Abstract
Scalable and genetically stable recombinant adeno-associated virus (rAAV) production systems combined with facile adaptability for an extended repertoire of AAV serotypes are required to keep pace with the rapidly increasing clinical demand. For scalable high-titer production of the full range of rAAV serotypes 1–12, we developed OneBac, consisting of stable insect Sf9 cell lines harboring silent copies of AAV1–12 rep and cap genes induced upon infection with a single baculovirus that also carries the rAAV genome. rAAV burst sizes reach up to 5×105 benzonase-resistant, highly infectious genomic particles per cell, exceeding typical yields of current rAAV production systems. In contrast to recombinant rep/cap baculovirus strains currently employed for large-scale rAAV production, the Sf9rep/cap cell lines are genetically stable, leading to undiminished rAAV burst sizes over serial passages. Thus, OneBac combines full AAV serotype options with the capacity for stable scale-up production, the current bottleneck for the transition of AAV from gene therapy trials to routine clinical treatment.
Introduction
Gene therapy with adeno-associated virus (AAV) vectors has witnessed enormous clinical progress in recent years. Exiting improvements in the treatment of diverse genetic diseases, including congenital blindness and hemophilia (Mingozzi and High, 2011; Nathwani et al., 2011), have been made possible by recombinant AAV (rAAV)-mediated gene delivery. Recently, Glybera, an AAV vector expressing lipoprotein lipase to treat metabolic lipid disorders, was approved as the first human gene therapy to become commercially available (Kastelein et al., 2013). The increasing request for clinical application requires robust and scalable production methods for an increasing range of AAV serotypes. At present, 12 major AAV serotypes have been isolated from humans and nonhuman primates, not counting the numerous engineered variants thereof. The AAV serotypes have distinct capsids, which interact with specific cell surface receptors. Whereas AAV2 or AAV1 have mostly been used in clinical trials so far, the extended cell- and tissue-specific transduction patterns of alternative AAV serotypes are being increasingly translated into clinical gene therapy (Mingozzi and High, 2011; Nathwani et al., 2011).
rAAV vectors are constructed as gene-deleted AAVs comprising the gene of interest flanked the AAV-ITRs, the only cis elements required for rAAV replication and packaging. The AAV gene rep codes for the regulatory proteins Rep78/68 and Rep52/40, while cap codes for the capsid proteins VP1, VP2, and VP3. For rAAV production, rep and cap have to be provided in trans together with the required helper virus genes. Plasmid cotransfection of the rAAV genome, AAVrep/cap, and adenovirus helper genes in 293 cells represents the most widely used protocol for rAAV production (Grimm et al., 1998, 2003; Xiao et al., 1998). However, inherent problems in growing and transfecting 293 cells in suspension culture limit the scalability of this system.
To develop a scalable rAAV production system, Urabe et al. (2002) exploited Sf9 insect cells that produced rAAV2 upon coinfection with three recombinant baculovirus (Bac) vectors, encoding the rep gene, the cap gene, and the rAAV genome, respectively. Production of three Bac-mediated rAAVs was subsequently scaled up to bioreactor size (Cecchini et al., 2011), and rep and cap were combined to reduce the number of coinfecting Bacs (Smith et al., 2009). Unfortunately, Bac instability, especially of Bac-Rep, limits rAAV yields after few successive passages (Kohlbrenner et al., 2005). Rep78 expression has long been known for its cell toxicity, a feature that has notoriously hampered efforts to generate stable Rep-expressing cell lines (Zolotukhin, 2005).
To circumvent the above problems, Aslanidi et al. (2009) did the next step and developed a two-component rAAV production system consisting of a stable Sf9 cell line with integrated but silent copies of AAV rep and cap, each controlled by the Bac-derived polyhedrin promoter polh and the cis-acting element hr2-0.9. Expression of AAV rep and cap is induced upon Bac infection by immediate-early (IE-1) gene-mediated transactivation of the hr2-0.9 enhancer. The Bac also harbors the rAAV genome as described before (Urabe et al., 2002; Aslanidi et al., 2009). Once expressed, Rep78 initiates a feed-forward loop that amplifies the integrated rep and cap genes by interaction with the incorporated cognate AAV Rep-binding site (RBE). This leads to rAAV2 burst sizes per cell exceeding those of the three-Bac infection system (Aslanidi et al., 2009).
In the present study, we have further developed this system to OneBac: Sf9 rep/cap producer cell lines for the entire range of rAAV1–12 serotypes produced upon infection with a single Bac (Bac-rAAV). rAAV production efficiencies were quantified in comparison to the 293-cell cotransfection protocol, the only other system offering easy access to an extended range of rAAV serotypes. High rAAV burst sizes per cell persist over successive cell passages surpassing the efficiency and stability of current rAAV production systems. OneBac can easily be adapted to production of novel serotypes by selection of additional Sf9 cell lines, as described here for rAAV serotypes 1–12. Taken together, OneBac is ideally suited for scale-up production of the rAAV serotype of choice as required for clinical application.
Materials and Methods
Plasmids and cloning
Plasmids pIR-VP-hr2-RBE and pIR-rep78-hr2-RBE were described previously (Aslanidi et al., 2009). AAV2 cap of pIR-VP-hr2-RBE was replaced by cap from other serotypes with the VP1 start codon mutated to ACG (Grimm et al., 1998, 2003; Gao et al., 2002, 2003; Mori et al., 2004; Schmidt et al., 2008). AAV2 rep of pIR-rep78-hr2-RBE was replaced by rep of AAV4 or by rep of AAV12 (Grimm et al., 2003; Schmidt et al., 2008). See Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/hum) for details on polymerase chain reaction (PCR) amplification.
Cell culture
HEK 293 and HeLa-derived C12 cells (Clark et al., 1996) were cultivated as adherent monolayers as described (Winter et al., 2012). Sf9 cells and cell lines derived thereof were maintained either as adherent monolayers or in suspension culture at 27°C under constant agitation in serum-free Spodopan medium (Pan-Biotech) supplemented with 200 μg/ml streptomycin, 200 U/ml penicillin, and 250 ng/ml amphotericin B (Invitrogen).
