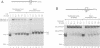Abstract
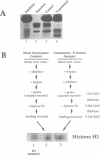
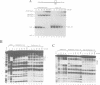
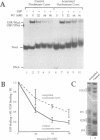
In order to investigate the interrelated roles of nucleosome cores and histone H1 in transcription repression, we have employed a purified system to analyze the function of H1 in the repression of transcription factor binding to nucleosomes. H1 binding to nucleosome cores resulted in the repression of USF binding to nucleosomes. By contrast, H1 only slightly inhibited the binding of GAL4-AH, indicating that H1 differentially represses the binding of factors with different DNA-binding domains. H1-mediated repression of factor binding was dependent on the core histone amino-terminal tails. Removal of these domains alleviated H1-mediated repression and increased acetylation of these domains partly alleviated repression by H1. H1 binding assays suggest a less stable interaction of histone H1 with the core particle in the absence of the amino termini.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Workman J. L. Nucleosome displacement in transcription. Cell. 1993 Feb 12;72(3):305–308. doi: 10.1016/0092-8674(93)90109-4. [DOI] [PubMed] [Google Scholar]
- Allan J., Cowling G. J., Harborne N., Cattini P., Craigie R., Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981 Aug;90(2):279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Harborne N., Rau D. C., Gould H. Participation of core histone "tails" in the stabilization of the chromatin solenoid. J Cell Biol. 1982 May;93(2):285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986 Feb 20;187(4):591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- Allegra P., Sterner R., Clayton D. F., Allfrey V. G. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J Mol Biol. 1987 Jul 20;196(2):379–388. doi: 10.1016/0022-2836(87)90698-x. [DOI] [PubMed] [Google Scholar]
- Annunziato A. T., Seale R. L. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 1983 Oct 25;258(20):12675–12684. [PubMed] [Google Scholar]
- Archer T. K., Cordingley M. G., Wolford R. G., Hager G. L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991 Feb;11(2):688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J. Structure and dynamics of transcriptionally active chromatin. J Cell Sci. 1992 May;102(Pt 1):1–5. doi: 10.1242/jcs.102.1.1. [DOI] [PubMed] [Google Scholar]
- Bone J. R., Lavender J., Richman R., Palmer M. J., Turner B. M., Kuroda M. I. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994 Jan;8(1):96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993 Apr;7(4):592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., Bustin M., Marsaud V., Richard-Foy H., Hager G. L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992 Jan 25;20(2):273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm L., Crane-Robinson C. Proteases as structural probes for chromatin: the domain structure of histones. Biosci Rep. 1984 May;4(5):365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- Chen H., Li B., Workman J. L. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 1994 Jan 15;13(2):380–390. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipev C. C., Wolffe A. P. Chromosomal organization of Xenopus laevis oocyte and somatic 5S rRNA genes in vivo. Mol Cell Biol. 1992 Jan;12(1):45–55. doi: 10.1128/mcb.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. L., Halay E. D., Lai E., Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993 Jul 29;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Croston G. E., Laybourn P. J., Paranjape S. M., Kadonaga J. T. Mechanism of transcriptional antirepression by GAL4-VP16. Genes Dev. 1992 Dec;6(12A):2270–2281. doi: 10.1101/gad.6.12a.2270. [DOI] [PubMed] [Google Scholar]
- Csordas A. On the biological role of histone acetylation. Biochem J. 1990 Jan 1;265(1):23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J., Quinn J., Workman J. L., Peterson C. L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994 Jul 1;265(5168):53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Dedon P. C., Soults J. A., Allis C. D., Gorovsky M. A. Formaldehyde cross-linking and immunoprecipitation demonstrate developmental changes in H1 association with transcriptionally active genes. Mol Cell Biol. 1991 Mar;11(3):1729–1733. doi: 10.1128/mcb.11.3.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S. I., Stefanovsky VYu, Karagyozov L., Angelov D., Pashev I. G. The enhancers and promoters of the Xenopus laevis ribosomal spacer are associated with histones upon active transcription of the ribosomal genes. Nucleic Acids Res. 1990 Nov 11;18(21):6393–6397. doi: 10.1093/nar/18.21.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumuis-Kervabon A., Encontre I., Etienne G., Jauregui-Adell J., Méry J., Mesnier D., Parello J. A chromatin core particle obtained by selective cleavage of histones by clostripain. EMBO J. 1986 Jul;5(7):1735–1742. doi: 10.1002/j.1460-2075.1986.tb04418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Gottesfeld J. M. Chromosomal footprinting of transcriptionally active and inactive oocyte-type 5S RNA genes of Xenopus laevis. Nucleic Acids Res. 1990 Oct 25;18(20):6031–6037. doi: 10.1093/nar/18.20.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., Burley S. K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994 Jan 1;13(1):180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ramirez M., Dong F., Ausio J. Role of the histone "tails" in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992 Sep 25;267(27):19587–19595. [PubMed] [Google Scholar]
