Abstract
It is well established that cells are more sensitive to ionizing radiation during the G2/M phase of the cell cycle when their chromatin is highly compacted. However, highly compacted chromatin is less susceptible to DNA Double Strand Breaks (DSBs) than relaxed chromatin. Therefore, it is now becoming apparent that it is the cell capacity to repair its damaged DNA and refold its chromatin into its original compacted status that primarily affects the overall cellular sensitivity to ionizing radiation. The Histone Deacetylase Inhibitors (HDACIs) are a new class of anticancer agents that relax chromatin structure by increasing the levels of histone acetylation. The effect of HDACIs on normal and cancer cells sensitivity to ionizing radiation differs. Reports have indicated that HDACIs can protect normal cells while simultaneously sensitize cancer cells to ionizing radiation. This difference may stem from the individual characteristic of the normal and cancer cells chromatin structure. This review discusses this possibility and addresses the role of HDACIs in radiation therapy.
Keywords: HDACI, HAT, Chromatin, Radiation
Chromatin
The DNA of eukaryotic cells is highly compacted and organized by histone and non-histone proteins into a nuclear structure known as the chromatin. Nucleosomes constitute the building bloc of chromatin. They are composed of core histones organized into a tetramer of histones (H3-H4)2 and two H2A–H2B dimers localized on each side of the tetramer. Stretches of DNA (146 bp) are wrapped twice around these histone octamers that are separated by short sections of linker DNA to constitute what is known as the beads on a string structure. The linker histone H1 promotes the folding of the chromatin into higher order structure (30 nm fibers) by anchoring its globular domain at the exit or entry end of the nucleosomes and its highly positively charged C-terminal domain at the linker DNA on either side of the nucleosomes (1). This association stabilizes this complex structure and prevents the DNA from peeling of the histone surface (2). Electrostatic interactions between the negatively charged phosphodiester backbone of the DNA and the basic (positively charged) residues of the core histone tails maintain the chromatin in to a well-organized tightly compacted structure. However, this organization is not static but rather dynamic. Because the core histone tails protrude outside the nucleosomes they are susceptible to a variety of Post Translational Modifications (PTMs) including acetylation, methylation, phosphorylation, ubiquitination, sumoylation, proline isomerization, and ADP ribosylation (3). Any combinations of these PTMs can to some extent increase or decrease the interactions between the DNA and the core histones and consequently affect chromatin accessibility at a particular locus. From a quantitative point of view histone acetylation is the most important modification occurring in all eukaryotes (3). The histone acetyltransferases (HATs) are the enzymes mediating histone acetylation and the reverse reaction, histone deacetylation, is catalyzed by histone deacetylases (HDACs). By deacetylating the core histones, HDACs neutralize the positive charges on histones tails and consequently compact the chromatin structure into a conformation that is repressive to most cellular processes (4).
Histone Acetyl Transferases (HATs)
HATs are evolutionary conserved from yeast to human and generally contain multiple protein subunits. These diverse proteins complexes are grouped in to two large families based on their catalytic domains. The GNATs family is named after its founding member, Gcn5 N-acetytransferase, and the MYST family is named for the founding members, Morf-Ybf2-Sas2-Tip60. Other proteins which do not contain consensus HAT domains also carry HAT activity such as p300/CBP (CREB-binding protein), Taf1 and some nuclear receptor co-activators. HATs enzymes take advantage of the proteins they are associated with for their recruitment to a particular location in the genome in order to carry a specific function. This mechanism also allows for substrate selection. For example PCAF associates with five different TAF proteins and acetylates histones H3 and H4 while TIP60 associates with a different set of proteins and acetylates histones H2A and H4 (5). In addition, a growing number of non-histones proteins are also acetylated by HATs. It is now well established that histone acetylation increases chromatin accessibility and the importance of this histone PTM in transcription has been demonstrated in several systems (6). Acetylation of the core histones has been associated with a looser, more open, chromatin structure that facilitates accessibility not only to the transcriptional machinery but also to other important cellular processes such as replication and DNA repair (4).
