Abstract
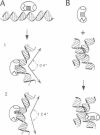
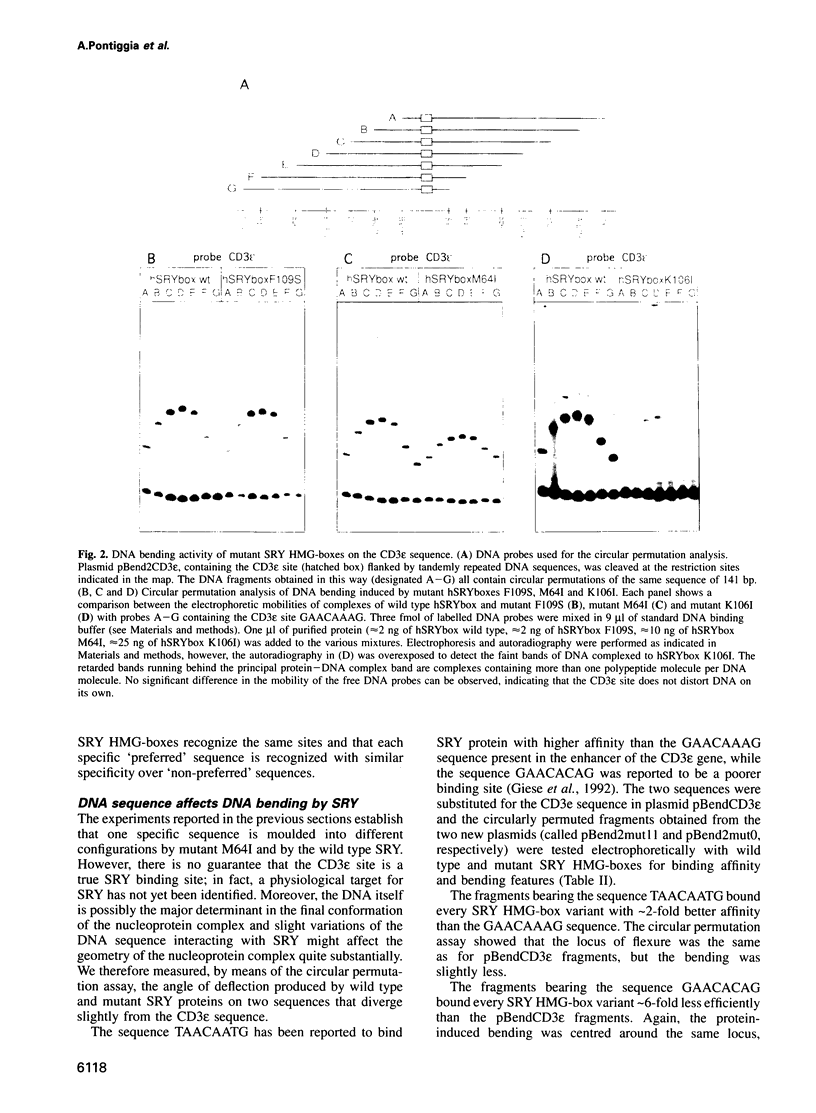
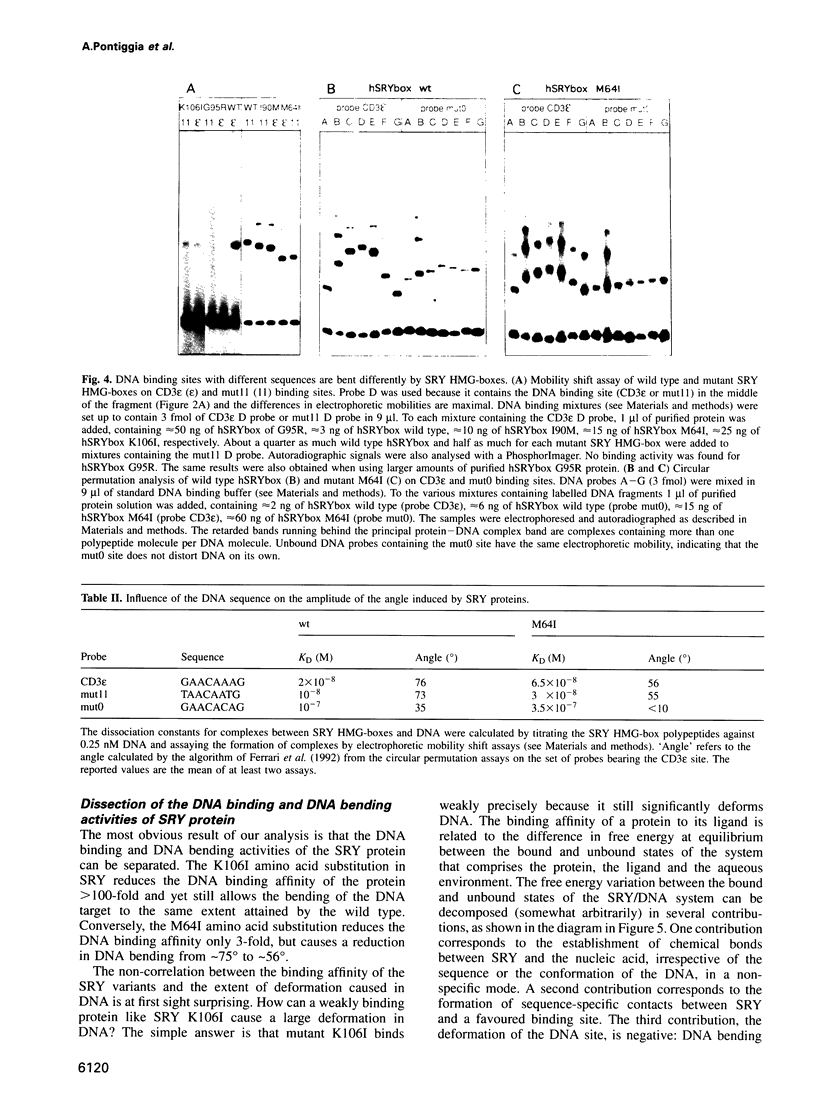
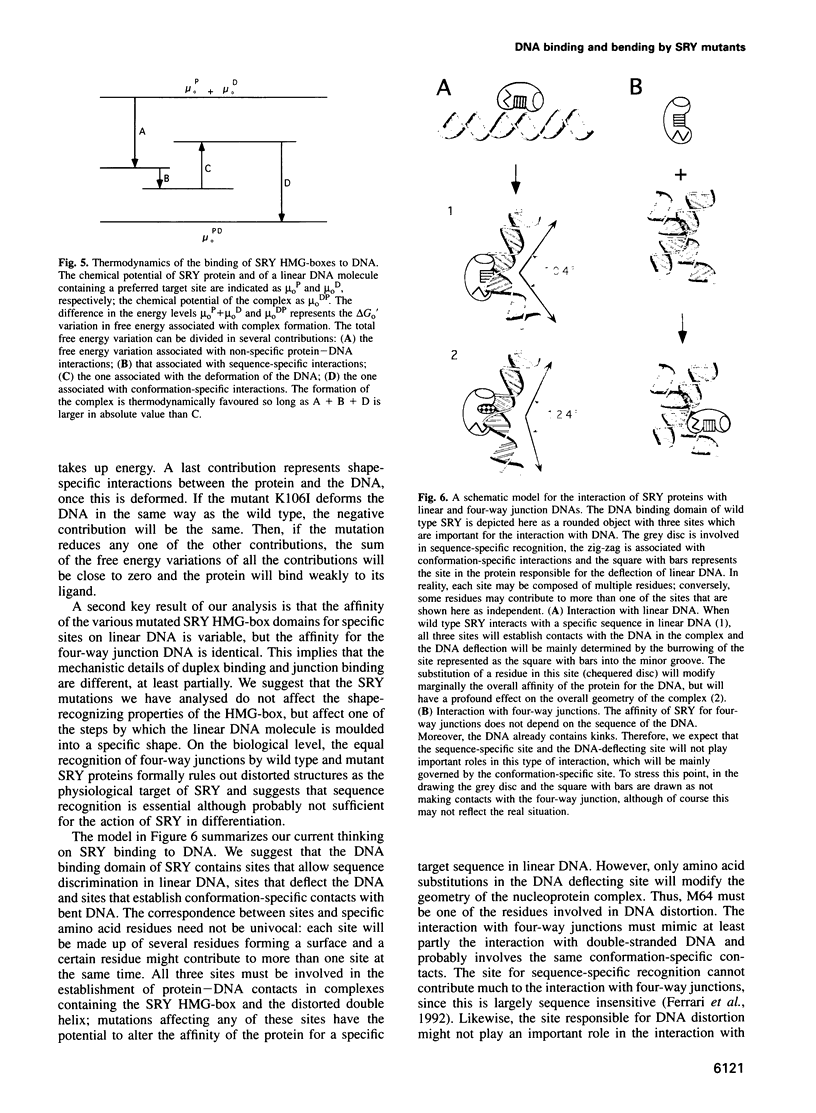
The testis determining factor, SRY, is a DNA binding protein that causes a large distortion of its DNA target sites. We have analysed the biochemical properties of the DNA binding domains (HMG-boxes) of mutant SRY proteins from five patients with complete gonadal dysgenesis. The mutant proteins fall into three categories: two bind and bend DNA almost normally, two bind inefficiently but bend DNA normally and one binds DNA with almost normal affinity but produces a different angle. The mutations with moderate effect on complex formation can be transmitted to male progeny, the ones with severe effects on either binding or bending are de novo. The angle induced by SRY depends on the exact DNA sequence and thus adds another level of discrimination in target site recognition. These data suggest that the exact spatial arrangement of the nucleoprotein complex organized by SRY is essential for sex determination.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berta P., Hawkins J. R., Sinclair A. H., Taylor A., Griffiths B. L., Goodfellow P. N., Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990 Nov 29;348(6300):448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Falciola L., Ferrari S., Lilley D. M. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992 Mar;11(3):1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny P., Swift S., Brand N., Dabhade N., Barton P., Ashworth A. A conserved family of genes related to the testis determining gene, SRY. Nucleic Acids Res. 1992 Jun 11;20(11):2887–2887. doi: 10.1093/nar/20.11.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny P., Swift S., Connor F., Ashworth A. An SRY-related gene expressed during spermatogenesis in the mouse encodes a sequence-specific DNA-binding protein. EMBO J. 1992 Oct;11(10):3705–3712. doi: 10.1002/j.1460-2075.1992.tb05455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciola L., Murchie A. I., Lilley D. M., Bianchi M. Mutational analysis of the DNA binding domain A of chromosomal protein HMG1. Nucleic Acids Res. 1994 Feb 11;22(3):285–292. doi: 10.1093/nar/22.