derived by physical resection of primary cutaneous lesions. Cell suspensions were plated in media (DMEM) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Once established, cell lines were passaged 2 times prior to starting the experiments. Cell survival, was monitored using ATP-Lite.
Activation of the Tyr::CreERT2 transgene
Topical administration of 4-hydroxytamoxifen (4-HT) was performed by preparing 50 mg/ml solution of 4-HT in DMSO and applying 10 µl with a small paint brush on the back skin on post natal days 1, 3 and 5. For localized melanoma induction on the back skin, adult (3 weeks of age) mice were shaved and treated topically with 1.5 µl of 7.8 mg/ml of 4-HT in ethanol vehicle using a pipette. We estimate that 15–20% of cells within the tumor are macrophages, lymphocytes and endothelial cells.
Histological analyses
Tumors and sections were fixed in Z-fix (Anatech) overnight, washed twice with PBS and processed for paraffin embedding. Sections were sliced at 5 µm and stained with hematoxylin and eosin.
Antibodies and reagents
Antisera against TYRP1 was a kind gift from Dr. Hearing (National Cancer Institute). Anti-cleaved caspase 3, pNDRG1, pPDK1, pAKT308, pAKT473, pGSK3β (Ser 9), pPRAS40 (Thr 246), pFOXO3a (Thr 32), pERK1/2, AKT, FOXO3a, pPKC pan (γ Thr514), pS6K (Thr389), pRSK (Ser 227) were purchased from Cell Signaling Technologies. Monoclonal anti β-actin, anti-lamin A/C, and tubulin were purchased from Santa Cruz. Antibodies against Ki-67, and 4-hydroxytamoxifen were purchased from Sigma. Antibodies to S100 were obtained from Dako.
Gene silencing and transfection
Pre-designed siRNA for mouse FOXO3a (NM_019740) was purchased from Ambion and siRNA was transfected with Lipofectamine RNAiMAX (Invitrogen) according to protocol supplied by the vendor. Mouse FOXO3a shRNA constructs were from Open Biosystems (TRCN0000071614 and TRCN0000071617). pLKO.1 empty vector was used for control cells. The cells were selected in full media containing 1.5 µg/ml puromycin.
Western blotting
To extract whole cell lysate, cells were harvested using RIPA buffer. Cell lysates were subjected to SDS-PAGE and proteins transferred onto a nitrocellulose membrane (Osmonics Inc.). The membrane was probed with primary antibodies followed by a secondary antibody conjugated with fluorescent dye and detected by the Odyssey detecting system (Amersham/Bioscience).
Nuclear and cytoplasmic lysate extraction
The fractionation was done using the nuclear extract kit from Active Motif following the manufacturer’s protocol.
Immunohistochemistry and immunofluorescence
All sections were deparaffinized, rehydrated, washed in PBS, and blocked with Dako protein block for 30 min at room temperature. Antigen retrieval was performed in a pressure cooker (Decloaking chamber, Biocare Medical) in citrate buffer (pH 6.0) and used for Tyrp1, cleaved caspase3, Ki-67, Pdk1, pAKT (308), and FOXO3a immunostaining. Sections immunostained for biotinylated-bromodeoxyuridine (BrdUrd; Abcam) were pretreated with 2 N HCl for 15 min at room temperature and 0.01% protease (type XXIV, Sigma) for 15 s. Antibodies/dilutions for the following markers were used in Dako antibody diluent and applied overnight at 4°C. Secondary antibodies labeled with Alexa Fluor 488, Alexa Fluor 594, or Alexa Fluor 594 streptavidin conjugate were placed on tissue sections for 1 h at room temperature (1:400, Molecular Probes). Nuclei were counterstained using SlowFade Gold Anti-fade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Vector). For immunohistochemistry analysis the staining was visualized using either an immunoperoxidase technique (Vectastain ABC kit, Vector Laboratories), and diaminobenzadine (Dako) or an alkaline phosphatase technique (Vector red alkaline substrate kit I, Vector Laboratories).