Construction of stable Sf9 cell lines
Adherent Sf9 cells in 6-cm-diameter dishes were transfected with Cellfectin II Reagent (Invitrogen) at a confluency of 70%. A total of 15 μg of pIR-Rep-hr2-RBE and pIR-VP-hr2-RBE as needed for the AAV serotype of choice was transfected at a molar ratio of 1:2.5. For selection and isolation of single-cell clones, transfected cells were re-plated on 6-cm-diameter dishes at 48 hr posttransfection in Spodopan medium with 10% fetal calf serum (FCS) and 25 μg/ml Blasticidin S (Invitrogen) at dilutions from 1:20 to 1:500. After 1 week, the medium was replaced to remove dead cells. Single-cell colonies become visible after 2–3 weeks. Up to 50 cell clones were picked and expanded on cell culture dishes of stepwise increasing diameters. rAAV production efficiency was screened by infection with Bac-rAAV-GFP (multiplicity of infection [MOI]=3). Increasing GFP expression in infected Sf9 cells leads to green coloration of the suspension culture, the extent of which served as rough estimate of rAAV replication efficiency. Genomic rAAV titers (genomic particles [gp]/ml) of the most promising cell clones were determined as outlined below.
Recombinant Bac
Recombinant Bac carrying the rAAV cassette for GFP expression under the control of the chicken β-actin-CMV hybrid (CBA) promoter (Bac-rAAV-GFP) was generated by the MultiBac system (Berger et al., 2004). Recombinant Bac replication was identified by GFP expression combined with detachment of Sf9 cells. Bac stocks were prepared and titers were quantified by plaque assays on monolayer Sf9 cells as described (Kohlbrenner et al., 2005). For short-term storage, stocks were kept at 4°C in the dark. For long-term storage, Bac-infected Sf9 cells were frozen 24 hr postinfection (p.i.), and stored in liquid nitrogen.
rAAV production in 293 cells
HEK 293 cells were seeded at 25–33% confluency. Cells were transfected 24 hr later using the calcium phosphate cotransfection method as described (Winter et al., 2012). AAV vectors were produced using plasmids for AAV rep, cap, Ad5 helper genes, and the rAAV cassette expressing GFP under the control of the CBA promoter (pTR-UF26) by two- or three-plasmid transfection as described (Grimm et al., 1998, 2003; Xiao et al., 1998; Gao et al., 2002; Mori et al., 2004) For the production of rAAV12, vector transfection of four plasmids, pTR-UF26, pAAV12-Rep, pAAV12-Cap, and pHelper, was required (Schmidt et al., 2008). The medium was replaced 12 hr later by a medium with 2% FCS. Cultures were harvested 72 hr after transfection by three freeze–thaw cycles of cell lysis. Crude lysates were digested with 250 U/ml benzonase (Merck) at 37°C for 1 hr to degrade input and unpackaged AAV DNA until centrifugation at 8,000×g for 30 min to pellet cell debris.
rAAV purification
rAAV vectors of the serotypes 1, 2, 3, 4, 5, 6, 7, 8, and rh.10 and 12 were purified from benzonase-treated, cleared freeze–thaw supernatants by one-step AVB sepharose affinity chromatography using 1 ml prepacked HiTrap columns on an ACTA purifier (GE Healthcare) as follows: Freeze–thaw supernatants were diluted 1:1 in 1× phosphate-buffered saline (PBS) supplemented with 1 mM MgCl2 and 2.5 mM KCl (1×PBS-MK) before sample loading on the column at 0.5 ml/min. The column was washed with 20 ml of 1×PBS-MK at a rate of 1 ml/min. AAV vectors were eluted with 0.1 M sodium acetate and 0.5 M NaCl pH 2.5 at a rate of 1 ml/min and neutralized immediately with 1/10 volume of 1 M Tris-HCl pH 10. rAAV9 and rAAV11 vectors were purified by two-step ultracentrifugation through a 20% (w/v) sucrose cushion followed by a sucrose-step gradient [5–40% (w/v)] at 150,000×g for 3 hr at 4°C as described (Mitchell et al., 2009). The AAV band at 25–30% sucrose was removed with a syringe. All rAAV preparations were dialyzed against 1×PBS-MK using Slide-A-Lyzer dialysis cassettes (10,000 MWCO; Thermo Scientific).
Quantification of rAAV vector preparations
Highly purified rAAV vector preparations or benzonase-treated rAAV freeze–thaw supernatants were spiked with 1 μg pBluescript carrier plasmid and digested with Proteinase K for 2 hr at 56°C. DNA was purified by repeated extractions with phenol and chloroform and precipitated with ethanol. Varying dilutions of capsid-released AAV genomes were analyzed by quantitative Light-Cycler PCR, using the Fast Start DNA Master SYBR Green kit (Roche). PCR primers were specific for the bovine growth hormone gene-derived polyA site of the vector backbone (5′-CTAGAGCTCGCTGATCAGCC-3′ and 5′-TGTCTTCCCAATCCTCCCCC-3′). Nonvector AAV rep-cap packaging was detected by primers specific for AAV2 rep and serotype-specific cap (Supplementary Table S1).
Southern blot analysis
Southern blot analysis of vector genomes packaged in AAV capsids was performed with AVB sepharose-purified AAV preparations. Capsid-released AAV genomes were quantified by Light-Cycler PCR, and 1×107 gp were loaded per lane of a 0.8% agarose gel. AAV vector genomes were hybridized for 48 hr with a 0.7 kb GFP-specific probe labeled with biotin-11-dUTP according to the manual of the DecaLabel DNA Labeling Kit (Fermentas), incubated for 20 min with horseradish-peroxidase-conjugated streptavidin (1:10,000), and detected by ECL (Pierce) (Winter et al., 2012).
Protein analysis
Western blot analysis was performed as described (Winter et al., 2012) using 1:10 dilutions of hybridoma supernatants monoclonal antibody (mAb) B1 (anti-VP1–3), mAb A1 (anti-VP1), or mAb 303.9 antibody (anti-Rep) (Wistuba et al., 1995, 1997). For quantitative Western blot analysis, near-infrared dye-labeled antimouse secondary antibodies were used. Signals were quantified with an Odyssey imager (Licor). For silver-staining gels, purified rAAV preparations, corresponding to 1010 gp, were lysed and gel-separated as described (Winter et al., 2012).
