Summary
Hunger is a hard-wired motivational state essential for survival. Agouti-related peptide (AgRP)-expressing neurons in the arcuate nucleus (ARC) at the base of the hypothalamus are crucial to its control. They are activated by caloric deficiency and, when naturally or artificially stimulated, they potently induce intense hunger and subsequent food intake1-5. Consistent with their obligatory role in regulating appetite, genetic ablation or pharmacogenetic inhibition of AgRP neurons decreases feeding3,6,7. Excitatory input to AgRP neurons is key in caloric-deficiency-induced activation, and is notable for its remarkable degree of caloric state-dependent synaptic plasticity8-10. Despite the important role of excitatory input, its source(s) has been unknown. Here, through the use of Cre-recombinase-enabled, cell-specific neuron mapping techniques, we have discovered strong excitatory drive that, unexpectedly, emanates from the hypothalamic paraventricular nucleus, specifically from subsets of neurons expressing Thyrotropin-releasing hormone (TRH) and Pituitary adenylate cyclase-activating polypeptide (PACAP). Pharmaco-genetic stimulation of these afferent neurons in sated mice markedly activates AgRP neurons and induces intense feeding. Conversely, acute inhibition in mice with caloric deficiency-induced hunger decreases feeding. Discovery of these afferent neurons capable of triggering hunger advances understanding of how this intense motivational state is regulated.
To identify monosynaptic inputs to AgRP neurons, we used a modified rabies virus SADΔG-EGFP (EnvA)11 in combination with Cre-dependent helper adeno-associated viruses (AAVs) expressing TVA (receptor for the avian sarcoma leucosis virus glycoprotein EnvA; AAV8-FLEX-TVA-mCherry) and RG (rabies envelope glycoprotein;AAV8-FLEX-RG). When used with Agrp-ires-Cre mice, TVA and RG, respectively, allow for rabies infection of AgRP neurons and subsequent retrograde transynaptic spread11,12 (Fig. 1a). Importantly, AAV targeting of the helper viruses was specific to AgRP neurons (Supplementary Fig. 1). Three weeks post-AAV transduction, we injected SADΔG-EGFP (EnvA) into the same area and examined brains 7 days later for EGFP+ signal. We detected the largest number of EGFP+ cells in the ARC (38%), likely representing the initially infected AgRP neurons, and possibly local afferents (Fig. 1b; Supplementary Fig. 2). We next evaluated distant upstream anatomical areas for EGFP+ neurons and found that the overwhelming majority were located in two hypothalamic nuclei, the dorsal medial hypothalamus (DMH; 26%) which contains both glutamatergic and GABAergic neurons13 and the paraventricular hypothalamus (PVH;18%) consisting primarily of glutamatergic neurons13 (Fig. 1b; Supplementary Fig. 2). Finally, we also observed a smaller number of EGFP+ cells in other hypothalamic sites (Supplementary Fig. 2).
Figure 1. Mapping and evaluating connectivity of inputs to AgRPARC neurons.
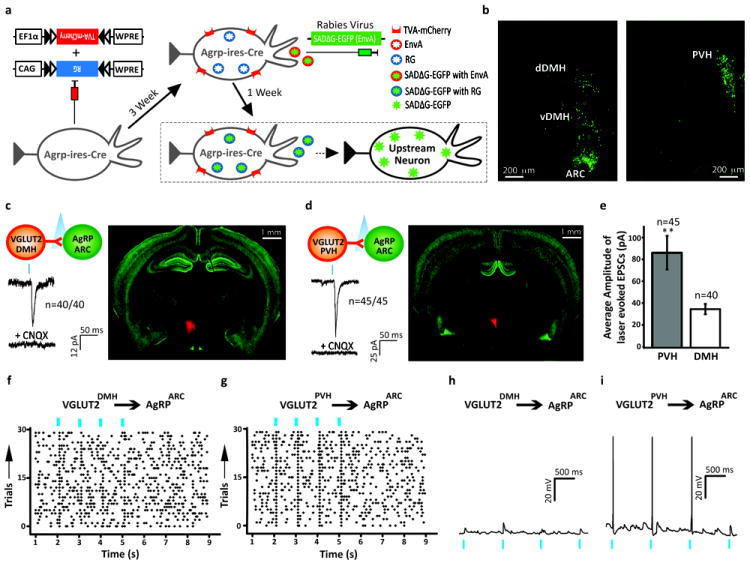
a, Rabies schematic.
b, EGFP detected in the ARC and DMH (Left) and PVH (Right) in Agrp-ires-Cre mice.
c,d, Top, schematic shows connections being tested. Right, representative brains from mice stereotaxically injected with AAV8-DIO-ChR2-mCherry (red = ChR2-mCherry, green = hrGFP from Npy-hrGFP. Bottom, representative traces showing assessment of light-evoked excitatory postsynaptic currents (EPSCs) with blue tic indicating the light pulse (473 nm wavelength, 2 msec). Mice used were Vglut2-ires-Cre; Npy-hrGFP. ChR2 was targeted to (c) DMH and (d) PVH. CNQX = AMPA receptor blocker.
e, Average amplitude of light-evoked EPSCs (pA) in AgRP neurons (n = 40 for VGLUT2DMH→AgRPARC group; n = 45 for VGLUT2PVH→AgRPARC group). Results are shown as mean ± SEM, **=p<0.001; see Supplementary Information for statistical analyses.
f,g, Representative raster plots of EPSCs for (f) VGLUT2DMH→AgRPARC and (g) VGLUT2PVH→AgRPARC.
h,i, Representative traces showing light-evoked changes in membrane potentials in AgRP neurons for (h) VGLUT2DMH→AgRPARC and (i) VGLUT2PVH→AgRPARC.
e. n = 45 for VGLUT2PVH→AgRPARC group; n = 40 for VGLUT2DMH→AgRPARC group. Data represent mean ± SEM. P values for unpaired comparisons were calculated by two-tailed Student’s t-test. *p< 0.05, **p <0.001.
We next employed channelrhodopsin(ChR2)-assisted circuit mapping (CRACM)14,15 to both confirm and determine valence of functional monosynaptic connectivity between afferents and AgRP neurons. CRACM involves in vivo targeted expression of ChR2, a photoexcitable cation channel, in presumptive presynaptic upstream neurons (and their terminals), followed by ex vivo electrophysiologic assessment in acute brain slices of light-evoked postsynaptic currents in candidate downstream neurons. To investigate excitatory input to AgRP neurons, we stereotaxically injected Cre-dependent AAV expressing ChR2-mCherry (AAV8-DIO-ChR2-mCherry) (Supplementary Fig. 3a) into brain sites of Vglut2-ires-Cre; Npy-hrGFP mice13. Of note, VGLUT2 (official gene symbol, Slc17a6) is the glutamate synaptic vesicle transporter expressed in the hypothalamus, hence Vglut2-ires-Cre mice target relevant excitatory neurons13. As AgRP neurons co-express NPY, Npy-hrGFP mice allow for visualization of AgRP neurons16,17. Consistent with the rabies tracing, we detected light-evoked excitatory post-synaptic currents (EPSCs) in all VGLUT2DMH→AgRPARC neurons tested (latency between onset of light and EPSC=4.7 ± 0.2 ms; Fig. 1c; Supplementary Fig. 3f). These were blocked by CNQX, an AMPA receptor antagonist, confirming their glutamatergic nature. Next, we examined monosynaptic connections between VGLUT2PVH→AgRPARC neurons and again, consistent with the rabies mapping, we observed light-evoked EPSCs in all AgRP neurons tested (latency=4.9 ± 0.4 ms; (Fig. 1d; Supplementary Fig. 3g). These also were blocked by CNQX.