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Harley V. R., Pontiggia A., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992 Dec;11(12):4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K., Cox J., Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992 Apr 3;69(1):185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Giese K., Grosschedl R. LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. EMBO J. 1993 Dec;12(12):4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K., Pagel J., Grosschedl R. Distinct DNA-binding properties of the high mobility group domain of murine and human SRY sex-determining factors. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3368–3372. doi: 10.1073/pnas.91.8.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Giese K., Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994 Mar;10(3):94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Gubbay J., Collignon J., Koopman P., Capel B., Economou A., Münsterberg A., Vivian N., Goodfellow P., Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990 Jul 19;346(6281):245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Harley V. R., Jackson D. I., Hextall P. J., Hawkins J. R., Berkovitz G. D., Sockanathan S., Lovell-Badge R., Goodfellow P. N. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992 Jan 24;255(5043):453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- Harley V. R., Lovell-Badge R., Goodfellow P. N. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994 Apr 25;22(8):1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J. R., Taylor A., Berta P., Levilliers J., Van der Auwera B., Goodfellow P. N. Mutational analysis of SRY: nonsense and missense mutations in XY sex reversal. Hum Genet. 1992 Feb;88(4):471–474. doi: 10.1007/BF00215684. [DOI] [PubMed] [Google Scholar]
- Hawkins J. R., Taylor A., Goodfellow P. N., Migeon C. J., Smith K. D., Berkovitz G. D. Evidence for increased prevalence of SRY mutations in XY females with complete rather than partial gonadal dysgenesis. Am J Hum Genet. 1992 Nov;51(5):979–984. [PMC free article] [PubMed] [Google Scholar]
- Ho I. C., Leiden J. M. Regulation of the human T-cell receptor alpha gene enhancer: multiple ubiquitous and T-cell-specific nuclear proteins interact with four hypomethylated enhancer elements. Mol Cell Biol. 1990 Sep;10(9):4720–4727. doi: 10.1128/mcb.10.9.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger R. J., Harley V. R., Pfeiffer R. A., Goodfellow P. N., Scherer G. A familial mutation in the testis-determining gene SRY shared by both sexes. Hum Genet. 