Native immuno-dot blot
Dilutions ranging from 1010 gp down to 108 gp of purified rAAVs of serotypes 1–12 were spotted onto nitrocellulose membranes and fixed in a dot-blot apparatus. The membranes were probed with antibodies specific for conformational epitopes on intact AAV particles of particular serotypes as described (Wistuba et al., 1997; Kuck et al., 2007; Sonntag et al., 2010, 2011). Hybridoma supernatants were used at the following dilutions: ADK1a (1:200), A20 (1:10), ADK4 (1:10), ADK5a (1:50), ADK6 (1:10), ADK8 (1:10), and ADK9 (1:10). Polyclonal rabbit antibody VP51 was used at a 1:200 dilution (Kuck et al., 2007; Sonntag et al., 2011). The membranes were incubated with an antimouse IgG horseradish-peroxidase-linked secondary antibody (1:2,500) and visualized by ECL (PromoKine).
Electron microscopy
High-performance liquid chromatography (HPLC)-purified rAAVs (5 μl) of serotype 1, 2, 5, or 8, were loaded onto carbon-coated copper EM grids (Ted Pella Inc.; cat. no. 01754-f ) for 2 min, blotted with filter paper, and negatively stained with 5 μl of 2% uranyl acetate for 20 sec. The grids were air-dried and examined in an FEI-Spirit transmission electron microscope, operating at an accelerating voltage of 120 kV, at a magnification of 90,000.
Evaluation of transduction efficiency using fluorescence-activated cell sorting analysis
HeLa C12 cells were transduced by rAAV-GFP at MOI=1,000 (gp) and superinfected with adenovirus type 2 at MOI=10 (infectious titer). Cells were harvested at 48 hr posttransduction, washed with PBS, and fixed with 4% formaldehyde in PBS. For fluorescence-activated cell sorting (FACS) analysis, 100,000 cells per sample were counted. The proportion of GFP-positive cells was determined according to the manufacturer's protocol (Becton & Dickinson; FACS Calibur). Cells exceeding the cutoff of 30 in the FL1-H channel were counted as positive.
Results
Generation of Sf9 cell lines carrying integrated AAV rep and cap genes
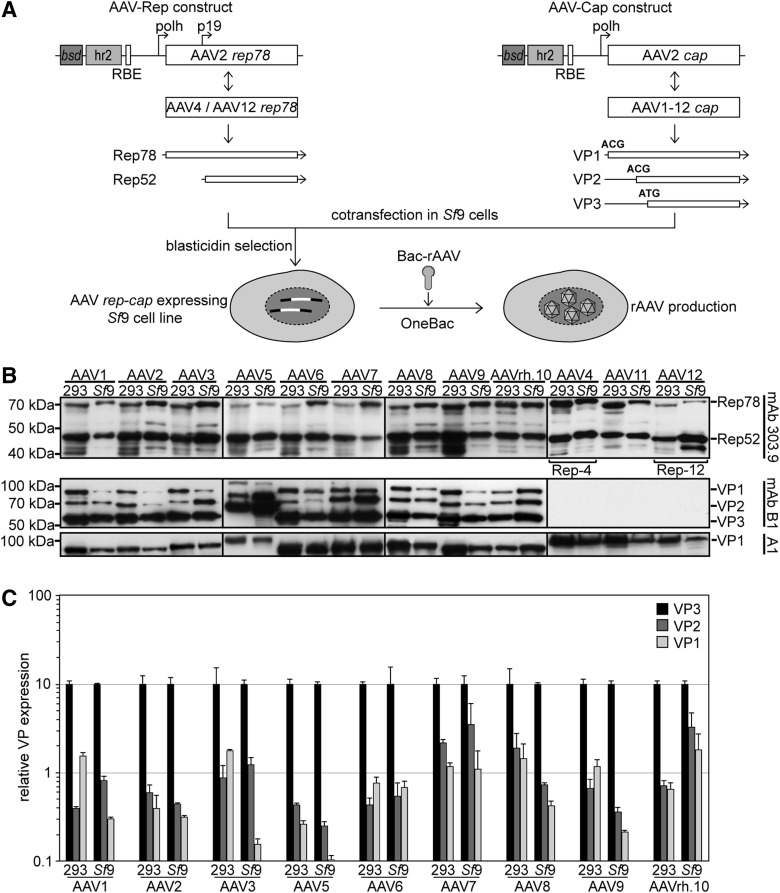
To extend the production range of rAAV serotypes, Sf9 cell lines were generated that carry integrated copies of AAV serotype cap genes combined with either AAV2 rep, or AAV4 rep or -12 rep in the case of rAAV4 or -12 production, as shown before for rAAV production by plasmid transfection in 293 cells (Grimm et al., 2003; Schmidt et al., 2008) (Fig. 1A). The start codon for VP1 was mutated to ACG to allow sufficient expression of VP2 and VP3 from the downstream start codons ACG (VP2) and ATG (VP3), a strategy proven useful for AAV2 and -1 cap expression (Urabe et al., 2002; Aslanidi et al., 2009). For selection of rep/cap-expressing cell clones, up to 50 single-cell clones per AAV serotype were isolated from transfected Sf9 cells grown in blasticidin and screened for Bac-induced rAAV production using Bac-rAAV-GFP. rAAV titers were quantified in cleared supernatants as benzonase-resistant gp per cell. The most efficient producer cell lines for each serotype were expanded for further testing.
FIG. 1.