In addition, we selectively expressed ChR2 in the ventral medial hypothalamus (VMH) and lateral hypothalamus (LH), two sites with few EGFP+ cells, and also the ARC, which could provide local afferents, and investigated possible connectivity to AgRP neurons. In agreement with the negative rabies data, no light-evoked EPSCs were detected in 36 of 37 VGLUT2VMH→AgRPARC neurons tested (Supplementary Fig. 3b,h) or in any VGLUT2LH→AgRPARC neurons tested (Supplementary Fig. 3c,i). Likewise, we failed to detect light-evoked EPSCs in any VGLUT2ARC→AgRPARC neurons tested (Supplementary Fig. 3d,j). On the other hand, and as previously noted18, glutamatergic VMH neurons were monosynaptically connected to nearby Pro-opiomelanocortin (POMC) neurons (VGLUT2VMH→POMCARC), as we observed light-evoked EPSCs in all POMC neurons tested (latency=4.4 ± 0.2 ms; Supplementary Fig. 3e).
The CRACM studies suggest marked differences in the strength of VGLUT2PVH→AgRPARC versus VGLUT2DMH→AgRPARC inputs. First, the amplitude of light-evoked EPSCs generated from VGLUT2PVH inputs were ~3-fold greater (Fig. 1e). Second, the effectiveness of light pulses in evoking EPSCs differed, with DMH inputs showing a much higher failure rate (~32% ;Fig. 1f; Supplementary Figure 4) compared with PVH inputs where time-locked EPSCs followed every light pulse (0% failure rate; Fig. 1g; Supplementary Figure 4). To test for consequences of these synaptic strength differences, we assessed the ability of VGLUT2DMH→AgRPARC or VGLUT2PVH→AgRPARC inputs to fire action potentials in AgRP neurons. Whereas we never detected VGLUT2DMH→AgRPARC light-evoked action potentials (Fig. 1h), we observed VGLUT2PVH→AgRPARC light-evoked action potentials approximately 75% of the time (Fig. 1i), demonstrating potent, excitatory input to AgRP neurons originating from the PVH.
To determine which PVH neurons monosynaptically drive AgRP neurons, we employed numerous lines of Cre-expressing mice, each targeting subsets of PVH neurons, and then used CRACM to determine connectivity. Sim1 encodes for the single minded 1 transcription factor required for developmental specification of the PVH, and is thus expressed in the majority of PVH neurons,19 as verified in the Sim1-Cre transgenic mouse20. Not surprisingly, we observed light-evoked EPSCs in all SIM1PVH→AgRPARC neurons tested (latency=4.6 ± 0.4 ms; Fig. 2a). We next surveyed the Allen Brain Atlas for mRNAs highly enriched in the PVH, and then assembled a series of ires-Cre knock-in mice targeting cells marked by Prodynorphin (Pdyn-ires-Cre; Supplementary Fig. 5), Oxytocin (Oxt-ires-Cre) 21, Arginine Vasopressin (Avp-ires-Cre; Supplementary Fig. 6), Corticotropin-Releasing Hormone (Crh-ires-Cre; Supplementary Fig. 7), TRH (Trh-ires-Cre; Supplementary Fig. 8) and PACAP (Pacap-ires-Cre; Supplementary Fig. 9). Following ChR2 delivery into the PVH, we failed to detect light-evoked EPSCs in all PDYNPVH→AgRPARC (Fig. 2b), OXYPVH→AgRPARC (Supplementary 10a), AVPPVH→AgRPARC (Supplementary 10b), or CRHPVH→AgRPARC (Supplementary 10c) neurons tested. On the other hand, we detected light-evoked EPSCs in all TRHPVH→AgRPARC neurons (latency=4.7 ± 0.4 ms; Fig. 2c) and all PACAPPVH→AgRPARC neurons tested (latency=4.8 ± 0.3 ms; Fig. 2d). These findings demonstrate that TRHPVH and PACAPPVH neurons provide excitatory drive to AgRP neurons. It is not known if these excitatory inputs come from two distinct classes of PVH neurons or from one which co-expresses TRH and PACAP. The latter is possible given that 37% of TRH mRNA-expressing neurons are marked by Pacap-ires-Cre (Supplementary Fig. 11).
Figure 2. TRHPVH and PACAPPVH neurons provide excitatory input to AgRP neurons.
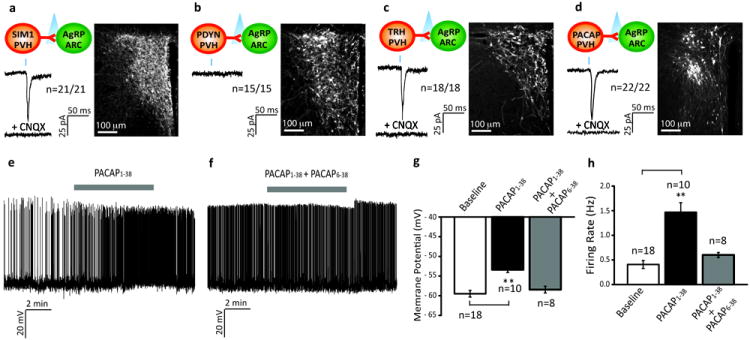
a-d, Top, schematic shows connections being tested. Right, representative brain sections of PVH injected with AAV8-DIO-ChR2-mCherry (white = ChR2-mCherry). Bottom, representative traces showing assessment of light-evoked EPSCs with blue tic indicating the light pulse (473 nm wavelength, 2 msec). Mice used were (a) Sim1-Cre; Npy-hrGFP, (b) Pdyn-ires-Cre; Npy-hrGFP, (c) Trh-ires-Cre; Npy-hrGFP, and (d) Pacap-ires-Cre; Npy-hrGFP. CNQX = AMPA receptor blocker.
e-h, Baseline and effects of PACAP (100 nM) with or without PAC1R blocker (PACAP6-38; 200 nM) on membrane potential and firing rate of AgRP neurons (n = 18, n = 10, n = 8, respectively). Results are shown as mean ± SEM, **=p<0.001; see Supplementary Information for statistical analyses.
g, h. n = 18 for baseline group; n = 10 for PACAP1-38 group; n = 8 for PACAP1-38 + PACAP6-38 group. P values for pair-wise comparisons (baseline versus PACAP1-38 group) and unpaired comparisons (baseline versus PACAP1-38 + PACAP6-38 group) were calculated by two-tailed Student’s t-test. *p< 0.05, **p <0.001.
