Abstract
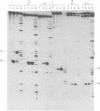
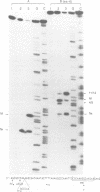
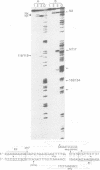
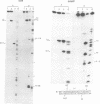
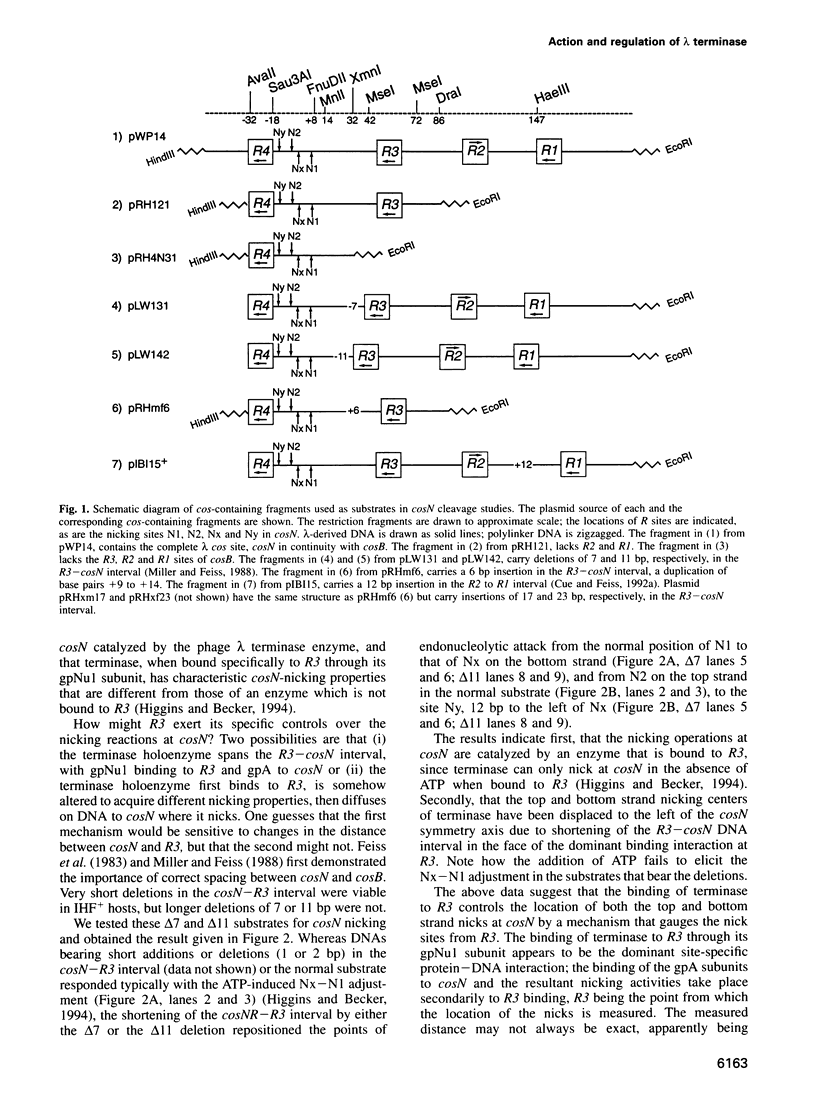
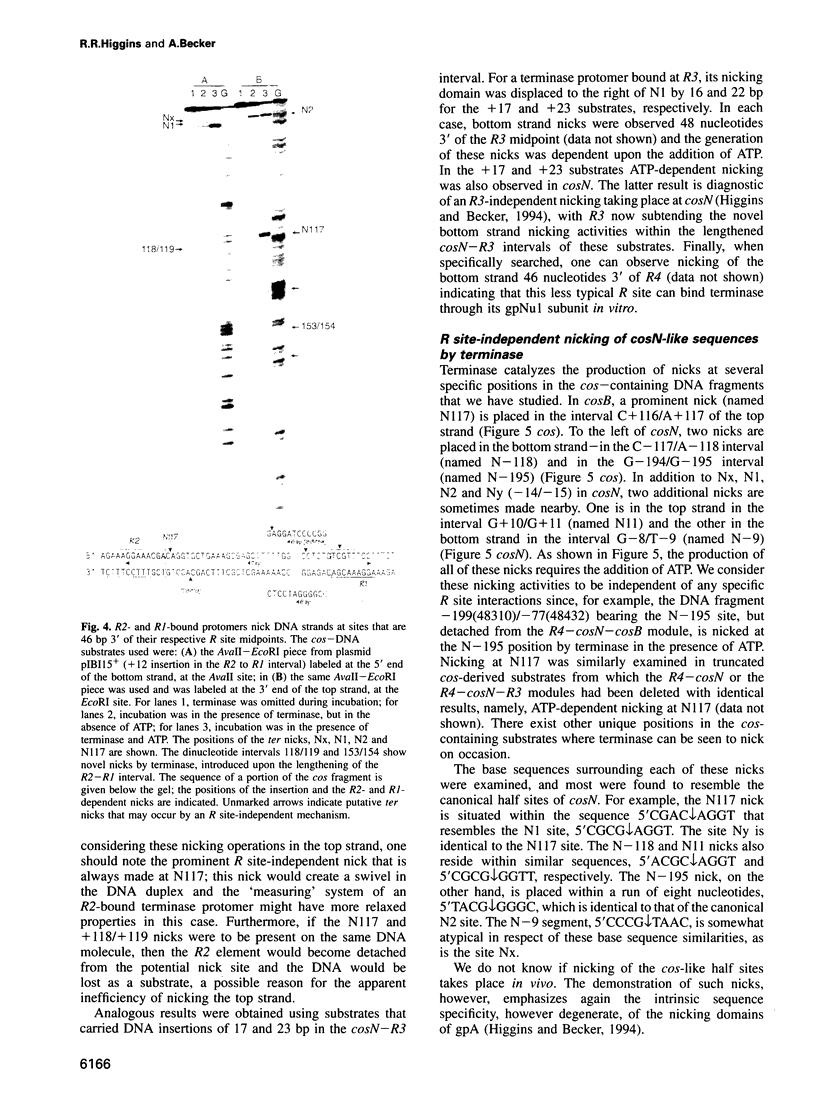
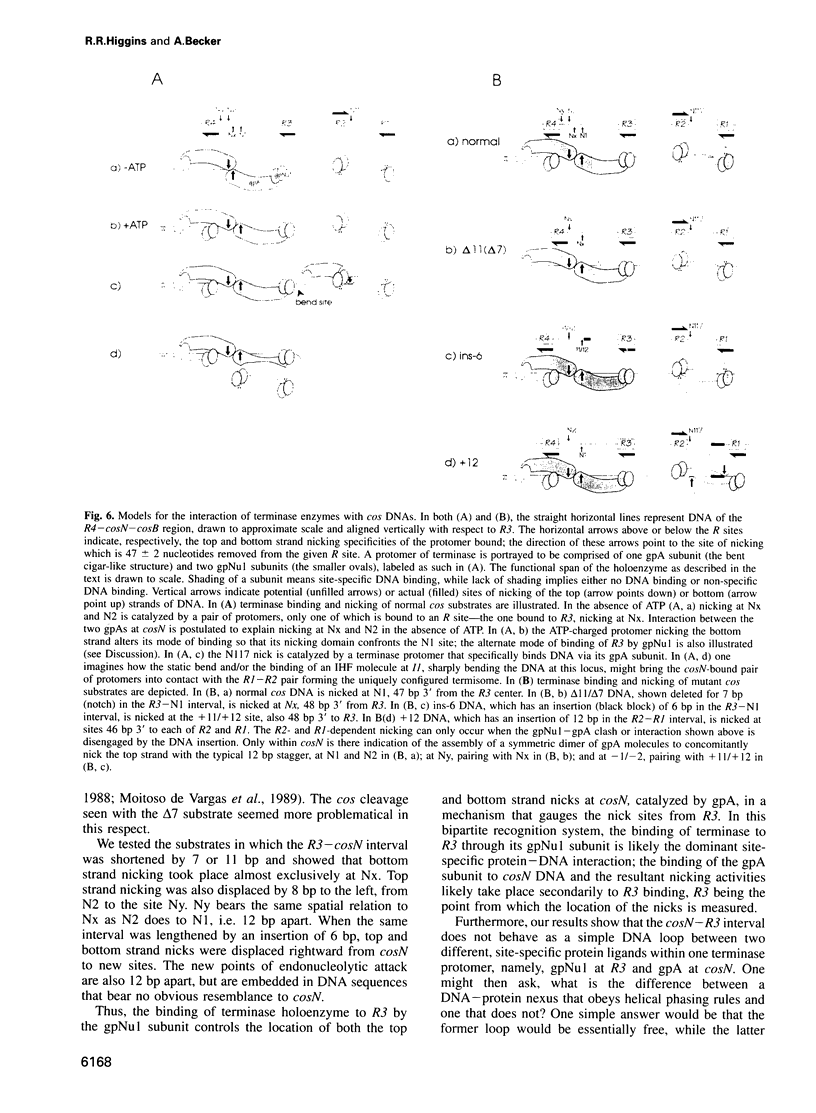
lambda terminase is an ATP-interactive, site-specific endonuclease comprising the products of lambda genes Nu1 and A. Terminase binds to cos, at the junction of two chromosomes in a concatemer, catalyzes cos cleavage and initiates the packaging of lambda DNA into proheads. cos consists of a nicking domain, cosN, where terminase cleaves to regenerate the 12 nucleotide cohesive ends of mature lambda chromosomes and a binding domain, cosB, where terminase binds to 16 bp repeat sequences called R3, R2 and R1. Evidence is presented that terminase is a single-strand endonuclease that can nick DNA by one of two mechanisms, both of which require ATP. (i) When bound to any R site, terminase nicks the strand which, within that R site, is purine-rich; the position of this nick is 47 +/- 2 nucleotides away from the mid-point of that R site, measured in the 3' direction; (ii) enzymes that are not bound to R sites nick DNA within certain specific sequences that resemble cosN half sites. These two modes of action are nicely combined for the R3-bound protomer that nicks the bottom strand at position N1 in cosN since the interval between N1 and the R3 midpoint is 47 nucleotides. Within cosN, the bottom and top strand nicks are generated by a rigid protein couple with a 2-fold rotational symmetry. The location of both of these nicks, however, is gauged asymmetrically from R3, 47 nucleotides away. Again, R1 and R2 are separated by 47 bp and orient bound protomers towards each other but, unless the DNA between these R sites is lengthened, the enzymes do not nick, indicating an inhibitory gpA-gpNu1 apposition.