Tissue microarray (TMA) analysis
TMAs consisting melanoma or nevi cohorts were previously described,30 and were stained using PDK1 antibodies (Abcam). Briefly, TMA consist of 747 neoplastic lesions (209 primary cutaneous, 312 metastatic or cutaneous origin and 226 nevi). PDK1 levels were measured by a dermatopathologist, who scored the intensity on a scale of 0–3. Expression in the nucleus and cytoplasm was concordant, and therefore a single score was included for each patient. The percent of cells staining positive for PDK1 expression was multiplied by the intensity score, which provided scores that ranged between 0 and 300. (i.e. intensity score of 2 seen in 75% of cells provides an H score of 150, intensity score of 1 in 50% of cells yields an H score 50).
Microarray analysis
RNA (500 ng) from each experimental group was pooled to enable four biological replicates and used for synthesis of biotin-labeled cRNA using an RNA amplification kit (Ambion). The cRNA labeled with streptavidin-Cy3 was used for hybridization with the Mouse Ref-8 v2.0 Expression BeadChip (Illumina) (~25K mouse transcripts). The BeadChips were scanned and data were pre-processed by Illumina GenomeStudio software without background correction or normalization (threshold detection p-values = 0.05). The redefined list consisted of 15,922 probes detected in the experimental samples.
Raw data were log2-transformed and quantile normalized using neqc function in limma package.31 To identify differentially expressed genes, the linear modeling approach and empirical Bayes statistics as implemented in the limma package were employed.21 The Benjamini-Hochberg method was used to correct for the multiple comparison errors.32 Principal component analysis (PCA) was performed with Partek Genomics Suite (Partek Inc.), and hierarchical clustering and other statistical analyses were performed using R/Bioconductor.33 Genes with at least a 2-fold change at the 95% confidence level were considered as significant. Functional enrichments were performed using (IPA) software (Ingenuity Systems Inc.). The microarray data were deposited in the NCBI GEO database, accession number GSE42187.
qRT-PCR analysis
Total RNA was extracted using a total RNA miniprep kit (Sigma) and digested with DNase I. cDNA was synthesized using oligo-dT and random hexomer primers for SYBR Green QPCR analysis. H3.3A were used as internal controls. At least triplicate biological samples were used for the QPCR analysis. The PCR primers were designed using Primer3 and their specificity was checked using BLAST. The PCR products were limited to 100–200 bp. The primers used for QPCR analysis are available upon request.
BrdUrd incorporation
Mice were injected ip with 100 mg/kg BrdUrd (Sigma). Tissue was collected 3 h after BrdUrd injection for measurement of cell replication. Incorporated BrdUrd was detected as described above.
Quantification of lymph node and lung metastasis
Metastases to lymph nodes were observed only 2–5 weeks following primary tumor induction, restricted to draining lymph nodes, supporting metastasis from a regional primary tumor. Fixed and embedded lung tissues were sliced to obtain five serial sections per lung. H&E stain was used to quantify the number and size of metastases under a histology microscope. For lymph node metastases, H&E stained lymph node were photographed and analyzed with Image-J. Percent lymph node involvement was defined as the surface area of the sub-capsular tumor metastasis relative to the surface area of the entire lymph node. For quantitation of S100 positive cells all slides were scanned at a magnification of 20X using the Aperio ScanScope FL system (Aperio Technologies) and quantified using the “Area Quantification FL” algorithm (version 11 Aperio Technologies) the algorithm was “tuned” using a preset procedure and the subsequent macro was saved and applied to all slides. For quantitation of BrdU, cleaved caspase 3, and S100 positive cells along with DAPI positive cells were counted in six random 20x fields per mouse.
Colony assay formation
Five hundred cells were plated into 6-well plates. After 7–12 days in culture viable colonies were stained with Cristal Violet (Sigma-Aldrich). Colony number and size were determined using ImageJ.
Statistical analysis
The data were analyzed by unpaired t-test and p < 0.05 was considered statistically significant. Kaplan-Meyer survival curve were compiled using Prism statistical analysis software. Significance was assessed using the Log-rank (Mantel-Cox) test.
Synthesis of PDK1 inhibitor GSK2334470
GSK2334470 was synthesized according to the reported procedure as shown in Figure S4.30 Purification of GSK2334470 was obtained in a HPLC Breeze from Waters Co. using an Atlantis T3 3 µm 4.6×150 mm reverse phase column. Synthesis of (3S, 6R)-1-[6-(3-amino-1H-indazol-6-yl)-2-methylamino)-4-pyrimidinyl]-N-cyclohexyl-6-methyl-3-piperidinecarboxamide (5, GSK2334470) was followed according to the published procedure 34, HRMS calculated for C25H35N8O (M+H) 463.2934, found 463.2936. Purity: 99% (HPLC).