Innervation by PACAPPVH neurons suggests that PACAP, in addition to glutamate, could also activate AgRP neurons. To assess this, we exogenously added PACAP1-38 (100 nM)22 and determined effects on activity of synaptically isolated AgRP neurons (in the presence of picrotoxin (PTX) and kynurenate). Of note, PACAP1-38 markedly depolarized and increased firing rate, and this was prevented by co-addition of the PAC1-receptor blocker, PACAP6-38 (200 nM) (Fig. 2e-h). Thus, in addition to glutamate, PACAP working through PAC1 receptors also likely plays an important role in the PACAPPVH→AgRPARC circuit.
POMC and AgRP neurons lie in close proximity within the ARC, but have opposing effects on feeding 2,23. If fidelity of the identified PVH→AgRPARC neuron hunger circuit is to be high, then it should not also engage satiety-promoting POMC neurons. We evaluated this and found that the vast majority of TRHPVH→POMCARC neurons (21 of 22) and all PACAPPVH→POMCARC neurons are not monosynaptically connected (Fig. 3a, b, respectively). However, consistent with our observation that VGLUT2VMH→POMCARC neurons are connected (Supplementary Fig. 3e), and as a positive control for studies investigating PACAP→POMCARC connections, we readily detected PACAPVMH→POMCARC monosynaptic connections (latency=4.5 ± 0.2 ms; Fig. 3c). These studies clearly demonstrate that the identified PVH→AgRPARC neuron circuit selectively drives hunger-promoting AgRP neurons, but not satiety-promoting POMC neurons.
Figure 3. Fidelity of TRHPVH/PACAPPVH→ARC and AgRPARC→PVH circuitry.
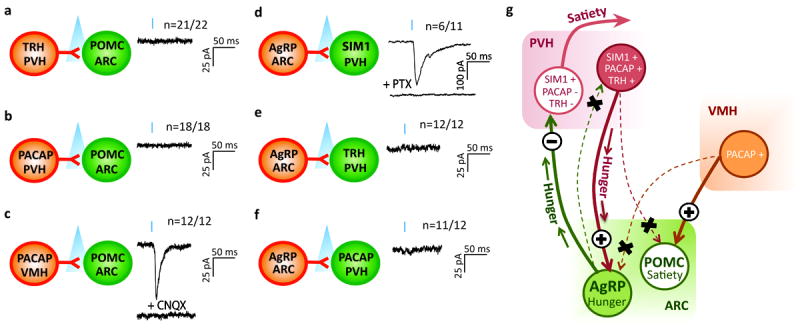
a-f, Left, schematic shows connections being tested. Right, representative traces showing assessment of light-evoked EPSCs (a-c) or IPSCs (d-f) with blue tic indicating the light pulse (473 nm wavelength, 2 msec). Mice used were (a) Trh-ires-Cre; Pomc-hrGFP, (b-c) Pacap-ires-Cre; Pomc-hrGFP, (d) Agrp-ires-Cre; Sim1-Cre; R26-loxSTOPlox-L10-GFP mice for visualization of SIM1 neurons, (e) Agrp-ires-Cre; Trh-ires-Cre injected with AAV8-DIO-mCherry into the PVH for visualization of TRH neurons and (f) Agrp-ires-Cre; Pacap-ires-Cre; R26-loxSTOPlox-L10-GFP mice for visualization of PACAP neurons. CNQX = AMPA receptor blocker. PTX = GABAA receptor blocker. (g) Model summarizing reciprocal circuitry and its fidelity.
Fidelity should also exist in the downstream connections of orexigenic AgRP neurons, which have reciprocal connections with the PVH. GABAergic AgRP neurons are monosynaptically connected with a subset of satiety-promoting neurons in the PVH, and importantly, this inhibitory connection drives feeding5. In the present study, we confirm this inhibitory AgRPARC→PVH connection following ChR2 expression in AgRP neurons, as we detected light-evoked IPSCs (latency=5.2 ± 0.3 ms) in a subset (55%) of AGRPARC→SIM1PVH neurons (Fig. 3d), which were blocked by PTX. If fidelity of this hunger-promoting, GABAergic reciprocal AgRPARC→PVH connection is to be high, then it should not also engage and consequently inhibit the TRHPVH and PACAPPVH neurons. As expected, we detected no AgRPARC→TRHPVH connections (Fig. 3e) and only rare AgRPARC→PACAPPVH connections (1 of 12 neurons tested) (Fig. 3f). In total, the above studies demonstrate marked fidelity in the reciprocal TRH/PACAPPVH glutamatergic → AgRPARC GABAergic → SatietyPVH neuron circuit that drives feeding (Fig 3g).
To confirm function of the TRHPVH and PACAPPVH neurons, we targeted DREADDs (Designer Receptors Exclusively Activated by Designer Drugs; AAV8-DIO-hM3Dq-mCherry)3 to the PVH. Upon binding of Clozapine-N-Oxide (CNO), hM3Dq activates neurons via the Gq signaling cascade24. After verifying in brain slices that CNO depolarized and increased firing frequency of TRHPVH and PACAPPVH neurons (Fig. 4a and 4d, respectively), we found that acutely stimulating upstream TRHPVH or PACAPPVH neurons markedly induced Fos activity in AgRP cells, ipsilateral to the DREADD-activated upstream PVH neurons (Fig. 4b and 4e, respectively).
Figure 4. DREADD-mediated manipulation of TRHPVH or PACAPPVH neurons mediates feeding through AgRP neurons.
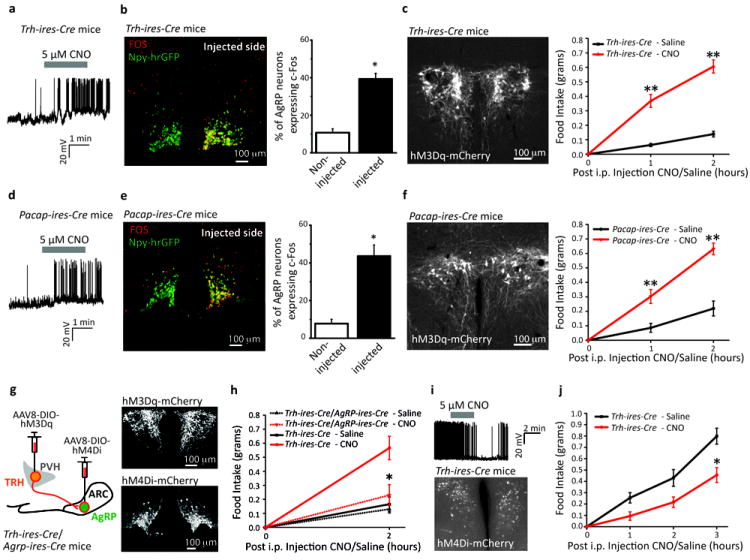
a,d, Membrane potential and firing rate of (a) TRHPVH or (d) PACAPPVH neurons transduced with AAV8-DIO-hM3Dq-mCherry upon CNO application.