In keeping with the general mechanism by which HATs carry their specificity, two different HATs complexes have been associated with the repair of two different type of DNA damage. The MYST HATs function at sites of DNA double strand breaks (DSBs) while the GNAT HATs are recruited at sites of Nucleotide Excision Repair (NER). DBSs can be generated by ionizing radiation and are predominantly repaired by the non-homologous end joining (NHEJ) DNA repair pathway. This repair mechanism requires remodeling of chromatin into an “open” state at the sites of DSBs to allow the repair machinery to access the DNA ends (7). Acetylation of histones by HATs at sites of DSBs is thus an important step to allow efficient repair (8). Consequently, decreasing DSBs repair efficiency by inhibiting HATs could increase cells radio- sensitivity. This assumption has recently been validated in human lung and cervical cancer cells treated with garcinol, a HAT inhibitor (9). However, inhibition of the reverse enzymes, HDAC, can also sensitize cells to radiation (see below). It thus appears that both histone acetylation and deacetylation are essential for efficient DSBs repair. This reflects the importance of the histones PTMs dynamics at the sites of DSBs and their influence on chromatin remodeling.
Histone Deacetylase (HDAC) and HDAC Inhibitors
There are 18 known human HDAC grouped into two broad families; one family containing HDAC1-11 requires Zn2+ for deacetylase activity and is subdivided into three classes based on sequence homology to yeast deacetylases. Class I enzymes share sequence homology with yeast deacetylase RPD3, they are all located in the nucleus and includes HDAC1, 2, 3 and 8. HDAC4, 5, 6, 7, 9 and 10 form the class II enzymes. They share sequence homology with yeast deacetylase HDA1. Within the class II enzymes, HDAC6 and 10 form a sub-class, IIb, because they possess two catalytic sites and are expressed only in the cytoplasm. All other class II enzymes can shuttle back and forth between the nucleus and the cytoplasm. HDAC11 constitutes the class IV enzyme and shares homology with both the yeast RPD3 and HDA1 enzymes. The second HDAC family consists of seven members, sirtuins 1–7 and requires the NAD+ co-factor for activity. This family constitutes the class III enzymes and acts primarily on non-histones proteins. HDACs expression and activity are altered in many cancers (10) (11) but the mechanisms that lead to HDAC activation in tumor cells are not well understood. Both transcriptional as well as post-transcriptional mechanisms have been reported (12) (13). It is also possible that the unique environment of the cancer cells promotes HDAC activation. In fact, Reactive Oxygen Species (ROS) and hypoxia, which are elevated in cancer compared to normal cells, can increase HDAC activities (12–16). HDACs are thus believed to promote carcinogenesis through modulation of chromatin at specific loci and interaction with key transcriptional regulators.
Given the increasing understanding of HDAC role in cancer biology, small molecules inhibitors of HDAC enzymes have recently been identified and developed for cancer treatments. The current HDAC Inhibitors (HDACIs) are divided into four major classes based on their structure: 1) small molecular weight carboxylates (Valproic Acid), 2) hydroxyamic acids (Vorinostat/SAHA, Trichostatin A, Panobinostat, Belinostat), 3) benzamides (Entinostat, MGD0103), and 4) cyclic peptides (Romidepsin). Vorinostat/SAHA and Romidepsin (Depsipeptide) are currently the only two HDACIs approved by the FDA for cancer treatment. Their indication is for refractory cutaneous T-cell lymphoma. Vorinostat/SAHA and Romidepsin (Depsipeptide) are pan-specific HDACIs, targeting the zinc molecule found in the active site of class I, II and IV HDAC enzymes. The development of more specific HDACI is actively being pursuit but so far the clinical relevance of such a pursuit has only been demonstrated for HDAC8 inhibitors for the treatment of neuroblastoma (17). The pan specificity of the current HDACIs may contribute to their efficiency in a variety of drug combination therapies and could account for their relatively low toxicity (18). At least 12 different HDACIs are currently in some phase of clinical trials as monotherapy or in combination with chemotherapy or radiation therapy in patients with hematologic and solid tumors. However, the mechanism of action of HDACIs is still not completely understood.