Animal studies and in vivo experiments
The Institutional Animal Care and Use Committee (IACUC) of Sanford-Burnham Medical Research Institute approved our study protocols. BrafV600E::Pten−/− and Pdk1−/− were generated as previously described.17, 35 Cohorts of six animals per group were used in each experimental group. GSK224470 was administered through IP injection (100 mg/kg) 3 times per week starting the same day of topical administration of 4-hydroxytamoxifen and ending at the time of mouse collection, based on earlier studies.34
Supplementary Material
Acknowledgments
We thank Dario Alessi for providing the conditional Pdk1−/− mice, Vincent Hearing for the Tyrp1 antibody, and Pedro Aza-Blanc for siRNA library work. Support by NIH grants CA099961 (to ZR) and CA128814 (to ZR and MP) and a Melanoma Research Alliance grant (to ZR) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
References
- 1.Choucair KA, Guerard KP, Ejdelman J, Chevalier S, Yoshimoto M, Scarlata E, et al. The 16p13.3 (PDPK1) Genomic Gain in Prostate Cancer: A Potential Role in Disease Progression. Translational oncology. 2012;5:453–460. doi: 10.1593/tlo.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen H, Zhu Y, Wu YJ, Qiu HR, Shu YQ. Genomic alterations in lung adenocarcinomas detected by multicolor fluorescence in situ hybridization and comparative genomic hybridization. Cancer genetics and cytogenetics. 2008;181:100–107. doi: 10.1016/j.cancergencyto.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Casamayor A, Morrice NA, Alessi DR. Phosphorylation of Ser-241 is essential for the activity of 3-phosphoinositide-dependent protein kinase-1: identification of five sites of phosphorylation in vivo. The Biochemical journal. 1999;342(Pt 2):287–292. [PMC free article] [PubMed] [Google Scholar]
- 4.Maurer M, Su T, Saal LH, Koujak S, Hopkins BD, Barkley CR, et al. 3- Phosphoinositide-dependent kinase 1 potentiates upstream lesions on the phosphatidylinositol 3- kinase pathway in breast carcinoma. Cancer research. 2009;69:6299–6306. doi: 10.1158/0008-5472.CAN-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finlay DK, Sinclair LV, Feijoo C, Waugh CM, Hagenbeek TJ, Spits H, et al. Phosphoinositide-dependent kinase 1 controls migration and malignant transformation but not cell growth and proliferation in PTEN-null lymphocytes. The Journal of experimental medicine. 2009;206:2441–2454. doi: 10.1084/jem.20090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayascas JR, Leslie NR, Parsons R, Fleming S, Alessi DR. Hypomorphic mutation of PDK1 suppresses tumorigenesis in PTEN(+/−) mice. Current biology : CB. 2005;15:1839–1846. doi: 10.1016/j.cub.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 8.Miao B, Skidan I, Yang J, Lugovskoy A, Reibarkh M, Long K, et al. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to pleckstrin homology domains. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20126–20131. doi: 10.1073/pnas.1004522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagashima K, Shumway SD, Sathyanarayanan S, Chen AH, Dolinski B, Xu Y, et al. Genetic and pharmacological inhibition of PDK1 in cancer cells: characterization of a selective allosteric kinase inhibitor. The Journal of biological chemistry. 2011;286:6433–6448. doi: 10.1074/jbc.M110.156463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, et al. Selective Requirement of PI3K/PDK1 Signaling for Kras Oncogene-Driven Pancreatic Cell Plasticity and Cancer. Cancer cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Bagrodia S, Smeal T, Abraham RT. Mechanisms of intrinsic and acquired resistance to kinase-targeted therapies. Pigment cell & melanoma research. 2012 doi: 10.1111/pcmr.12007. [DOI] [PubMed] [Google Scholar]
- 12.Vultur A, Herlyn M. Cracking the system: melanoma complexity demands new therapeutic approaches. Pigment cell & melanoma research. 2009;22:4–5. doi: 10.1111/j.1755-148X.2008.00527.x. [DOI] [PubMed] [Google Scholar]