b,e, AAV8-DIO-hM3Dq-mCherry was transduced unilaterally into the PVH of (b) Trh-ires-Cre or (e) Pacap-ires-Cre mice and ipsilateral induction of Fos (red) was assessed in AgRP neurons (marked by hrGFP, green) 3 hrs following CNO (0.3 mg kg-1) injection (n=3, n=3; respectively).
c,f, Left, AAV8-DIO-hM3Dq-mCherry (white) was transduced bilaterally into the PVH of (c) Trh-ires-Cre or (f) Pacap-ires-Cre mice. Right, light-cycle food intake after injection of saline (black) or CNO (red; 0.3 mg kg-1) (n=7-8 animals per condition; experiment replicated three times per animal).
g,h, Simultaneous inhibition of AgRPARC neurons with activation of TRHPVH neurons attenuates food intake. (g) Sagittal schematic of occlusion study, whereby AAV8-DIO-hM3Dq-mCherry (white) was transduced bilaterally into the PVH (Top) and AAV8-DIO-hM4Di-mCherry (white) was transduced bilaterally into the ARC (Bottom) of Trh-ires-Cre; Agrp-ires-Cre mice. (h) light-cycle food intake after injection of saline (black) or CNO (red; 0.3 mg kg-1) in Trh-ires-Cre; Agrp-ires-Cre (dotted; hM3Dq in PVH and hM4Di in ARC) and Trh-ires-Cre (solid; hM3Dq in PVH) mice (n=4 animals per condition; experiment replicated three times per animal).
i, (Top) Membrane potential and firing rate of TRHPVH neurons transduced with AAV8-DIO-hM4Di-mCherry upon CNO application. (Bottom) AAV8-DIO-hM4Di-mCherry (white) was transduced bilaterally into the PVH of Trh-ires-Cre mice.
j, Dark-cycle food intake after injection of saline (black) or CNO (red; 0.3 mg kg-1) (n=4 animals per condition; experiment replicated three times per animal). Results are shown as mean ± SEM, *=p<0.05, **=p<0.001; see Supplementary Information for statistical analyses.
b,e. n = 3 for each group. Data represent mean ± SEM. P values for unpaired comparisons were calculated by two-tailed Student’s t-test. *p< 0.05.
c, f. n = 8 for Trh-ires-Cre group; n = 7 for Pacap-ires-Cre group. Data represent mean ± SEM. Two-way repeated measures ANOVA detected significant interaction of ‘time’ and ‘treatment’. F3,20 = 56.13, **p <0.001; F3,18 = 0.056, **p <0.001; respectively. Sidak’s post-hoc test shows significant difference between ‘time’ and ‘treatment’ at 1 and 2 hrs as indicated; respectively.
h. n = 4 for each group. Data represent mean ± SEM. Two-way repeated measures ANOVA detected significant interaction of ‘genotype’ and ‘treatment’. F3,18 = 6.083, *p < 0.05. Sidak’s post-hoc test shows significant difference between ‘genotype’ and ‘treatment’ as indicated.
j. n = 4. Data represent mean ± SEM. Two-way repeated measures ANOVA detected significant interaction of ‘time’ and ‘treatment’. F3,18 = 3.962, *p < 0.05. Sidak’s post-hoc test shows significant difference between ‘time’ and ‘treatment’ at 3 hrs as indicated.
We next assessed effects of bilateral h3MDq-DREADD-mediated TRHPVH or PACAPPVH neuron stimulation on feeding behavior during the light cycle, a time when food intake is usually low because of feeding during the preceding dark cycle (as observed following saline injections, Fig. 4c and 4f, respectively – black lines). Importantly, acute activation by CNO injection of either TRHPVH or PACAPPVH neurons caused robust feeding (Fig. 4c and 4f, respectively – red lines). Of note, this effect was absent following an overnight fast which itself elevates excitatory drive and consequently AgRP activity1,8,9 (Supplementary Fig. 12). To directly demonstrate that this enhanced feeding was mediated via AgRP neurons, we activated TRHPVH neurons (AAV8-DIO-hM3Dq-mCherry) while simultaneously inhibiting AgRPARC neurons (AAV8-DIO-hM4Di-mCherry3; Fig. 4g). Importantly, this significantly attenuated feeding (Fig. 4h). Finally, to examine whether endogenous activity of the TRHPVH→AgRPARC pathway is physiologically relevant for feeding, we bilaterally transduced TRHPVH neurons with inhibitory DREADDs, which led to rapid hyperpolarization and decreased firing rate upon CNO application (as assessed in slice studies, Fig. 4i), and found that this manipulation drastically reduced food intake during the dark cycle (Fig. 4j). These studies demonstrate both the sufficiency and necessity of this novel PVH→AgRPARC orexigenic circuit in regulating food intake.
Given the vast number of studies implicating an anorexigenic role for the PVH5,20,25-28, it is remarkable to now discover a subset of neurons that drive feeding, through a reciprocal circuit (Fig. 3g). Failure of prior studies to detect orexigenic activity in the PVH is likely due to the reciprocal nature of this hunger circuit (leaving and returning to the PVH) and the inhibitory aspect of its return arm (AgRPARC→SatietyPVH neurons).
Methods
Animals
All animal care and experimental procedures were approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. Mice were housed at 22_C–24_C with a 12 h light/12 h dark cycle with standard mouse chow (Teklad F6 Rodent Diet 8664; 4.05 kcal/g, 3.3 kcal/g metabolizable energy, 12.5% kcal from fat; Harlan Teklad, Madison, WI) and water provided ad libitum. All diets were provided as pellets. Mice were euthanized by CO2 narcosis.
Generation of Mice
Pdyn-ires Cre, Avp-ires-Cre, Crh-ires-Cre, Trh-ires-Cre and Pacap-ires-Cre mice were generated using recombineering techniques as previously described13,21. Briefly, a selection cassette containing an internal ribosomal entry sequence linked to Cre-recombinase and an Frt-flanked kanamycin resistance gene was targeted just downstream of the stop codon of the Prodynorphin, Arginine vasopressin, Corticotropin releasing hormone, Thyrotropin releasing hormone or Adenylate cyclase activating peptide 1 gene, respectively, in a bacterial artificial chromosome, so that Cre recombinase expression was driven by the endogenous genes. A targeting plasmid containing the Cre-containing selection cassette and 4 kb genomic sequence upstream and downstream of the Prodynorphin, Arginine vasopressin, Corticotropin releasing hormone, Thyrotropin releasing hormone or Adenylate cyclase activating peptide 1 stop codon, respectively was isolated and used for embryonic stem cell targeting. Correctly targeted clones were identified by long range PCR and injected into blastocysts. Chimeric animals generated from blastocyst implantation were then bred for germline transmission of the altered Prodynorphin, Arginine vasopressin, Corticotropin releasing hormone, Thyrotropin releasing hormone or Adenylate cyclase activating peptide 1-allele, respectively. Flp-deleter mice were then used to remove the neomycin selection cassette.