Generation of OneBac: AAV1–12 rep/cap carrying Sf9 cell lines and their baculovirus-induced expression profiles. (A) Plasmid pIR-rep78-hr2-RBE expressing rep of AAV2 (AAV-Rep construct) and the derivatives for AAV4 or AAV12 rep are depicted with the expressed proteins Rep78 and Rep52. Plasmid pIR-VP-hr2-RBE expressing cap of AAV2 (AAV-Cap construct) and the derived plasmids for cap of AAV serotypes 1–12 are depicted. The expressed proteins VP1 and VP2 start with an ACG codon and VP3 starts from an internal ATG. AcMNPVhr2, Autographa californica multiple nuclear polyhedrosis virus homologous region 2; bsd, blasticidin S deaminase; polh, baculovirus polyhedrin promoter; RBE, Rep-binding element (AAV2). (B) AAV serotype-dependent Rep- and Cap expression was analyzed by Western blot analysis of cell extracts harvested in parallel to rAAV production. Cell extracts from 293 cells were analyzed 72 hr posttransfection of the packaging plasmids. Equivalents of 5×104 293 cells were loaded per well. Cell extracts from rep/cap-expressing Sf9 cells were prepared 72 hr p.i. with the recombinant Bac-rAAV at an MOI=5. For Sf9 cells, an equivalent of 1–2×104 cells were loaded per well. Rep proteins were detected by mAb 303.9, initially generated for AAV2 Rep, but also detecting Rep of AAV4 and AAV12. VP proteins expressed from AAV serotype-specific cap genes were reacted with mAb B1, which detects all three capsid proteins of most AAV serotypes with the exception of AAV4, -11, and -12. mAb A1 binds to VP1 of all serotypes. (C) Cell extracts from three independent rAAV preparations for each serotype were analyzed by quantitative Western blot analysis to compare the ratios of VP1, VP2, and VP3 expressed in 293 or Sf9 cells, respectively. VP proteins expressed in either cell type were analyzed by mAb B1 and a secondary antibody emitting near-infrared fluorescence as quantified by an Odyssey imager (Licor). Expression levels of VP3 were arbitrarily set to 10, and expression levels of VP2 and VP1 were calculated as relative values thereof. The results of three experiments are given as mean±standard deviation. Please note the logarithmic scale of the y-axis. AAV, adeno-associated virus; mAb, monoclonal antibody; MOI, multiplicity of infection; p.i., postinfection; rAAV, recombinant AAV.
AAV rep and cap expression in Sf9 cell lines compared with plasmid-transfected 293 cells
To compare Bac-induced rAAV rep and cap expression patterns in Sf9 cell clones with those of plasmid-transfected 293 cells, the most widely used method for laboratory-scale rAAV production, cell extracts were analyzed side by side on Western blots (Fig. 1B). Rep showed comparable expression of Rep78 and Rep52 in Bac-infected Sf9 cell lines and plasmid-transfected 293 cells as detected by mAb 303.9 (Fig. 1B, upper panel). Sf9 cells were designed to express Rep78 and Rep52, but most of them appeared to also express small amounts of Rep68 and/or Rep40 similar to 293 cells. Expression patterns of cap analyzed by mAb B1 (VP1–3) (Wistuba et al., 1995) and mAb A1 (VP1) (Wistuba et al., 1997) were comparable among different serotypes and also between insect cells and 293 cells. This includes previously described variations in mobility, as in the case of AAV5 (Fig. 1B, mAb B1; A1) (Kohlbrenner et al., 2005). For wild-type AAV2, the ratio of VP1:VP2:VP3 was originally described as 1:1:10 (Becerra et al., 1988). VP1 is critical, since it comprises a phospholipase domain, which is required for AAV infectivity (Sonntag et al., 2006). Fluorescence intensity of the respective bands was quantified with near-infrared spectrometry. As depicted in Fig. 1C, most AAV serotypes expressed VP1:VP2:VP3 close to the prototypic 1:1:10 ratio.
AAV vector yield in Sf9 cells compared with 293 cells
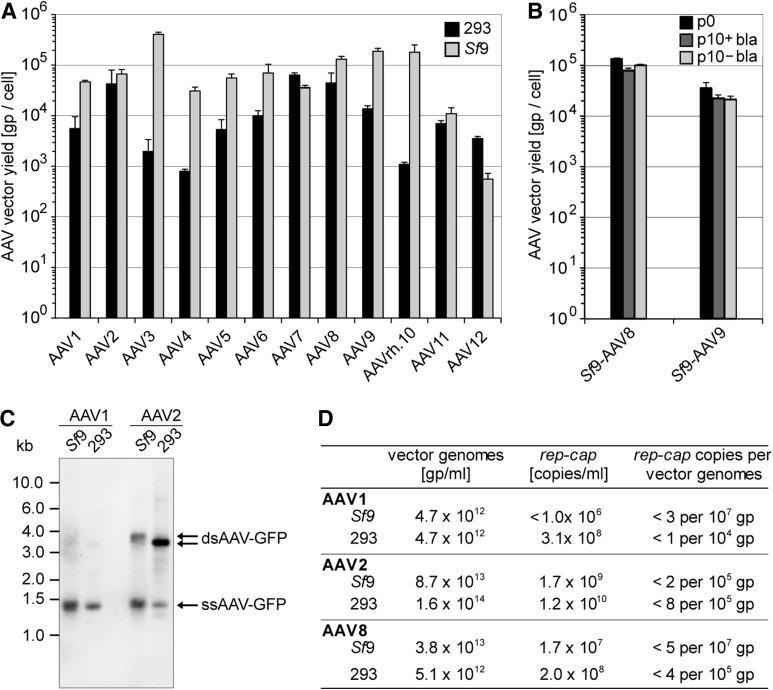
The major advantage of rAAV production in Sf9 cells is that the cells can grow in suspension culture to high densities and thus allow scale-up for rAAV production in bioreactors as required for clinical-scale production. Without extensive efforts to optimize growth conditions, rAAV preparations could be scaled up to 1-liter suspensions with rAAV yields of up to 2×1014 gp/liter. Using small-scale culture formats, the burst sizes (rAAV yields per cell) were compared in triplicate 30 ml cultures of Sf9 cells and plasmid-transfected 293 cells (6 cm dishes). rAAV titers were quantified as gp per cell in benzonase-treated, cleared freeze–thaw supernatants of cells and medium (Fig. 2A). At this point, neither Sf9- nor 293 cell-based AAV production protocols were optimized as commonly done for large-scale, clinical-grade production (Lock et al., 2010). For the majority of AAV serotypes, vector yields per cell in Sf9 cells exceeded those of 293 cells, ranging from almost similar yields in case of AAV2 and AAV11 to up to 100-fold higher yields in the case of AAV3 and AAVrh.10. Only Sf9 cells producing AAV12 yielded lower titers compared with those in 293 cells. The previously described Sf9 cell lines for AAV2 and AAV1 reproduced the reported high rAAV yields after numerous cell passages over several years without selection (Aslanidi et al., 2009). rAAV production stability of the newly generated Sf9-AAV8 and Sf9-AAV9 cell lines was therefore also tested after serial passages at high dilutions of 1:10 for 10 successive passages, with or without blasticidin selection. After passage 10, rAAV yields (gp) per cell were determined and compared with the starting rAAV yields (passage 0). As can be seen in Fig. 2B, rAAV yields remained stable, even in the absence of selection. This further underscores the stability of the Sf9 producer cells, a prerequisite for successive scale-up. Comparative analysis of the integrity of packaged vector genomes from AVB sepharose-purified vector preparations by Southern blot hybridization showed intact AAV ssDNA and reassociated dsDNA genomes for Sf9- and 293-cell derived AAV vectors (Fig. 2C). To evaluate whether potentially replication-competent AAVwt was generated during the production process, quantitative PCR of packaged genomes was performed with primers spanning the rep/cap gene region (Supplementary Table S1). As displayed in Fig. 2D, purified rAAV1, -2, or -8 prepared from Sf9 cells displayed significantly reduced levels of packaged rep/cap sequences compared with vectors prepared in 293 cells. Comparable or higher percentages of packaged AAV2 rep or cap genes had been reported before (Nony et al., 2003; Martin et al., 2013). It is well established that AAV preparations also contain variable, low proportions of host cell or plasmid-derived DNA sequences (Hüser et al., 2003; Chadeuf et al., 2005). In summary, most Sf9 cell-derived AAVs show higher burst sizes and improved purity of packaged rAAV genomes, when compared with AAVs from 293 cells.