HDACIs can inhibit the proliferation of transformed cells in culture and tumor growth in animal models by inhibiting cell cycle progression, inducing differentiation and apoptosis. Less than 10% of transcribed genes are altered by HDACIs treatment (19) but the altered genes vary from cell line to cell line and between different HDACIs. Therefore, no consistent picture of a target(s) or pathway(s) modulated by HDACIs has emerged yet. The low specific activity of the first generation of HDACIs is probably responsible for the pleoitropic activities associated with these inhibitors herein the apparent lack of a common denominator for their actions. What is consistent though is the HDACIs preferential selectivity for cancer cells as compared to normal cells and, acetylation of lysine residues on histones protein from the Peripheral Blood Mononuclear Cells (PBMC) of patients treated with HDACIs (18,20). Several mechanisms have been proposed to explain the HDACIs preferential selectivity for cancer cells but none so far can fully account for this generalized effect that crosses multiple cell lines and tumor types irrespective of the different cell line specific control of gene transcription. For instance, Vorinostat/SAHA can decrease the expression of the ROS scavenger thioredoxin in transformed but not normal cells (16). This effect could probably contributes to the HDACIs selectivity for some cancer cells but sensitivity to ROS varies within given histologies, cancer cell lines and tumor xenografts. Decreased expression of thioredoxin is thus unlikely to solely account for HDACIs selectivity for cancer cells. DNA repair defects that are prominent in cancer cells were also recently proposed as a mechanism for HDACIs selectivity for cancer cells (21). However, if the selection mechanism was so simple, why other anticancer drugs that damage DNA that can be repaired in normal but not cancer cells do no show this selectivity? Clearly, a more fundamental mechanism that could account for the collective selectivity of HDACIs for cancer cells is likely to underlie this general phenomenon. We recently proposed that the altered chromatin structure of cancer cells predisposed them to HDACIs sensitivity and selectivity (22). Abnormal nuclear morphology including chromatin clumping, irregular parachromatin clearing, and variability of nuclear size and shape are still hallmarks of the progressive distortion of the nuclear structures accompanying neoplasia. In addition, alterations in cancer cells morphology is altering the order and positioning of chromatin. It is thus possible that altered chromatin structure in cancer cells increases accessibility to the histone tails and allow a more profound distribution of histone PTMs. Such broad mechanism would not be HDACI specific or cancer cell type specific but would provide HDACIs selectivity for cancer cells.
HDACIs: Radioprotectors or radiosensitizers ?
The preference of HDACIs for cancer cells might also be due to a protective role on normal cells. When used topically at a relatively high dose (1%), the HDACI phenylbutyrate was shown to reduce the number of skin tumors in response to a single dose of radiation (40 Gy) in a well-established animal model of cutaneous radiation syndrome (23). The reduction of tumor formation was correlated with down regulation of oncogenes such as c-Jun, Myc and Bcl-2 followed by a reduction of inflammatory cytokines such as interleukins, TNF-α and TGF-β. More recent evidence have indicated that HDACIs can also protect mice against total body irradiation (TBI)(24, 25). HDACIs given either 24h before or 1h after a supralethal dose (7 Gy) TBI considerably reduced lethality. Without HDACI no mice survived a TBI dose of 7 Gy but half the mice that received HDCAI prior TBI were still alive 2 months after being exposed to 7 Gy (TBI) (24). Pathological analysis and spleen colony-forming assay indicated that intestinal and bone marrow cells recovered significantly from radiation-induced damage in the animals treated with HDACIs (24, 25). These studies are in good agreement with the stimulatory effect of HDACI on stem cells proliferation (26, 27) and may again reflect the different effect of HDACIs on the normal and cancer cells overall chromatin structure.
Changes on chromatin structure are expected to affect radiosensitivity. It is well established that cells are more radiosensitive when their chromatin is at its highest state of compaction during the G2/M phase of the cell cycle (28). However, G2/M cells do not get more DNA DSBs. In fact, elegant studies performed with 11 Mbp-long chromatin regions and with whole chromosome territories indicate that genetically inactive condensed chromatin is much less susceptible to radiation-induced DNA DSBs than decondensed chromatin (29). This is consistent with studies performed on chromatin isolated from V-79 Chinese hamster lung fibroblast where the amount of DNA breaks generated by ionizing radiation doubled on the relaxed acetylated chromatin (30). Therefore, even though a more relaxed chromatin structure favors the production of radiation induced DNA strand breaks this is not translated into increased radiosensitivity in cells. This is probably because DNA repair, an important component of radiosensitivity, is not measured on isolated chromatin. Initially it was thus believed that the compacted chromatin of the G2/M cells restricted DNA access to the repair enzymes and consequently increased radiosensitivity (31). More recent studies have shown that global chromatin compaction has moderate impact on the access and repair of DNA double strand breaks (DSBs) (32). However, dynamic local chromatin remodeling is required at the sites of DSBs. Studies have shown that DNA damage is repaired more efficiently on open chromatin (29). Chromatin needs to be remodeled at the sites of DNA DBS to first allow access of the repair machinery and then the removal of marks that signal DNA damage to subsequently allow chromatin refolding into its original more compacted structure once the repair is done (8). These events directly impact DNA repair efficiency and consequently the overall cellular radiosensitivity.