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bear S. E., Court D. L., Friedman D. I. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol. 1984 Dec;52(3):966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Murialdo H. Bacteriophage lambda DNA: the beginning of the end. J Bacteriol. 1990 Jun;172(6):2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Feiss M. Genetic analysis of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage lambda. J Mol Biol. 1992 Nov 5;228(1):58–71. doi: 10.1016/0022-2836(92)90491-2. [DOI] [PubMed] [Google Scholar]
- Cue D., Feiss M. The role of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage lambda, in the nicking reaction. J Mol Biol. 1993 Dec 5;234(3):594–609. doi: 10.1006/jmbi.1993.1614. [DOI] [PubMed] [Google Scholar]
- Feiss M., Widner W. Bacteriophage lambda DNA packaging: scanning for the terminal cohesive end site during packaging. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3498–3502. doi: 10.1073/pnas.79.11.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiss M., Widner W., Miller G., Johnson G., Christiansen S. Structure of the bacteriophage lambda cohesive end site: location of the sites of terminase binding (cosB) and nicking (cosN). Gene. 1983 Oct;24(2-3):207–218. doi: 10.1016/0378-1119(83)90081-1. [DOI] [PubMed] [Google Scholar]
- Higgins R. R., Becker A. Chromosome end formation in phage lambda, catalyzed by terminase, is controlled by two DNA elements of cos, cosN and R3, and by ATP. EMBO J. 1994 Dec 15;13(24):6152–6161. doi: 10.1002/j.1460-2075.1994.tb06962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R. R., Lucko H. J., Becker A. Mechanism of cos DNA cleavage by bacteriophage lambda terminase: multiple roles of ATP. Cell. 1988 Sep 9;54(6):765–775. doi: 10.1016/s0092-8674(88)91021-5. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller G., Feiss M. The bacteriophage lambda cohesive end site: isolation of spacing/substitution mutations that result in dependence on Escherichia coli integration host factor. Mol Gen Genet. 1988 Apr;212(1):157–165. doi: 10.1007/BF00322459. [DOI] [PubMed] [Google Scholar]
- Miwa T., Matsubara K. Lambda phage DNA sequences affecting the packaging process. Gene. 1983 Oct;24(2-3):199–206. doi: 10.1016/0378-1119(83)90080-x. [DOI] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Kim S., Landy A. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science. 1989 Jun 23;244(4911):1457–1461. doi: 10.1126/science.2544029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinder G., Gold M. The Nul subunit of bacteriophage lambda terminase binds to specific sites in cos DNA. J Virol. 1988 Feb;62(2):387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Snyder U. K., Landy A. Helical-repeat dependence of integrative recombination of bacteriophage lambda: role of the P1 and H1 protein binding sites. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6323–6327. doi: 10.1073/pnas.85.17.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomka M. A., Catalano C. E. Physical and kinetic characterization of the DNA packaging enzyme from bacteriophage lambda. J Biol Chem. 1993 Feb 15;268(5):3056–3065. [PubMed] [Google Scholar]
- Xin W. N., Feiss M. The interaction of Escherichia coli integration host factor with the cohesive end sites of phages lambda and 21. Nucleic Acids Res. 1988 Mar 25;16(5):2015–2030. doi: 10.1093/nar/16.5.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin W., Feiss M. Function of IHF in lambda DNA packaging. I. Identification of the strong binding site for integration host factor and the locus for intrinsic bending in cosB. J Mol Biol. 1993 Mar 20;230(2):492–504. doi: 10.1006/jmbi.1993.1166. [DOI] [PubMed] [Google Scholar]