FIG. 2.
Burst sizes and integrity of AAV1–12 vectors produced in 293 cells compared with Sf9 cells. (A) The yields of rAAV1–12 preparations were quantified as benzonase-resistant genomic particles (gp) per cell by LightCycler PCR using rAAV genome-specific primers. Benzonase-treated freeze–thaw supernatants were prepared 72 hr posttransfection of 293 cells or infection of Sf9 cells, respectively. rAAV burst sizes of three experiments are displayed as mean±standard deviation. Please note the logarithmic scale of the y-axis. (B) Analysis performed similar to (A). The burst sizes of rAAVs from AAV8 and -9 Sf9 producer cell lines after serial passages with or without blasticidin selection were quantified at passage 0 (p0) and passage 10 (p10±basticidin). (C) Southern blot analysis of vector genomes from AVB sepharose-purified AAV1 and -2 vector preparations. Vector genomes are detected by a biotin-labeled probe specific for GFP. The size of the packaged rAAV genomes varies between Sf9- (pTR-UF26: 3,794 bp) and 293-cell-derived (pTR-UF5: 3,388 bp) vectors. The arrows point to ssDNA and dsDNA AAV genomes that are detected because of partial reassociation of packaged (+) and (−) strand genomes. (D) Quantitative analysis of the AAV vector genomes (vg) and copackaged wild-type rep/cap sequences of AAV1, -2, and -8.
Integrity of AAV capsids produced in Sf9 cells
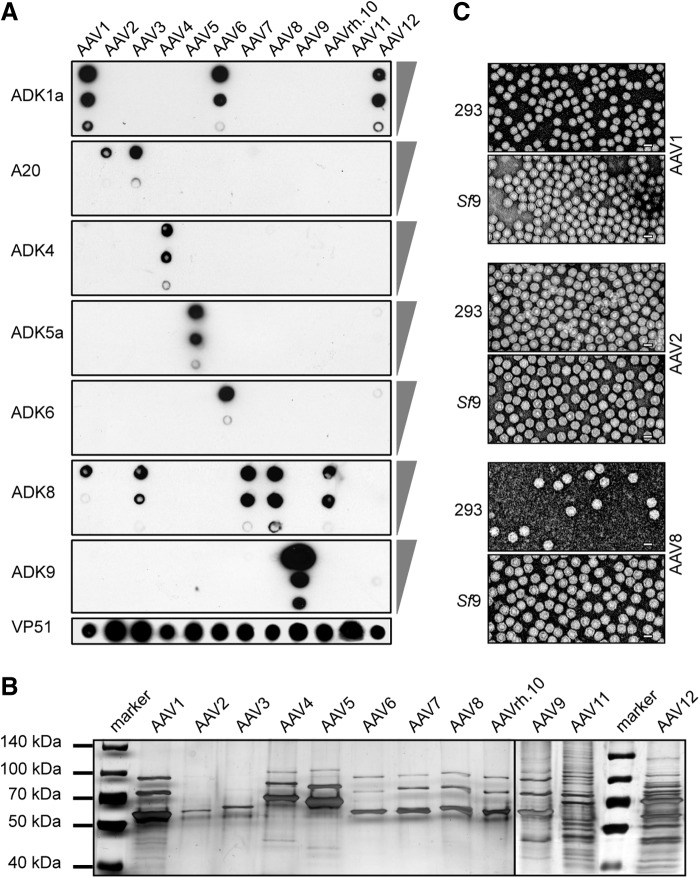
To analyze the capsid composition and integrity of AAV vectors prepared from Sf9 cells, large-scale rAAV preparations from 1 liter of Sf9 cell suspensions were highly purified by HPLC using AVB sepharose chromatography for all serotypes except for AAV9 and AAV11. These were refractory to HPLC purification and were therefore purified by sucrose gradient centrifugation (Mitchell et al., 2009). rAAV titers of the peak fractions exhibited genomic titers between 4×1012 and 7×1013 gp/ml. The resulting rAAVs of serotypes 1–12 were analyzed with a series of antibodies specific for conformational epitopes present on the surfaces of intact AAV capsids as described for rAAVs purified from transfected 293 cells (Wistuba et al., 1997; Kuck et al., 2007; Sonntag et al., 2010) (Fig. 3A). Since the proportions of genome-containing (gp) to physical (empty) particles are expected to vary among serotypes, equal amounts of gp of the purified rAAV preparations were also separated on a polyacrylamide gel, and the VP1–3 bands visualized by silver staining (Fig. 3B). Intact, background-free VP1–3 bands were detected for all HPLC-purified AAVs. Sucrose-banded AAV-9 and AAV11 displayed more background, as expected. Furthermore, the morphologies of rAAVs prepared in Bac-infected Sf9 and plasmid-transfected 293 cells were compared by transmission electron microscopy of highly purified AAV vector preparations of serotypes 1, 2, and 8. Clusters of uniform, icosahedral capsids with a diameter of ∼25 nm characteristic of AAV were observed (Fig. 3C). There were no discernible differences between the capsids produced in Sf9 and 293 cells, respectively.