- 13.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes & development. 2012;26:1131–1155. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Bergami P, Habelhah H, Bhoumik A, Zhang W, Wang LH, Ronai Z. RACK1 mediates activation of JNK by protein kinase C [corrected] Molecular cell. 2005;19:309–320. doi: 10.1016/j.molcel.2005.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Bergami P, Huang C, Goydos JS, Yip D, Bar-Eli M, Herlyn M, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Bergami P, Kim H, Dewing A, Goydos J, Aaronson S, Ronai Z. c-Jun regulates phosphoinositide-dependent kinase 1 transcription: implication for Akt and protein kinase C activities and melanoma tumorigenesis. The Journal of biological chemistry. 2010;285:903–913. doi: 10.1074/jbc.M109.075630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nature genetics. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, et al. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis. 2006;44:262–267. doi: 10.1002/dvg.20205. [DOI] [PubMed] [Google Scholar]
- 19.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Seminars in cell & developmental biology. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Bayascas JR. PDK1: the major transducer of PI 3-kinase actions. Current topics in microbiology and immunology. 2010;346:9–29. doi: 10.1007/82_2010_43. [DOI] [PubMed] [Google Scholar]
- 21.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 22.You H, Jang Y, You-Ten AI, Okada H, Liepa J, Wakeham A, et al. p53-dependent inhibition of FKHRL1 in response to DNA damage through protein kinase SGK1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14057–14062. doi: 10.1073/pnas.0406286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H, Tindall DJ. Dynamic FoxO transcription factors. Journal of cell science. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 24.Najafov A, Sommer EM, Axten JM, Deyoung MP, Alessi DR. Characterization of GSK2334470, a novel and highly specific inhibitor of PDK1. The Biochemical journal. 2011;433:357–369. doi: 10.1042/BJ20101732. [DOI] [PubMed] [Google Scholar]
- 25.Najafov A, Shpiro N, Alessi DR. Akt is efficiently activated by PIF-pocket- and PtdIns(3,4,5)P3-dependent mechanisms leading to resistance to PDK1 inhibitors. The Biochemical journal. 2012;448:285–295. doi: 10.1042/BJ20121287. [DOI] [PubMed] [Google Scholar]
- 26.Axten JM, Grant SW, Heerding DA, Medina JR, Romeril SP, Tang J. In: Chemical Compounds. A1 ed. Office USP, editor. USA: 2011. [Google Scholar]
- 27.Skurk C, Izumiya Y, Maatz H, Razeghi P, Shiojima I, Sandri M, et al. The FOXO3a transcription factor regulates cardiac myocyte size downstream of AKT signaling. The Journal of biological chemistry. 2005;280:20814–20823. doi: 10.1074/jbc.M500528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellwood-Yen K, Keilhack H, Kunii K, Dolinski B, Connor Y, Hu K, et al. PDK1 attenuation fails to prevent tumor formation in PTEN-deficient transgenic mouse models. Cancer research. 2011;71:3052–3065. doi: 10.1158/0008-5472.CAN-10-2282. [DOI] [PubMed] [Google Scholar]
- 29.Gagliardi PA, di Blasio L, Orso F, Seano G, Sessa R, Taverna D, et al. 3-phosphoinositide-dependent kinase 1 controls breast tumor growth in a kinase-dependent but akt-independent manner. Neoplasia. 2012;14:719–731. doi: 10.1593/neo.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aziz SA, Davies M, Pick E, Zito C, Jilaveanu L, Camp RL, et al. Phosphatidylinositol-3-kinase as a therapeutic target in melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:3029–3036. doi: 10.1158/1078-0432.CCR-08-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi W, Oshlack A, Smyth GK. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic acids research. 2010;38:e204. doi: 10.1093/nar/gkq871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B, Statistical methodology. 1995;57:289–300. [Google Scholar]
- 33.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medina JR, Becker CJ, Blackledge CW, Duquenne C, Feng Y, Grant SW, et al. Structure-based design of potent and selective 3-phosphoinositide-dependent kinase-1 (PDK1) inhibitors. Journal of medicinal chemistry. 2011;54:1871–1895. doi: 10.1021/jm101527u. [DOI] [PubMed] [Google Scholar]
- 35.Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, et al. Essential role of PDK1 in regulating cell size and development in mice. The EMBO journal. 2002;21:3728–3738. doi: 10.1093/emboj/cdf387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.