Several studies have indicated that HDACIs can sensitize cancer cells to radiation (33, 34). As mentioned earlier, the overall effect of HDACIs on chromatin structure is to increase the levels of histone acetylation which results in a more relaxed chromatin structure. HDACIs do not significantly affect the number of DSBs inflicted by ionizing radiation. HDACIs rather maintain the histones in a hyperacetylated status which prevents the refolding of chromatin into a more condensed structure following repair (35). In fact, previous studies have shown that increased histone acetylation in the region of DSBs are soon replaced by histone PTMs associated with a more condensed chromatin structure, such as decreased aceylation of histone H4-K5 and increased methylation of histone H3-K9, following DNA repair (35). HDACIs thus prevent these rapid epigenetic exchanges and lead to radiosensitivity. HDACIs can also decrease DNA repair efficiency by down regulating the expression of several DNA repair proteins such as Ku70, Ku86, Rad51 and DNA-PKc (36). In addition, HDACIs can prolong the expression of the DNA damage µarker γH2AX (36,37) thus preventing DNA DSB repair and potentiating the effects of radiation-induced killing. Moreover, histone acetylation could also facilitate the recruitment of chromatin remodeling complexes and other chromosomal proteins that might modify higher order chromatin structure to influence DNA repair efficiency. Conversely, chromatin remodeling proteins could influence the level of histone acetylation. For example, the nucleosome binding protein HMGN1 is required to mediate global increase in histone H3-K14 acetylation following ionizing radiation exposure (38). HMGN1 modulates the interaction of ATM with chromatin before and after DSB formation by reducing ATM binding to the chromatin (38). Similarly, HDACIs reduce the levels of chromatin-bound ATM both prior to and following IR exposure.
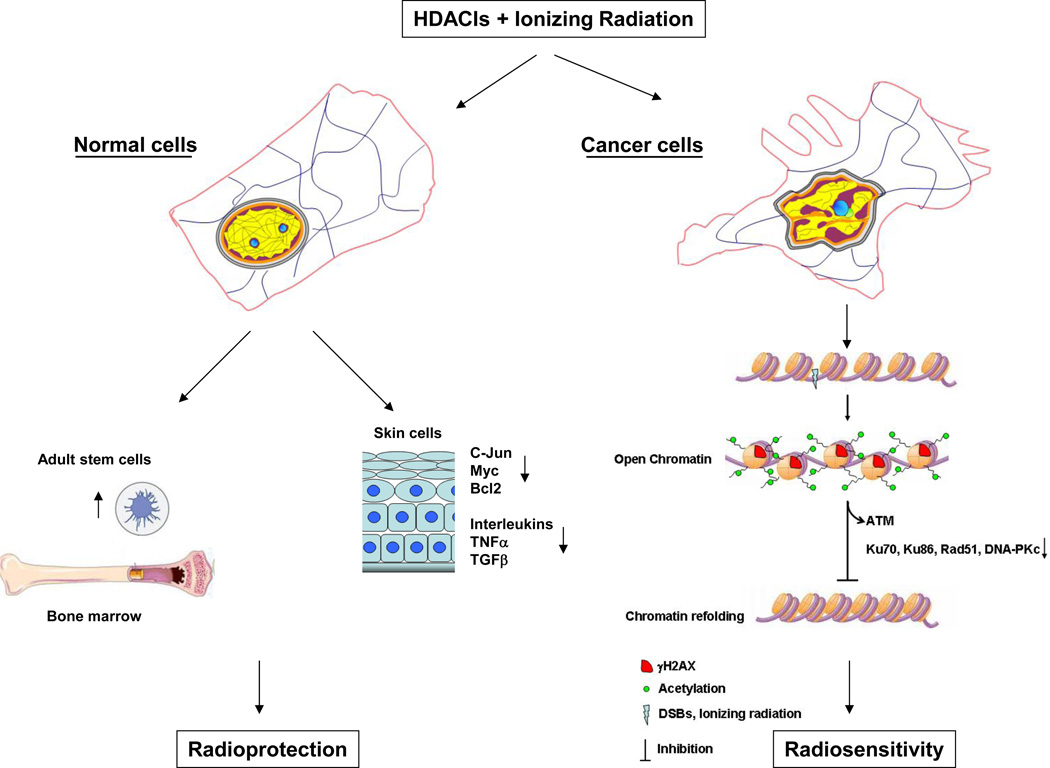
HDACIs can thus simultaneously protect normal cells while causing tumor cells to be more radiosensitive. The events leading to this distinctive effect on radiosensitivity are summarized in Fig. 1. As mentioned earlier, the chromatin of normal and cancer cells is dramatically different. To this day, changes in chromatin texture are still used as important criterion for cancer diagnosis. Increased heterochromatin (chromatin coarsening) and loss of heterochromatin aggregates (exaggerated open chromatin) are two significant changes observed in chromatin texture. These changes can have important effects on the strength of DNA damage signaling occurring at each DNA breaks (32). In fact, the impact of chromatin compaction on DNA damage signaling seems to be more important for low doses of radiation. Cells depleted of the DNA linker binder histone H1 have a more relaxed chromatin structure than the wild type cells and harbor hypersensitive G2/M checkpoint at radiation doses as low as 0.25 Gy (32). This dose had no effect in the wild type cells. Although higher order chromatin structure is important