Generation of an enhanced Cre-dependent GFP reporter mice (R26-loxSTOPlox-L10-GFP) were generated using recombineering techniques as previously described20. A transgene containing a lox-flanked transcriptional blocking cassette followed by eGFP fused to the L10-ribosomal subunit31 was placed under the control of a CMV-enhancer/chicken beta-actin promoter and targeted to the Rosa26 locus using standard techniques32. Correctly targeted blastocysts were identified by long range PCR and confirmed by southern blotting and injected into blastocysts.
Characterization of mice
Pdyn-ires Cre, Avp-ires-Cre, Crh-ires-Cre, and Pacap-ires-Cre mice were crossed to R26-loxSTOPlox-L10-GFP mice, sacrificed and sectioned at 30 μm. Brain sections were washed in 0.1 M phosphate-buffered saline with Tween 20, pH 7.4 (PBST, 2 changes) and then incubated in the primary antiserum (anti-GFP, Abcam (1:1000), rabbit anti-hrGFP. After several washes in PBS, sections were incubated in Alexa fluorophore secondary antibody (Molecular Probes, 1:200) for 2 hours at room temperature. After several washes in PBS, sections were mounted onto gelatin-coated slides and fluorescent images were captured with Olympus VS120 Slide Scanner microscope.
Trh-ires-Cre mice were stereotaxically injected with Cre-dependent AAV8-DIO-mCherry,8 due to transient, early embryonic expression and subsequent deletion of floxed alleles by this mouse line, sacrificed and sectioned at 30 μm. Native fluorescence was used and fluorescent images were captured with Olympus VS120 Slide Scanner microscope. All digital images were processed in the same way between experimental conditions to avoid artificial manipulation between different data sets.
In situ mRNA hybridization experiments were performed for Pdyn-ires-Cre, Trh-ires-Cre and Pacap-ires-Cre mice (details below). Antibody staining experiments were performed for Avp-ires-Cre and Crh-ires-Cre mice. Briefly, Avp-ires-Cre; R26-loxSTOPlox-L10-GFP mice were co-localized with anti-AVP, Sigma (1:500), rabbit anti-AVP and anti-GFP, Abcam (1:1000), chicken anti-GFP. 48 hrs ICV colchicine treatment (Sigma, 10 ug), Crh-ires-Cre; R26-loxSTOPlox-L10-GFP mice were co-localized with anti-CRF, Peninsula Laboratories (1:2500), rabbit anti-CRF and anti-GFP, Abcam (1:1000), chicken anti-GFP.
All images were subsequently compared to in situ mRNA expression profiles generated by ©2013 Allen Institute for Brain Science; Allen Mouse Brain Atlas [Internet].
Breeding strategies
Vglut2-ires-Cre, Sim1-Cre, Pdyn-ires-Cre, Oxy-ires-Cre Avp-ires-Cre, Crh-ires-Cre, Trh-ires-Cre and Pacap-ires-Cre mice were bred to Npy-hrGFP transgenic mice to generate heterozygous Vglut2-ires-Cre; Npy-hrGFP, Sim1-Cre; Npy-hrGFP, Pdyn-ires-Cre; Npy-hrGFP, Oxy-ires-Cre; Npy-hrGFP, Avp-ires-Cre; Npy-hrGFP, Crh-ires-Cre; Npy-hrGFP, Trh-ires-Cre; Npy-hrGFP and Pacap-ires-Cre; Npy-hrGFP mice, respectively as previously reported.8 Vglut2-ires-Cre and Pacap-ires-Cre mice were bred to Pomc-hrGFP transgenic mice to generate heterozygous Vglut2-ires-Cre; Pomc-hrGFP and Pacap-ires-Cre; Pomc-hrGFP mice, respectively as previously reported13.
Agrp-ires-Cre, Pdyn-ires-Cre, Avp-ires-Cre, Crh-ires-Cre and Pacap-ires-Cre mice were bred to R26-loxSTOPlox-L10-GFP mice to generate heterozygous Agrp-ires-Cre; R26-loxSTOPlox-L10-GFP, Pdyn-ires-Cre; R26-loxSTOPlox-L10-GFP, Avp-ires-Cre; R26-loxSTOPlox-L10-GFP, Crh-ires-Cre; R26-loxSTOPlox-L10-GFP and Pacap-ires-Cre; R26-loxSTOPlox-L10-GFP mice, respectively. Agrp-ires-Cre; R26-loxSTOPlox-L10-GFP mice were then bred to Sim1-Cre and Pacap-ires-Cre to generate heterozygous Agrp-ires-Cre; Sim1-Cre R26-loxSTOPlox-L10-GFP and Agrp-ires-Cre; Pacap-ires-Cre; R26-loxSTOPlox-L10-GFP mice, respectively. All mice are on a mixed background.
Viral injections
Stereotaxic injections were performed as previously described.3 Mice were anesthetized with xylazine (5 mg/kg) and ketamine (75 mg/kg) diluted in saline (350 mg/kg) and placed into a stereotaxic apparatus (KOPF Model 963). After exposing the skull via small incision, a small hole was drilled for injection. A pulled-glass pipette with 20–40 mm tip diameter was inserted into the brain and virus was injected by an air pressure system. A micromanipulator (Grass Technologies, Model S48 Stimulator) was used to control injection speed at 25 nl/min and the pipette was withdrawn 5 min after injection. For retrograde rabies tracing, AAV-FLEX-TVA-mCherry, serotype 8 (titer 1.1 × 1012 genomes copies per ml) and AAV-FLEX-RG, serotype 8 (titer 1.4 × 1012 genomes copies per ml)12 were injected unilaterally into the ARC (200 nl, coordinates, bregma: AP:-1.50 mm, DV:-5.80 mm, L: -0.20 mm) of 5-6 weeks old mice. 21 days later SADΔG-EGFP (EnvA) rabies (titer 107) was unilaterally injected into the ARC (400 nl, coordinates, bregma: AP:-1.50 mm, DV:-5.80 mm, L: -0.20 mm). For electrophysiology experiments, AAV-DIO-ChR2(H134R)-mCherry, serotype 8 (titer 1.3 × 1012 genomes copies per ml)29 was injected unilaterally into the DMH (20 nl, coordinates, bregma: AP:-1.70 mm, DV:-5.00 mm, L: -0.25 mm), PVH (25 nl, coordinates, bregma: AP:-0.70 mm, DV:-4.75 mm, L: -0.20 mm), VMH (25 nl, coordinates, bregma: AP:-1.40 mm, DV:-5.60 mm, L: -0.40 mm) or ARC (20 nl, coordinates, bregma: AP:-1.50 mm, DV:-5.80 mm, L: -0.20 mm) of 5-6 weeks old mice. For feeding and Fos studies, AAV-DIO-hM3Dq-mCherry, serotype 8 (titer 1.2 × 1012 genomes copies per ml)3 or AAV-DIO-hM4Di-mCherry, serotype 8 (titer 1.7 × 1012 genomes copies per ml)3 was injected bilaterally or unilaterally, respectively into the PVH (25 nl, coordinates, bregma: AP:-0.70 mm, DV:-4.75 mm, L: -0.20 mm) of 5-6 weeks old mice. For validation of Trh-ires-Cre mice, AAV-DIO-mCherry, serotype 8 (titer 1.2 × 1012 genomes copies per ml)8 or AAV-DIO-GFP, serotype 8 (titer 1.4 × 1012 genomes copies per ml) was injected bilaterally into the PVH (25 nl, coordinates, bregma: AP:-0.70 mm, DV:-4.75 mm, L: -0.20 mm) of 5-6 weeks old mice. All viruses were packaged at the Gene Therapy Center at the University of North Carolina. For postoperative care, mice were injected intraperitoneally with meloxicam (0.5 mg/kg). All stereotaxic injection sites were verified under electrophysiological microscopy (for electrophysiology-related studies) or by immunohistochemistry (for anatomy and in vivo studies). All “misses” or “partial hits” were excluded from data analyses.