FIG. 3.
Comparative capsid analysis of AAV serotypes 1–12 generated in 293 or Sf9 cells. (A) Immuno-dot blot analysis of native rAAV capsids of the panel of AAV serotypes 1–12. All rAAV subtypes were purified by AVB chromatography except for rAAV9 and rAAV11, which were purified by sucrose gradient centrifugation. About 1010, 109, and 108 gp of serotype-specific rAAVs were spotted on a nitrocellulose membrane as indicated by the triangles to the right of the panels. The membranes were incubated with a panel of mAbs as depicted to the left of the membrane. Each mAb detects specific conformational epitopes on the surface of defined AAV capsids. Polyclonal antibody VP51 detects all AAV serotypes and served as an internal loading control for 109 gp of each rAAV serotype. (B) Silver-staining gel analysis of the rAAV preparations used in (A) separated on 9% sodium dodecyl sulfate polyacrylamide gel electrophoresis with 1010 gp loaded per lane. (C) Electron microscopic analysis of negative-stained rAAV-GFP vectors of AAV serotypes 1, 2, and 8 generated from either transfected 293 cells or baculovirus-infected Sf9 cells. AAV vectors were highly purified by AVB chromatography. Titers ranged between 3×1012 and 4×1013 gp/ml. Scale bar=25 nm.
Transduction efficiencies of Sf9- and 293-derived AAV vectors
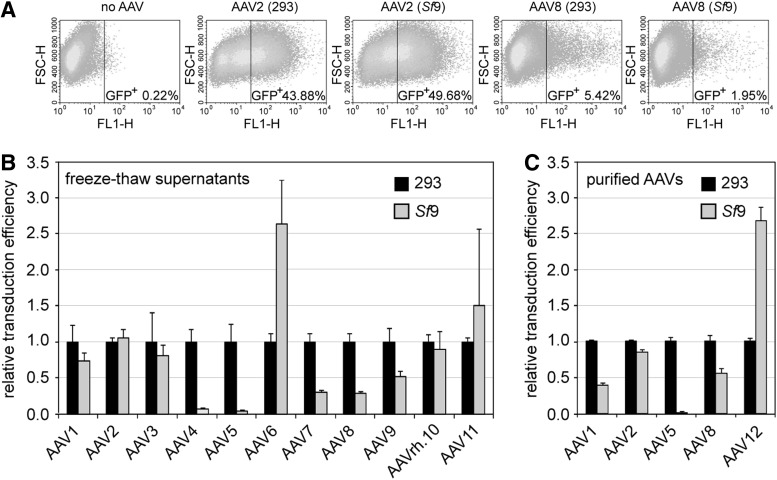
Functional validation of gene therapy vectors commonly scores the efficiency of gene transfer through analysis of successful expression of the gene of interest. Since capsids of variant AAV serotypes target specific cell types, the absolute transduction efficiencies vary markedly depending on the target cell line. We therefore decided to compare only cell transduction of identical amounts of gp from the same AAV serotype produced in 293 and in Sf9 cells. As target cells, we chose C12 cells (Clark et al., 1996), which express integrated AAV2rep/cap and replicate transduced rAAVs after Ad2 infection, thereby boosting expression of GFP levels even in the case of low transduction efficiency. Using FACS analysis as displayed in Fig. 4A, for each serotype, the percentage of GFP-positive cells after transduction of Sf9-derived rAAV-GFP was compared with that of 293 cell-derived rAAV-GFP. The transduction efficiencies of the latter were arbitrarily set to 1.0. The analysis was performed for rAAV preparations from cleared freeze–thaw supernatants (Fig. 4B), and also for a selection of AAV serotypes purified by HPLC-AVB column chromatography (Fig. 4C). Irrespective of the degree of purification, AAV transduction efficiencies of Sf9-derived vectors were mostly comparable with those of 293 cell-derived vectors, with minor 2–3-fold variance (Fig. 4B and C). Sf9-derived rAAV4 and rAAV5 transduced at a 10-fold reduced rate compared with vectors of the same serotype produced in 293 cells (Fig. 4B and C). This effect is compensated by more than 10-fold higher burst sizes in Sf9 compared with 293 cells (Fig. 2A). In all cases where the analysis of vectors from cleared extracts was repeated with HPLC-purified preparations, that is, AAV1, -2, -5, and -8, the observed differences in transduction remained similar. Transduction efficiencies of rAAV1, -2, -3, -6, -8, -9, and -rh.10 were also analyzed in HEK 293 cells, confirming the above results (data not shown). In summary, AAV vectors prepared and purified from the majority of newly generated Sf9 cell lines transduce comparably well as vectors produced in plasmid-transfected 293 cells.
FIG. 4.
Cell transduction efficiencies of rAAV serotype 1–12 vectors produced in 293 cells compared with Sf9 cells. The transduction efficiencies of rAAV-GFP vectors of various serotypes derived from either 293 or Sf9 cells, respectively, were compared. AAV2 rep/cap-expressing HeLa C12 cells were transduced with 1,000 gp of rAAV-GFP vector of the chosen serotype. Cells were coinfected with adenovirus type 2 at an MOI of 10 (infectious titer). At 48 hr p.i., cells were harvested and the percentage of GFP-positive cells was determined by FACS analysis (GFP+: FL1-H >30). (A) FACS read-outs displaying the transduction efficiencies for rAAV vectors of serotype 2 and produced either in 293 cells or in Sf9 cells. Control cells (designated “no AAV”) were infected with adenovirus only. (B) Comparative analysis of the transduction efficiencies of rAAV vectors with serotype 1–11 capsids derived from freeze–thaw supernatants. The various AAV serotypes display variable absolute transduction efficiencies in C12 cells. To compare transduction efficiencies for each serotype, transduction of rAAVs prepared in 293 cells was set to 1.0. The transduction efficiency of rAAV vectors derived from Sf9 cells from the same serotype is displayed as percentage thereof. The experiments were performed in triplicates and are displayed as mean±standard deviation (n=3). (C) Analysis of rAAV-1, -2, -5, -8, and -12 transduction as described in (B), except that highly purified (AVB-column) rAAV vectors were used. FACS, fluorescence-activated cell sorting.