for ATM activation in response to radiation (38), the strength of the ATM signal is not affected by H1 depletion (32). It seems rather that the ATR pathway is most sensitive to chromatin modulation. Phosphorylation of the ATR substrate ChK1 has been observed at doses as low as 0.1 Gy in cells harboring a more relaxed chromatin structure (32). This is probably because the ATR pathway relies on chromatin remodeling for resection of DSB into single strand breaks and this process can be limited by chromatin compaction (39, 40). These observations are consistent with the radiosensitization effect of HDACIs at low doses of radiation (0.2 Gy) (41) and support the idea that one of the main mechanisms by which HDACIs affect radiosensitivity is through chromatin modulation.
Figure 1. Potential mechanisms by which HDACIs could simultaneously protect normal cells and sensitize cancer cells to ionizing radiation.
The chromatin of normal and cancer cells is dramatically different. In cancer cells, virtually all subcomponents of the nuclear and cellular matrix are altered. The shape of cancer cells nuclei is irregular and heterochromatin (purple) that usually surrounds nucleoli appears as coarse aggregates. Alterations in the cytomatrix also distort the overall cellular shape. In normal skin HDACIs reduce the number of radiation-induced skin tumors by down regulating oncogenes (c-Jun, Myc and Bcl-2) and inflammatory cytokines (interleukins, TNFα, TGF-β). HDACIs can also induce radioprotection by stimulating stem cells proliferation. In cancer cells, HDACIs acetylate core histones in the distorted chromatin and maintain the chromatin into an open state wich prevents the refolding of the chromatin into its original compacted structure following DNA repair. The acetylated chromatin prevents the binding of ATM and decreases the expression of repair proteins (KU70, Ku86, Rad51 and DNA-PKc).
Clinical trials
There are currently several clinical trials evaluating the combination of HDACIs with radiation therapy, some of which are listed in Table 1. Most of the trials are using the pan specific HDACI Vorinostat/SAHA which can cross the blood brain barrier. These trials reflect the potential benefit of HDACIs as radiosensitizers. The maximum tolerated dose of Vorinostat/SAHA has been determined at 300 mg once daily in combination with 30Gy radiation over 2 weeks in a Phase 1 trial for short term palliative pelvic radiotherapy for patients with gastrointestinal carcinoma (42). This small trial (16 evaluable patients) demonstrated that combination of HDACIs with radiation therapy is possible. Although a mean tumor reduction of 26% was observed, tumor reduction was highly variable and seven patients experienced grade 3 adverse events. Nonetheless, no grade 4 toxicities were associated with the treatment. It is expected that the outcomes of the currently undergoing clinical trials will contribute to effectively evaluate the efficacy and safety of HDACIs combined with radiation therapy.
Table 1.
Clinical trials combining HDACIs with radiation therapy.