SADΔG-EGFP (EnvA) rabies cell counting
One week after SADΔG-EGFP (EnvA) rabies injection, mice were perfused and brains were sectioned and mounted as described above. For each brain (n=6), 10X images were taken throughout 1 entire brain series using a Zeiss AxioImager Z.1 microscope and EGFP+ cells were quantified using these images. Each EGFP+ cell was assigned to a specific anatomical structure of the hypothalamus using The Mouse Brain in Stereotaxic Coordinates (Franklin &Paxinos). As the number of labeled cell varied over multiple animals depending on the transduction rate of the viruses, the counted cells in each anatomical structure were expressed as a percentage of the total number of cells counted throughout each mouse brain. The percentages for each anatomical structure in a given mouse were then averaged over the entire cohort of mice (Supplementary Fig. 2).
Electrophysiology and circuit mapping
For brain slice preparation, 6-10 weeks old mice were anesthetized with isoflurane before decapitation and removal of the entire brain. Brains were immediately submerged in ice-cold, carbogen-saturated (95% O2, 5% CO2) high sucrose solution (238 mM sucrose, 26 mM NaHCO3, 2.5 mM KCl, 1.0 mM NaH2PO4, 5.0 mM MgCl2, 10.0 mM CaCl2, 11 mM glucose). Then, 300 μM thick coronal sections were cut with a Leica VT1000S Vibratome and incubated in oxygenated aCSF (126 mM NaCl, 21.4 mM NaHCO3, 2.5 mM KCl, 1.2 mM NaH2PO4, 1.2 mM MgCl2, 2.4 mM CaCl2, 10 mM glucose) at 34°C for 30 minutes. Then, slices were maintained and recorded at room temperature (20–24°C). The intracellular solution for voltage clamp recording contained the following (in mM): 140 CsCl, 1 BAPTA, 10 HEPES, 5 MgCl2, 5 Mg-ATP, 0.3 Na2GTP, and 10 lidocaine N-ethyl bromide (QX-314), pH 7.35 and 290 mOsm. The intracellular solution for current clamp recordings contained the following (in mM): 128 K gluconate, 10 KCl, 10 HEPES, 1 EGTA, 1 MgCl2, 0.3 CaCl2, 5 Na2ATP, 0.3 NaGTP, adjusted to pH 7.3 with KOH.
Photostimulation-evoked EPSCs and IPSCs were recorded in the whole cell voltage clamp mode, with membrane potential clamped at -60 mV. Photostimulation –evoked EPSCs was recorded in presence of picrotoxin (100 μM) to block inhibitory postsynaptic currents. All recordings were made using multiclamp 700B amplifier, and data were filtered at 2 kHz and digitized at 10 kHz. To photostimulate Channelrhodopsin2-positive fibers, a laser or LED light source (473 nm; Opto Engine LLC; Thorlabs) was used. The blue light was focused on to the back aperture of the microscope objective, producing a wide-field exposure around the recorded cell of 10–15 mW/mm2. The light power at the specimen was measured using an optical power meter PM100D (ThorLabs). The light output is controlled by a programmable pulse stimulator, Master-8 (AMPI Co. ISRAEL) and the pClamp 10.2 software (AXON Instruments). Photostimulation-evoked EPSCs/IPSCs detection protocol constitutes four blue light laser pulses administered 1 second apart during the first 4 seconds of a 8 second sweep, repeating for a total of 30 sweeps. When recording photostimulation-evoked action potentials in AgRP neurons, current was injected into cells to keep the base line membrane potential at ~-60 mV. Mice with total misses, partial expression or expression outside the intended area were excluded from analysis after examination of mCherry expression.
VGLUT2DMH→AgRPARC n=4; VGLUT2PVHH→AgRPARC n=5; VGLUT2VMH→AgRPARC n=4; VGLUT2LH→AgRPARC n=3; VGLUT2ARC→AgRPARC n=3; VGLUT2VMH→POMCARC n=3; SIM1PVH→AgRPARC n=3; PDYNPVH→AgRPARC n=3; OXYPVH→AgRPARC n=3; AVPPVH→AgRPARC n=3; CRHPVH→AgRPARC n=3; TRHPVH→AgRPARC n=3; PACAPPVH→AgRPARC n=4; TRHPVH→POMCARC n=4; PACAPPVH→POMCARC n=3; PACAPVMH→POMCARC n=2; AgRPARC→SIM1PVH n=2; AgRPARC→TRHPVH n=3; AgRPARC→PACAPPVH n=3.
To assess the effect of PACAP1-38 (100 nM)22 and PACAP6-38 (200 nM) onto AgRP neurons, we performed whole cell current clamp recordings on to 5-8 weeks old Npy-hrGFP mice. Synaptic blockers (1 mM kynurenate and 100 μM picrotoxin) were added in aCSF to synaptically isolate AgRP neurons.
To assess the effect of CNO on to TRH and PACAP neurons, 5- to 7-week-old Pacap-ires-Cre and Trh-ires-Cre mice were injected with AAV8-DIO-hM3Dq-mCherry into the PVH 2-3 weeks prior to recording. CNO was applied to bath solution through perfusion as previously reported.3 After acquisition of stable whole-cell recordings for 2–5 min, aCSF solution containing 5 μM CNO was perfused into the brain slice preparation.