Discussion
For bioreactor-size rAAV production, high-titer, scalable, and genetically stable systems are required to keep pace with the rapid increase of rAAV application in the clinic. The extended range of AAV serotypes currently being introduced for transduction of particular cell types or tissues requires the uncomplicated transfer of AAV production systems to novel AAV serotypes. Here we show the development of OneBac, a platform comprising a series of genetically stable insect Sf9 cell lines that allow single Bac-induced, scalable production of the full range of AAV serotype 1–12 vectors. The described methods for cell line generation can be transferred to novel AAV serotypes to come. Moreover, existing Bac-rAAV recombinants developed for the three- or two-Bac infection system (Urabe et al., 2002; Smith et al., 2009) can be directly employed without modifications. The key features of OneBac are (i) high-titer, scalable production of an extended range of AAV serotypes; (ii) high infectivity-to-particle ratios of rAAVs produced in Bac-infected Sf9 cell lines; (iii) high stability of rep/cap-carrying Sf9 cell lines with undiminished Bac-induced rAAV burst sizes upon serial cell passage.
Composition and VP ratios of AAV capsids
Ever since rAAV production systems have been designed, obtaining the correct stoichiometry of VP1:VP2:VP3 has been a challenge. The strategy of wild-type AAV2 relies on the use of overlapping and partially spliced ORFs combined with the use of ACG as noncanonical start codon for VP2 translation. This is maintained in the pDG/pDP series (Grimm et al., 2003) of plasmids for AAV production in transfected 293 cells, but is difficult to mimic in insect cells. Because of the limited functionality of mammalian introns, the unmodified cap ORF would predominantly lead to VP1 expression. In wild-type AAV2, the ratio of VP1–3 proteins was originally determined as 1:1:10 (Becerra et al., 1988), which approximates to 3–6 VP1 molecules per capsid (Excoffon et al., 2009). To maintain the wild-type capsid composition, reduced but sufficient VP1 expression is critical, which was achieved by mutating the VP1 ATG start codon to ACG (Aslanidi et al., 2009). The resulting capsid protein ratios are close to those seen for prototype AAV2. Burst sizes in Sf9 cells are mostly higher than in 293 cells and correlate with infectivity. AAV5 produced in Sf9 cells is an exception with calculated mean VP1 levels of one per capsid, which is obviously not sufficient for full infectivity. Insufficient VP1 levels resulting in reduced rAAV transduction rates have previously been described for AAV5 and AAV8 capsids generated from Bac-cap constructs that also used an ACG start codon for VP1 expression (Kohlbrenner et al., 2005; Urabe et al., 2006). Reduced AAV5 infectivity is compensated by increased burst sizes, making the Sf9 cell line proficient for scale-up production. Alternative expression strategies for AAV5 cap may be considered to further improve Sf9 AAV5 producer cells to decrease the ratio of noninfectious AAV5 vectors.
rAAV burst sizes and particle to infectivity ratios in Sf9 produced rAAV 1–12
The burst sizes for the majority of rAAV serotypes in Sf9 cells are at least one log higher than those of 293 cells except for recently discovered AAV12 (Schmidt et al., 2008). In the case of AAV12, the increased infectivity almost compensates the reduced burst size. To generate the respective Sf9 cells, the AAV12 rep gene instead of that of AAV2 was expressed. In the absence of data on AAV12-derived Rep interaction with the AAV2 ITR, the reduced replication efficiency is not that surprising. Exchange of either rep or the RBE may improve burst sizes. All cell lines produced reproducible burst sizes upon successive cell passages, and AAV particle-to-infectivity ratios remained similarly high as those described for 293 cells.
Comparison of scalable rAAV production systems
Production of AAV vectors has long been a challenge. Whereas AAV's high physicochemical stability has allowed the development of sophisticated downstream purification schemes leading to near-homogeneous, stable AAV vector preparations, the upstream scale-up is more demanding. The ideal rAAV production system leads to high vector burst sizes per cell of highly infectious virus stocks. To render a system proficient for production in bioreactors, additional aspects have to be considered: (i) simplicity of scale-up, (ii) modest requirements for cell growth, (iii) efficacy of gene transfer combined with (iv) genetic stability of the components, and (v) flexibility to adapt to an increasing range of AAV serotypes and variants thereof.
The first AAV production systems to be developed consisted of AAV2 producer cell lines of various formats harboring the required components for AAV production to be induced upon adenovirus infection as reviewed by Zolotukhin (2005). Since Rep78 is cell toxic, strict silencing of rep is required in the noninduced state. The best-studied producer cell format was generated by extensive multiple-round selection of cell lines with integrated high copy numbers of rAAV and p5-promoter-driven rep/cap. Rep silencing in the absence of adenovirus infection appears to be mediated by Rep-mediated negative feedback, as is the case in AAV latency. The AAV2 producer cell system has recently been further optimized to yield 5×104 to 2×105 DNAse-resistant particles per cell (Martin et al., 2013). However, new cell lines have to be constructed for every new vector to be produced. It will be seen how readily the system is adaptable to alternative serotypes or novel vectors.
Production of rAAV by plasmid cotransfection of adherent 293 cells (Grimm et al., 1998, 2003; Xiao et al., 1998) leads to high burst sizes and infectivity rates for virtually all AAV serotypes tested so far. Mammalian 293 cells are robust, and plasmid DNA transfection is a standard technique in research laboratories. The genetic stability of the plasmids and the ease of swapping AAV rep and cap genes or domains thereof by standard cloning technology have made this system a laboratory standard for rAAV research. Its major limitations are scalability and effectiveness of gene transfer. HEK-293 cells do not grow well in suspension, and plasmid cotransfection is troublesome in suspension cells.