- Hayes J. J., Wolffe A. P. Histones H2A/H2B inhibit the interaction of transcription factor IIIA with the Xenopus borealis somatic 5S RNA gene in a nucleosome. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1229–1233. doi: 10.1073/pnas.89.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Wolffe A. P. Preferential and asymmetric interaction of linker histones with 5S DNA in the nucleosome. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6415–6419. doi: 10.1073/pnas.90.14.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T. R., Clayton A. L., Thorne A. W., Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994 Apr 15;13(8):1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T. R., Thorne A. W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988 May;7(5):1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Thomas J. O. Core histone-DNA interactions in sea urchin sperm chromatin. The N-terminal tail of H2B interacts with linker DNA. Eur J Biochem. 1990 Jan 12;187(1):145–153. doi: 10.1111/j.1432-1033.1990.tb15288.x. [DOI] [PubMed] [Google Scholar]
- Hong L., Schroth G. P., Matthews H. R., Yau P., Bradbury E. M. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 "tail" to DNA. J Biol Chem. 1993 Jan 5;268(1):305–314. [PubMed] [Google Scholar]
- Imbalzano A. N., Kwon H., Green M. R., Kingston R. E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994 Aug 11;370(6489):481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Jeppesen P., Turner B. M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993 Jul 30;74(2):281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Johnson E. M., Sterner R., Allfrey V. G. Altered nucleosomes of active nucleolar chromatin contain accessible histone H3 in its hyperacetylated forms. J Biol Chem. 1987 May 25;262(15):6943–6946. [PubMed] [Google Scholar]
- Juan L. J., Walter P. P., Taylor I. C., Kingston R. E., Workman J. L. Nucleosome cores and histone H1 in the binding of GAL4 derivatives and the reactivation of transcription from nucleosome templates in vitro. Cold Spring Harb Symp Quant Biol. 1993;58:213–223. doi: 10.1101/sqb.1993.058.01.026. [DOI] [PubMed] [Google Scholar]
- Kamakaka R. T., Thomas J. O. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990 Dec;9(12):3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov V. L., Preobrazhenskaya O. V., Mirzabekov A. D. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5' region. Cell. 1984 Feb;36(2):423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991 Jul 26;66(2):317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Irresistible force meets immovable object: transcription and the nucleosome. Cell. 1991 Nov 29;67(5):833–836. doi: 10.1016/0092-8674(91)90354-2. [DOI] [PubMed] [Google Scholar]
- Kwon H., Imbalzano A. N., Khavari P. A., Kingston R. E., Green M. R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994 Aug 11;370(6489):477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Lambert S. F., Thomas J. O. Lysine-containing DNA-binding regions on the surface of the histone octamer in the nucleosome core particle. Eur J Biochem. 1986 Oct 1;160(1):191–201. doi: 10.1111/j.1432-1033.1986.tb09957.x. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993 Jan 15;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Li B., Adams C. C., Workman J. L. Nucleosome binding by the constitutive transcription factor Sp1. J Biol Chem. 1994 Mar 11;269(10):7756–7763. [PubMed] [Google Scholar]
- Li Q., Wrange O. Translational positioning of a nucleosomal glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev. 1993 Dec;7(12A):2471–2482. doi: 10.1101/gad.7.12a.2471. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Carey M. F., Ptashne M., Green M. R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988 Aug 26;54(5):659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Initiation on chromatin templates in a yeast RNA polymerase II transcription system. Genes Dev. 1992 Dec;6(12A):2282–2287. doi: 10.1101/gad.6.12a.2282. [DOI] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Meersseman G., Pennings S., Bradbury E. M. Chromatosome positioning on assembled long chromatin. Linker histones affect nucleosome placement on 5 S rDNA. J Mol Biol. 1991 Jul 5;220(1):89–100. doi: 10.1016/0022-2836(91)90383-h. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Norton V. G., Imai B. S., Yau P., Bradbury E. M. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989 May 5;57(3):449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- Norton V. G., Marvin K. W., Yau P., Bradbury E. M. Nucleosome linking number change controlled by acetylation of histones H3 and H4. J Biol Chem. 1990 Nov 15;265(32):19848–19852. [PubMed] [Google Scholar]