| Clinical Trial No | Title | Sponsor |
|---|---|---|
| NCT00946673 | A Phase I Trial of Vorinostat Concurrent With Stereotactic Radiotherapy in Treatment of Brain Metastases From Non-Small Cell Lung Cancer |
Stanford University |
| NCT00983268 | Phase I Trial of Chemoradiation With Capecitabine and Vorinostat in Pancreatic Cancer. |
Vanderbilt-Ingram Cancer Center |
| NCT00731731 | Phase I/II Study of Vorinostat (Suberoylanilide Hydroxamic Acid [SAHA]), Temozolomide, and Radiation Therapy in Patients With Newly Diagnosed Glioblastoma |
National Cancer Institute (NCI) |
| NCT01236560 | A Randomized Phase II/III Study of Vorinostat and Local Irradiation OR Temozolomide and Local Irradiation OR Bevacizumab and Local Irradiation Followed by Maintenance Bevacizumab and Temozolomide in Children With Newly Diagnosed High-Grade Gliomas |
National Cancer Institute (NCI) |
| NCT01189266 | A Phase I-II Study of Suberoylanilide Hydroxamic Acid (SAHA, Vorinostat) and Local Irradiation, Followed by Maintenance SAHA in Children With Newly Diagnosed Diffuse Intrinsic Pontine Gliomas (DIPG) |
National Cancer Institute (NCI) |
| NCT01064921 | A Phase I Trial of Vorinostat in the Treatment of Advanced Oropharyngeal Carcinoma of the Head and Neck |
Ohio State University Comprehensive Cancer Center |
| NCT00831493 | Phase I/II Trial of Vorinostat and Radiation Therapy in Patients With Locally Advanced Pancreatic Cancer |
M.D. Anderson Cancer Center |
| NCT00662311 | Phase I/II Clinical Trial Evaluating the Use of Vorinostat Combined With Paclitaxel and Radiotherapy in Patients With Inoperable Stage III Non-Small Cell Lung Cancer Unable to Tolerate Cisplatin |
Fred Hutchinson Cancer Research Center |
| NCT01378481 | High-Dose Vorinostat With Radiation Therapy in the Treatment of Recurrent Glioma |
Thomas Jefferson University |
| NCT00838929 | Phase I Study of the Combination of Vorinostat and Radiation Therapy for the Treatment of Patients With Brain Metastases |
Thomas Jefferson University |
| NCT00404326 | A Phase II Study of Transcriptional Therapy With the DNA Demethylating Hydralazine and the HDAC Inhibitor Valproate Associated to Concomitant Cisplatin Chemoradiation in FIGO Stage III Cervical Cancer. |
National Institute of Cancerología |
| NCT01384799 | A Phase I Dose Escalation Study to Investigate the Safety and Pharmacokinetics of Intravenous CUDC- 101 With Concurrent Cisplatin and Radiation Therapy in Subjects With Locally Advanced Human Papillomavirus Negative Head and Neck Cancer |
Curis, Inc. |
| NCT00455351 | Phase I Study on Suberoylanilide Hydroxyamic Acid (Vorinostat) a Histone Deacetylase Inhibitor, in Palliative Radiotherapy for Advanced Tumors. |
Oslo University Hospital |
| NCT00302159 | A Phase II Clinical Trial of the Histone Deacetylase Inhibitor Valproic Acid in Combination With Temodar and Radiation Therapy in Patients With High Grade Gliomas: Multi-Institutional Trial |
National Institutes of Health Clinical Center (CC) |
| NCT00948688 | Phase 1/2 Study of Vorinostat in Combination With Radiation Therapy and Infusional 5-FU in Patients With Locally Advanced Adenocarcinoma of the Pancreas |
Massachusetts General Hospital |
| NCT00437957 | Phase I Trial Of Temozolomide Combined With The Histone Deacetylase Inhibitor Valproic Acid (VPA) And Whole Brain Radiation Therapy (WBR) For Brain Metastases From Solid Tumors In Adults |
H. Lee Moffitt Cancer Center and Research Institute |
| NCT00821951 | A Dose Escalation Study of Vorinostat in Combination With Palliative Radiotherapy for Patients With Non- Small Cell Lung Cancer |
Yale University |
Concluding remarks
HDACIs have generated a lot of interest in the field of cancer research. They offer the possibility to potentiate a variety of conventional anticancer treatments while at the same time spare or even protect normal tissues. This pan specific potentiation effect probably stems from the low specificity of the first generation of HDACIs and thus brings in to question whether the quest for more specific HDACIs is the right direction to take. Nonetheless, there are some instances where this seems to be an obvious decision. For example, over expression of HDAC8 in neuroblastoma and reversal of the phenotype with HDAC8 inhibition clearly indicates that a specific inhibitor for HDAC8 would be greatly desirable (17). However, for most other cases it rather seems that the relatively low toxicity associated with HDACIs in the clinic is key to the added benefit of drug combination. The low toxicity of HDACIs may also suggest that low doses of HDACIs may be used as maintenance therapy. This could potentially be particularly beneficial after combing HDACIs with anticancer drugs that target DNA or enzymes acting on the DNA. By preventing the refolding of cancer cells chromatin through increased histone acetylation HDACIs could prevent repair of cancer cells and prolong the effect of the anticancer drug. The effect of HDACIs on chromatin refolding could also be exploited for low dose fractionated radiation therapy (LDFRT). LDFRT has been shown to potentiate a number of anticancer drugs due to a phenomenon known as hyper radiosensivity whereby cells are more susceptible to radiation killing at doses below 0.5 Gy where the conventional linear-quadratic model would predict survival (43, 44). We are just beginning to understand the clinical benefits of HDACIs for cancer treatments. As it is often the case when a new class of anticancer drugs is identified and investigated, expectations are high for HDACIs but as always we are gaining valuable knowledge along the way that bring us closer to the goal of curing or at least managing cancer progression.