Fos analysis
Animals (Trh-ires-Cre; Npy-hrGFP; n=3 and Pacap-ires-Cre; Npy-hrGFP; n=3 mice), were handled for 10 consecutive days before the assay to reduce stress response, and then injected with CNO (0.3 mg kg-1; i.p.) at 9am. 150 min later, the animals were sacrificed with 7% chloral hydrate diluted in saline (350 mg/kg) for histological assay. The mice were perfused and brains were sectioned as described above. Assessment of Fos induction was performed using a previously developed method8 modified for fluorescent colocalization with hrGFP in AgRP neurons. Brain sections were processed for immunohistochemical detection of Fos and hrGFP and counting.
Brain sections were washed in 0.1 M phosphate-buffered saline with Tween 20, pH 7.4 (PBST, 2 changes) and then incubated in the primary antiserum (anti-GFP, Abcam (1:1000), rabbit anti-hrGFP and anti-Fos, Calbiochem (1:10000). After several washes in PBS, sections were incubated Alexa fluorophore secondary antibodies (Molecular Probes, 1:200) for 2 hours at room temperature. After several washes in PBS, sections were mounted onto gelatin-coated slides and fluorescent 10X images were taken initially using a Zeiss AxioImager Z.1 microscope. Colocalization and quantification was further determined using fluorescent 20X images taken using a Zeiss AxioImager Z.1 microscope. Data are expressed as the percentage of all AgRP neurons (i.e., all hrGFP-positive neurons) that were double-positive for Fos and hrGFP.
Food intake studies
Food intake studies on chow were performed as previously described.3,8,29 All animals (10- to 12-week-old male mice) were singly housed for at least 2.5 weeks following surgery and handled for 10 consecutive days before the assay to reduce stress response. Feeding studies were performed in home cages with ad lib food access. CNO was administered at 0.3 mg kg-1 of body weight. Saline was delivered at the same volume to maintain consistency in the studies. Previous publications suggest the duration of the drugs effect at the dosage used throughout the studies is approximately 8 hrs24. Mice with “missed” injections, incomplete “hits” or expression outside the area of interest were excluded from analysis after post hoc examination of mCherry expression. In this way, all food intake measurements were randomized and blind to the experimenter.
Light cycle food intake
Animals (Trh-ires-Cre; n=8, Pacap-ires-Cre; n=7 mice; Fig. 4c,f, or Trh-ires-Cre; Agrp-ires-Cre; n=4, Trh-ires-Cre; n=4 mice; Fig. 4h) were injected with either saline or CNO (0.3 mg kg-1; i.p.) at 9 am and food intake was monitored 1 and/or 2 hrs after i.p. injection from 9 am-11 am. A full trial consisted of assessing food intake from the study subjects after they received injections of saline on day 1 and CNO on day 2. Animals received a day “off” between trials before another trial was initiated. The food intake data from all days following saline/CNO (n=24 trials for Trh-ires-Cre; n=21 trials for Pacap-ires-Cre; Fig. 4c,f; n=12 for Trh-ires-Cre; Agrp-ires-Cre; n=12 trials for Trh-ires-Cre; Fig. 4h) injections was then averaged and combined for analysis.
Dark cycle food intake
Animals (Trh-ires-Cre; n=4 mice; Fig. 4j) were injected with either saline or CNO (0.3 mg kg-1; i.p.) at 6 pm and food intake was monitored 1, 2 and 3 hrs after i.p. injection from 6 pm-9 pm. A full trial consisted of assessing food intake from the study subjects after they received injections of saline on day 1 and CNO on day 2. Animals received a day “off” between trials before another trial was initiated. The food intake data from all days following saline/CNO (n=12 trials for Trh-ires-Cre; Fig. 4j) injections was then averaged and combined for analysis.
Fast-Refeed food intake
Animals (Trh-ires-Cre; n=8, Pacap-ires-Cre; n=7 mice; Supplementary Fig. 12) were fasted overnight at 5 pm and the following day were injected with either saline or CNO (0.3 mg kg-1; i.p.) at 8:30 am and food intake was monitored 1.5, 2.5 and 4.5 hrs after i.p. injection from 9 am-1 pm. A full trial consisted of assessing food intake from the study subjects after they received injections of saline on week 1 and CNO on week 2. Animals received a week “off” between trials before another trial was initiated. The food intake data from all days following saline/CNO (n=16 trials for Trh-ires-Cre; n=14 trials for Pacap-ires-Cre; Supplementary Fig. 12) injections was then averaged and combined for analysis.
In situ hybridization
Pacap-ires-Cre; R26-loxSTOPlox-L10-GFP and Pdyn-ires-Cre; R26-loxSTOPlox-L10-GFP mice were sacrificed and brains were frozen fresh. Trh-ires-Cre mice with bilateral AAV-DIO-GFP injections into the PVH were perfused with 4% PFA, post-fixed for 2 hours and equilibrated in 20% sucrose overnight. 14 uM cryosections were analyzed by in situ hybridization as previously described30 with minor modifications. Slides were fixed (4% PFA), washed (2x RNase-free PBS), coverslipped, and imaged for intrinsic Cre-mediated GFP fluorescence. Digoxigenin labeled anti-sense cRNA probes were generated by T3 (Roche) in vitro transcription reactions using the complete coding sequence of Pdyn (5’:ATGGCGTGGTCCAGGCTGATGC 3’:TCAAACATCTAAATCTTCAGAATAGG) and a 914 bp fragment of Pacap cDNA (5’:CTGCGTGACGCTTACGCCCT 3’:TTGCCCCTGCAACCAGTGGG). A previously described murine cDNA33 (gift of Masanobu Yamada) was used to generate a Trh probe with SP6 RNA polymerase (Roche). Following hybridization, slides were incubated with anti-digoxigenin antibody conjugated to alkaline phosphatase (Roche, 1:200, 1hr RT), washed, and incubated (3-6 hr) in NBT/BCIP chromogenic substrate according to manufacturer’s specifications (Roche). Slides were coverslipped and brightfield images were captured. Images were pseudocolored and compared with intrinsic Cre-mediated GFP fluorescence for colocalization and quantification.
Statistical Analysis
Statistical analyses were performed using Origin Pro 8.6 and Prism 6.0 (GraphPad) Software.
Supplementary Material
Supplementary Figure 4. n = 45 for VGLUT2PVH→AgRPARC group; n = 40 for VGLUT2DMH→AgRPARC group. Results are shown as mean ± SEM. P values for unpaired comparisons were calculated by two-tailed Student’s t-test. **p<0.001.
Supplementary Figure 12. n = 8 for Trh-ires-Cre group; n = 7 for Pacap-ires-Cre group. Data represent mean ± SEM. Two-way repeated measures ANOVA detected no significant interaction of ‘time’ and ‘treatment’. F3,24 = 0.142, p = 0.933; F3,20 = 0.056, p = 0.981; respectively. Sidak’s post-hoc test shows no significant difference between ‘time’ and ‘treatment’.