- Paranjape S. M., Kamakaka R. T., Kadonaga J. T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- Perry C. A., Annunziato A. T. Histone acetylation reduces H1-mediated nucleosome interactions during chromatin assembly. Exp Cell Res. 1991 Oct;196(2):337–345. doi: 10.1016/0014-4827(91)90269-z. [DOI] [PubMed] [Google Scholar]
- Perry C. A., Annunziato A. T. Influence of histone acetylation on the solubility, H1 content and DNase I sensitivity of newly assembled chromatin. Nucleic Acids Res. 1989 Jun 12;17(11):4275–4291. doi: 10.1093/nar/17.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña B., Brüggemeier U., Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990 Mar 9;60(5):719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Pognonec P., Kato H., Sumimoto H., Kretzschmar M., Roeder R. G. A quick procedure for purification of functional recombinant proteins over-expressed in E.coli. Nucleic Acids Res. 1991 Dec 11;19(23):6650–6650. doi: 10.1093/nar/19.23.6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikov Y. V., Shick V. V., Belyavsky A. V., Khrapko K. R., Brodolin K. L., Nikolskaya T. A., Mirzabekov A. D. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 1991 Feb 25;19(4):717–725. doi: 10.1093/nar/19.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993 Mar 18;362(6417):219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Laskey R. A. Assembly of nucleosomes and chromatin in vitro. Methods Enzymol. 1989;170:575–585. doi: 10.1016/0076-6879(89)70065-3. [DOI] [PubMed] [Google Scholar]
- Ridsdale J. A., Davie J. R. Chicken erythrocyte polynucleosomes which are soluble at physiological ionic strength and contain linker histones are highly enriched in beta-globin gene sequences. Nucleic Acids Res. 1987 Feb 11;15(3):1081–1096. doi: 10.1093/nar/15.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale J. A., Hendzel M. J., Delcuve G. P., Davie J. R. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J Biol Chem. 1990 Mar 25;265(9):5150–5156. [PubMed] [Google Scholar]
- Sandaltzopoulos R., Blank T., Becker P. B. Transcriptional repression by nucleosomes but not H1 in reconstituted preblastoderm Drosophila chromatin. EMBO J. 1994 Jan 15;13(2):373–379. doi: 10.1002/j.1460-2075.1994.tb06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990 Jan 25;343(6256):387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Staynov D. Z., Crane-Robinson C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 1988 Dec 1;7(12):3685–3691. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Mitchell M. Generation of different nucleosome spacing periodicities in vitro. Possible origin of cell type specificity. J Mol Biol. 1988 Oct 20;203(4):1029–1043. doi: 10.1016/0022-2836(88)90127-1. [DOI] [PubMed] [Google Scholar]
- Svaren J., Hörz W. Histones, nucleosomes and transcription. Curr Opin Genet Dev. 1993 Apr;3(2):219–225. doi: 10.1016/0959-437x(93)90026-l. [DOI] [PubMed] [Google Scholar]
- Svaren J., Klebanow E., Sealy L., Chalkley R. Analysis of the competition between nucleosome formation and transcription factor binding. J Biol Chem. 1994 Mar 25;269(12):9335–9344. [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991 Jul;5(7):1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. M. Decoding the nucleosome. Cell. 1993 Oct 8;75(1):5–8. [PubMed] [Google Scholar]
- Turner B. M. Histone acetylation and control of gene expression. J Cell Sci. 1991 May;99(Pt 1):13–20. doi: 10.1242/jcs.99.1.13. [DOI] [PubMed] [Google Scholar]
- Vettese-Dadey M., Walter P., Chen H., Juan L. J., Workman J. L. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994 Feb;14(2):970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman J. L., Abmayr S. M., Cromlish W. A., Roeder R. G. Transcriptional regulation by the immediate early protein of pseudorabies virus during in vitro nucleosome assembly. Cell. 1988 Oct 21;55(2):211–219. doi: 10.1016/0092-8674(88)90044-x. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Buchman A. R. Multiple functions of nucleosomes and regulatory factors in transcription. Trends Biochem Sci. 1993 Mar;18(3):90–95. doi: 10.1016/0968-0004(93)90160-o. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G., Kingston R. E. An upstream transcription factor, USF (MLTF), facilitates the formation of preinitiation complexes during in vitro chromatin assembly. EMBO J. 1990 Apr;9(4):1299–1308. doi: 10.1002/j.1460-2075.1990.tb08239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman J. L., Taylor I. C., Kingston R. E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991 Feb 8;64(3):533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Taylor I. C., Kingston R. E., Roeder R. G. Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell Biol. 1991;35:419–447. doi: 10.1016/s0091-679x(08)60582-8. [DOI] [PubMed] [Google Scholar]
- Zhao K., Käs E., Gonzalez E., Laemmli U. K. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993 Aug;12(8):3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J., Van Holde K. Histone H1 and transcription: still an enigma? J Cell Sci. 1992 Dec;103(Pt 4):889–895. doi: 10.1242/jcs.103.4.889. [DOI] [PubMed] [Google Scholar]