Acknowledgements
This work was supported in part by the Marlene and Stewart Greenebaum Cancer center, University of Maryland, Baltimore, the Maryland Cigarette restitution fund and the National Cancer Institute, NIH/NCI, RO1 1CA116491.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Rupp RA, Becker PB. Gene regulation by histone H1: new links to DNA methylation. Cell. 2005;123:1178–1179. doi: 10.1016/j.cell.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Wolffe A. Chromatin. New York: Academic Press; 1998. [Google Scholar]
- 3.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 4.Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271:2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 5.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 6.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 7.Ogiwara H, Ui A, Otsuka A, Satoh H, et al. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30:2135–2146. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
- 8.Rossetto D, Truman AW, Kron SJ, Cote J. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin Cancer Res. 2010;16:4543–4552. doi: 10.1158/1078-0432.CCR-10-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oike T, Ogiwara H, Torikai K, et al. A Histone Acetyltransferase Inhibitor, Radiosensitizes Cancer Cells by Inhibiting Non-Homologous End Joining. Int J Radiat Oncol Biol Phys. 2012;84:815–821. doi: 10.1016/j.ijrobp.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa M, Oda Y, Eguchi T, et al. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep. 2007;18:769–774. [PubMed] [Google Scholar]
- 11.Jin KL, Pak JH, Park JY, et al. Expression profile of histone deacetylases 1, 2 and 3 in ovarian cancer tissues. J Gynecol Oncol. 2008;19:185–190. doi: 10.3802/jgo.2008.19.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim MS, Kwon HJ, Lee YM, et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat Med. 2001;7:437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 13.Pluemsampant S, Safronova OS, Nakahama K, Morita I. Protein kinase CK2 is a key activator of histone deacetylase in hypoxia-associated tumors. Int J Cancer. 2008;122:333–341. doi: 10.1002/ijc.23094. [DOI] [PubMed] [Google Scholar]
- 14.Miura K, Taura K, Kodama Y, Schnabl B, Brenner DA. Hepatitis C virus-induced oxidative stress suppresses hepcidin expression through increased histone deacetylase activity. Hepatology. 2008;48:1420–1429. doi: 10.1002/hep.22486. [DOI] [PubMed] [Google Scholar]
- 15.Zhu H, Shan L, Schiller PW, Mai A, Peng T. Histone deacetylase-3 activation promotes tumor necrosis factor-alpha (TNF-alpha) expression in cardiomyocytes during lipopolysaccharide stimulation. J Biol Chem. 2010;285:9429–9436. doi: 10.1074/jbc.M109.071274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler LM, Zhou X, Xu WS, et al. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci U S A. 2002;99:11700–11705. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oehme I, Deubzer HE, Wegener D, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15:91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 18.Schrump DS. Cytotoxicity mediated by histone deacetylase inhibitors in cancer cells: mechanisms and potential clinical implications. Clin Cancer Res. 2009;15:3947–3957. doi: 10.1158/1078-0432.CCR-08-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MS, Blake M, Baek JH, Kohlhagen G, Pommier Y, Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- 21.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci U S A. 2010;107:14639–14644. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nalabothula N, Carrier F. Cancer cells’ epigenetic composition and predisposition to histone deacetylase inhibitor sensitization. Epigenomics. 2011;3:145–155. doi: 10.2217/epi.11.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung YL, Wang AJ, Yao LF. Antitumor histone deacetylase inhibitors suppress cutaneous radiation syndrome: Implications for increasing therapeutic gain in cancer radiotherapy. Mol Cancer Ther. 2004;3:317–325. [PubMed] [Google Scholar]