1992 Dec;90(4):350–355. doi: 10.1007/BF00220457. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991 Nov 22;254(5035):1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- King C. Y., Weiss M. A. The SRY high-mobility-group box recognizes DNA by partial intercalation in the minor groove: a topological mechanism of sequence specificity. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11990–11994. doi: 10.1073/pnas.90.24.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P., Gubbay J., Vivian N., Goodfellow P., Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991 May 9;351(6322):117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Voit R., Stefanovsky V., Evers R., Bianchi M., Grummt I. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 1994 Jan 15;13(2):416–424. doi: 10.1002/j.1460-2075.1994.tb06276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene S. D., Zimm B. H. Understanding the anomalous electrophoresis of bent DNA molecules: a reptation model. Science. 1989 Jul 28;245(4916):396–399. doi: 10.1126/science.2756426. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. DNA--protein interactions. HMG has DNA wrapped up. Nature. 1992 May 28;357(6376):282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Nasrin N., Buggs C., Kong X. F., Carnazza J., Goebl M., Alexander-Bridges M. DNA-binding properties of the product of the testis-determining gene and a related protein. Nature. 1991 Nov 28;354(6351):317–320. doi: 10.1038/354317a0. [DOI] [PubMed] [Google Scholar]
- Sinclair A. H., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., Smith M. J., Foster J. W., Frischauf A. M., Lovell-Badge R., Goodfellow P. N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990 Jul 19;346(6281):240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Tucker P. K., Lundrigan B. L. Rapid evolution of the sex determining locus in Old World mice and rats. Nature. 1993 Aug 19;364(6439):715–717. doi: 10.1038/364715a0. [DOI] [PubMed] [Google Scholar]
- Whitfield L. S., Lovell-Badge R., Goodfellow P. N. Rapid sequence evolution of the mammalian sex-determining gene SRY. Nature. 1993 Aug 19;364(6439):713–715. doi: 10.1038/364713a0. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992 Aug;11(8):3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]