Acknowledgments
This research was funded by the following NIH grants to BBL: R01 DK096010, R01 DK089044, R01 DK071051, R01 DK075632, R37 DK053477, BNORC Transgenic Core P30 DK046200 and BADERC Transgenic Core P30 DK0547521; to MJK: F32 DK089710, to DPO: K08 DK071561, to LV: F32 DK078478, to NU: R01 MH095953, and an ADA Mentor-Based Fellowship to BPS and BBL. We thank Daniel Cusher and Yanfang Li for mouse genotyping and Jenna Carroll and Yikun Guo for perfusions and tissue removal. Bryan L. Roth and Sarah C. Rogan for generating the AAV-DIO-hM3Dq-mCherry plasmid, Karl Deisseroth for generating the AAV-DIO-ChR2-mCherry plasmid, and Anthony N. Hollenberg for the TRH riboprobe.
Footnotes
Author Contributions
M.J.K, B.P.S. and B.B.L. designed the experiments and analysed data. M.J.K., B.P.S., J.C.M., D.S., A.S.G., L.V., and H.P. performed experiments. M.J.K. generated Pdyn-ires-Cre, Trh-ires-Cre and Pacap-ires-Cre mice, D.P.O. generated Oxy-ires-Cre, Avp-ires-Cre, Crh-ires-Cre and R26-loxSTOPlox-L10-GFP mice and L.V. generated Vglut2-ires-Cre mice. M.W-U. and N.U. generated, provided and advised on use of AAV-FLEX-TVA-mCherry, AAV-FLEX-RG and SADΔG-EGFP (EnvA) viruses. S.D.L. advised on in situ hybridization experiments. M.J.K and B.B.L. wrote the manuscript with comments from all of the authors.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Takahashi KA, Cone RD. Fasting induces a large, leptin-dependent increase in the intrinsic action potential frequency of orexigenic arcuate nucleus neuropeptide Y/Agouti-related protein neurons. Endocrinology. 2005;146:1043–1047. doi: 10.1210/en.2004-1397. [DOI] [PubMed] [Google Scholar]
- 2.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krashes MJ, Shah BP, Koda S, Lowell BB. Rapid versus Delayed Stimulation of Feeding by the Endogenously Released AgRP Neuron Mediators GABA, NPY, and AgRP. Cell Metab. 2013;18:588–595. doi: 10.1016/j.cmet.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atasoy D, Betley JN, Su HH, Sternson SM. Deconstruction of a neural circuit for hunger. Nature. 2012;488:172–177. doi: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luquet S, Perez FA, Hnasko TS, Palmiter RD. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310:683–685. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- 7.Gropp E, et al. Agouti-related peptide-expressing neurons are mandatory for feeding. Nat Neurosci. 2005;8:1289–1291. doi: 10.1038/nn1548. [DOI] [PubMed] [Google Scholar]
- 8.Liu T, et al. Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron. 2012;73:511–522. doi: 10.1016/j.neuron.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Atasoy D, Su HH, Sternson SM. Hunger states switch a flip-flop memory circuit via a synaptic AMPK-dependent positive feedback loop. Cell. 2011;146 doi: 10.1016/j.cell.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinto S, et al. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science. 2004;304:110–115. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- 11.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Vong L, et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nature neuroscience. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 15.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn TM, Breininger JF, Baskin DG, Schwartz MW. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat Neurosci. 1998;1:271–272. doi: 10.1038/1082. [DOI] [PubMed] [Google Scholar]
- 17.van den Pol AN, et al. Neuromedin B and gastrin-releasing peptide excite arcuate nucleus neuropeptide Y neurons in a novel transgenic mouse expressing strong Renilla green fluorescent protein in NPY neurons. J Neurosci. 2009;29:4622–4639. doi: 10.1523/JNEUROSCI.3249-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sternson SM, Shepherd GM, Friedman JM. Topographic mapping of VMH --> arcuate nucleus microcircuits and their reorganization by fasting. Nature neuroscience. 2005;8:1356–1363. doi: 10.1038/nn1550. [DOI] [PubMed] [Google Scholar]
- 19.Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–3275. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balthasar N, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 21.Wu Z, et al. An obligate role of oxytocin neurons in diet induced energy expenditure. PLoS One. 2012;7:e45167. doi: 10.1371/journal.pone.0045167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukushima Y, et al. Role of endogenous PACAP in catecholamine secretion from the rat adrenal gland. American journal of physiology Regulatory, integrative and comparative physiology. 2001;281:R1562–1567. doi: 10.1152/ajpregu.2001.281.5.R1562. [DOI] [PubMed] [Google Scholar]
- 23.Zhan C, et al. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J Neurosci. 2013;33:3624–3632. doi: 10.1523/JNEUROSCI.2742-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander GM, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leibowitz SF, Hammer NJ, Chang K. Hypothalamic paraventricular nucleus lesions produce overeating and obesity in the rat. Physiol Behav. 1981;27:1031–1040. doi: 10.1016/0031-9384(81)90366-8. [DOI] [PubMed] [Google Scholar]
- 26.Gold RM, Jones AP, Sawchenko PE. Paraventricular area: critical focus of a longitudinal neurocircuitry mediating food intake. Physiol Behav. 1977;18:1111–1119. doi: 10.1016/0031-9384(77)90019-1. [DOI] [PubMed] [Google Scholar]
- 27.Stanley BG, Leibowitz SF. Neuropeptide Y injected in the paraventricular hypothalamus: a powerful stimulant of feeding behavior. Proc Natl Acad Sci U S A. 1985;82:3940–3943. doi: 10.1073/pnas.82.11.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowley MA, et al. Integration of NPY, AGRP, and melanocortin signals in the hypothalamic paraventricular nucleus: evidence of a cellular basis for the adipostat. Neuron. 1999;24:155–163. doi: 10.1016/s0896-6273(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 29.Kong D, et al. GABAergic RIP-Cre neurons in the arcuate nucleus selectively regulate energy expenditure. Cell. 2012;151:645–657. doi: 10.1016/j.cell.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- 31.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satoh T, Yamada M, Monden T, Iizuka M, Mori M. Cloning of the mouse hypothalamic preprothyrotropin-releasing hormone (TRH) cDNA and tissue distribution of its mRNA. Brain Res Mol Brain Res. 1992;14:131–135. doi: 10.1016/0169-328x(92)90020-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 4. n = 45 for VGLUT2PVH→AgRPARC group; n = 40 for VGLUT2DMH→AgRPARC group. Results are shown as mean ± SEM. P values for unpaired comparisons were calculated by two-tailed Student’s t-test. **p<0.001.
Supplementary Figure 12. n = 8 for Trh-ires-Cre group; n = 7 for Pacap-ires-Cre group. Data represent mean ± SEM. Two-way repeated measures ANOVA detected no significant interaction of ‘time’ and ‘treatment’. F3,24 = 0.142, p = 0.933; F3,20 = 0.056, p = 0.981; respectively. Sidak’s post-hoc test shows no significant difference between ‘time’ and ‘treatment’.