- 24.Brown SL, Kolozsvary A, Liu J, Ryu S, Kim JH. Histone deacetylase inhibitors protect against and mitigate the lethality of total-body irradiation in mice. Radiat Res. 2008;169:474–478. doi: 10.1667/RR1245.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konsoula Z, Velena A, Lee R, Dritschilo A, Jung M. Histone deacetylase inhibitor: antineoplastic agent and radiation modulator. Adv Exp Med Biol. 2011;720:171–179. doi: 10.1007/978-1-4614-0254-1_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young JC, Wu S, Hansteen G, et al. Inhibitors of histone deacetylases promote hematopoietic stem cell self-renewal. Cytotherapy. 2004;6:328–336. doi: 10.1080/14653240410004899. [DOI] [PubMed] [Google Scholar]
- 27.De Felice L, Tatarelli C, Mascolo MG, et al. Histone deacetylase inhibitor valproic acid enhances the cytokine-induced expansion of human hematopoietic stem cells. Cancer Res. 2005;65:1505–1513. doi: 10.1158/0008-5472.CAN-04-3063. [DOI] [PubMed] [Google Scholar]
- 28.Chapman JD, Stobbe CC, Gales T, et al. Condensed chromatin and cell inactivation by single-hit kinetics. Radiat Res. 1999;151:433–441. [PubMed] [Google Scholar]
- 29.Falk M, Lukasova E, Kozubek S. Chromatin structure influences the sensitivity of DNA to gamma-radiation. Biochim Biophys Acta. 2008;1783:2398–2414. doi: 10.1016/j.bbamcr.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Nackerdien Z, Michie J, Bohm L. Chromatin decondensed by acetylation shows an elevated radiation response. Radiat Res. 1989;117:234–244. [PubMed] [Google Scholar]
- 31.Oleinick NL, Chiu SM, Friedman LR. Gamma radiation as a probe of chromatin structure: damage to and repair of active chromatin in the metaphase chromosome. Radiat Res. 1984;98:629–641. [PubMed] [Google Scholar]
- 32.Murga M, Jaco I, Fan Y, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camphausen K, Scott T, Sproull M, Tofilon PJ. Enhancement of xenograft tumor radiosensitivity by the histone deacetylase inhibitor MS-275 and correlation with histone hyperacetylation. Clin Cancer Res. 2004;10:6066–6071. doi: 10.1158/1078-0432.CCR-04-0537. [DOI] [PubMed] [Google Scholar]
- 34.Karagiannis TC, El-Osta A. The paradox of histone deacetylase inhibitor-mediated modulation of cellular responses to radiation. Cell Cycle. 2006;5:288–295. doi: 10.4161/cc.5.3.2421. [DOI] [PubMed] [Google Scholar]
- 35.Falk M, Lukasova E, Gabrielova B, Ondrej V, Kozubek S. Chromatin dynamics during DSB repair. Biochim Biophys Acta. 2007;1773:1534–1545. doi: 10.1016/j.bbamcr.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Munshi A, Kurland JF, Nishikawa T, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res. 2005;11:4912–4922. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 37.Camphausen K, Burgan W, Cerra M, et al. Enhanced radiation-induced cell killing and prolongation of gammaH2AX foci expression by the histone deacetylase inhibitor MS-275. Cancer Res. 2004;64:316–321. doi: 10.1158/0008-5472.can-03-2630. [DOI] [PubMed] [Google Scholar]
- 38.Kim YC, Gerlitz G, Furusawa T, et al. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol. 2009;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 41.Zaskodova D, Rezacova M, Vavrova J, Vokurkova D, Tichy A. Effect of valproic acid, a histone deacetylase inhibitor, on cell death and molecular changes caused by low-dose irradiation. Ann N Y Acad Sci. 2006;1091:385–398. doi: 10.1196/annals.1378.082. [DOI] [PubMed] [Google Scholar]
- 42.Ree AH, Dueland S, Folkvord S, et al. Vorinostat, a histone deacetylase inhibitor, combined with pelvic palliative radiotherapy for gastrointestinal carcinoma: the Pelvic Radiation and Vorinostat (PRAVO) phase 1 study. Lancet Oncol. 2010;11:459–464. doi: 10.1016/S1470-2045(10)70058-9. [DOI] [PubMed] [Google Scholar]
- 43.Gupta S, Koru-Sengul T, Arnold SM, Devi GR, Mohiuddin M, Ahmed MM. Low-dose fractionated radiation potentiates the effects of cisplatin independent of the hyper-radiation sensitivity in human lung cancer cells. Mol Cancer Ther. 2011;10:292–302. doi: 10.1158/1535-7163.MCT-10-0630. [DOI] [PubMed] [Google Scholar]
- 44.Dey S, Spring PM, Arnold S, et al. Low-dose fractionated radiation potentiates the effects of Paclitaxel in wild-type and mutant p53 head and neck tumor cell lines. Clin Cancer Res. 2003;9:1557–1565. [PubMed] [Google Scholar]