The need for plasmid transfection was overcome when HSV infection-based rAAV production systems were developed. There, AAV's requirement for helper HSV functions is exploited by construction of rHSV strains, one harboring the AAV rep/cap cassette, and another harboring the rAAV vector genome (Conway et al., 1999; Kang et al., 2008). Scalability was achieved by adapting rAAV production to HSV infection of BHK cells grown in suspension. The range of rHSV/AAV recombinants comprises rAAV serotypes 1, 2, 5, 8, and 9 (Clement et al., 2009; Thomas et al., 2009). Although expression from the integrated rep gene modestly inhibits HSV replication (Heilbronn et al., 2003), rHSVs are genetically stable and lead to rAAV burst sizes of around 8×104 DNAse-resistant particles per cell (Thomas et al., 2009).
The insect Sf9 cell-based rAAV production system, first developed by the Kotin group (Urabe et al., 2002; Virag et al., 2009), exploits the ease of growth and scalability of Sf9 cells. Sf9 cells grow in suspension not only to high densities but also at modest growth requirements. Compared with mammalian cells, simple culture media without FCS and growth at 27°C without need for CO2 are sufficient. Furthermore, only insect cells offer the possibility to grow rAAVs expressing proteins toxic to mammalian cells (Chen, 2012). Gene transfer by Bac infection per se is efficient. However, separate propagation of three Bac strains, Bac-rep, Bac-cap, and Bac-rAAV, before coinfection at individually optimized MOI ratios reduces effectiveness. Most importantly, Bac-rep and Bac-cap helpers are genetically unstable. As a consequence, Rep/Cap expression drops upon more than six successive Bac passages (Kohlbrenner et al., 2005; Smith et al., 2009; Virag et al., 2009). The introduction of artificial introns for collinear Rep and Cap expression appears to improve stability, but the necessity for coinfection with three separate Bac strains remains (Chen, 2008). An additional improvement was the combination of rep and cap into one Bac, thus reducing the number Bac strains to two and extending the number of stable virus passages to seven (Smith et al., 2009). The initial three-Bac coinfection system was developed for AAV2, leading to burst sizes of 5×104 gp per cell (Urabe et al., 2002). AAV serotypes 1, 6, 8, and 9 followed with burst sizes reported between 7×103 and 2×104 gp per cell (Cecchini et al., 2011; Galibert and Merten, 2011).
The here-described most recent development of Bac-based rAAV production systems reduces the number of Bac strains to the one that carries the rAAV backbone. AAV rep and cap are stably integrated in the genome of the host Sf9 insect cell but transcriptionally silent. As described before, infection with a single Bac leads to amplification and expression of rep and cap to levels sufficient for effective rAAV replication and packaging (Aslanidi et al., 2009). The resulting burst sizes of up to 5×105 benzonase-resistant rAAV genomes per cell exceed those of competitive production systems. The Sf9-rep/cap producer cells for AAV1–12 are genetically stable even in the absence of blasticidin, used initially for cell selection. Sf9-rep/cap producer cells can be grown and passaged similar to unmodified Sf9 cells. The OneBac system requires only a single Bac, Bac-rAAV, which can infect over 95% of Sf9 cells at a typical MOI of 3. Bac-rAAVs constructed for the two- or three-Bac Sf9 production system can be immediately transferred to the OneBac system, to produce rAAVs with the panel of AAV1–12 rep/cap packaging cells. These share the convenience of cell growth and upscaling capabilities of unmodified Sf9 cells. The major advance of OneBac for rAAV production lies in the combination of the high genetic stability of Sf9 rep/cap producer cells, and the simplicity of infection with a single virus. These features, combined with the here-described versatility in the construction of novel AAV serotype-specific Sf9 cell lines, make the new OneBac system readily applicable for bioreactor-scale rAAV production.
Supplementary Material
Acknowledgments
The authors thank Jürgen Kleinschmidt and Martin Müller (German Cancer Research Center, Heidelberg, Germany) for the series of mAbs directed against AAV capsids and for pDG/pDP-plasmids, and John Chiorini (National Institutes of Health, Bethesda, MD) and Tadahito Kanda (National Institute of Infectious Diseases, Tokyo, Japan) for plasmids for AAV10, -11, and -12. The authors also thank Kerstin Winter for advice with HPLC-based AAV purification, Kristina von Kietzell for help with FACS, and Catrin Stutika for critical reading of the manuscript (Charité, Berlin, Germany). This project was partially funded by a grant from the German Academic Exchange Service, DAAD (Promos program), and by NIH R01 AI1081961.
Author Disclosure Statement
M.A.-M., N.M., S.Z., and R.H. are inventors of patents related to rAAV technology; S.Z. is inventor in a pending patent on the inducible insect cell-based system for highly efficient production of rAAV vectors (US 20120100606); N.M. and R.H. own equity in a company that is commercializing AAV for gene therapy.
References
- Aslanidi G., Lamb K., and Zolotukhin S. (2009). An inducible system for highly efficient production of recombinant adeno-associated virus (rAAV) vectors in insect Sf9 cells. Proc. Natl. Acad. Sci. USA 106, 5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra S.P., Koczot F., Fabisch P., et al. (1988). Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J. Virol. 62, 2745–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger I., Fitzgerald D.J., and Richmond T.J. (2004). Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 22, 1583–1587 [DOI] [PubMed] [Google Scholar]
- Cecchini S., Virag T., and Kotin R.M. (2011). Reproducible high yields of recombinant adeno-associated virus produced using invertebrate cells in 0.02- to 200-liter cultures. Hum. Gene Ther. 22, 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadeuf G., Ciron C., Moullier P., et al. (2005). Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol. Ther. 12, 744–753 [DOI] [PubMed] [Google Scholar]
- Chen H. (2008). Intron splicing-mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Mol. Ther. 16, 924–930 [DOI] [PubMed] [Google Scholar]
- Chen H. (2012). Exploiting the intron-splicing mechanism of insect cells to produce viral vectors harboring toxic genes for suicide gene therapy. Mol. Ther. Nucleic Acids 1, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.R., Voulgaropoulou F., and Johnson P.R. (1996). A stable cell line carrying adenovirus-inducible rep and cap genes allows for infectivity titration of adeno-associated virus vectors. Gene Ther. 3, 1124–1132 [PubMed] [Google Scholar]
- Clement N., Knop D.R., and Byrne B.J. (2009). Large-scale adeno-associated viral vector production using a herpesvirus-based system enables manufacturing for clinical studies. Hum. Gene Ther. 20, 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J., Rhys C., Zolotukhin I., et al. (1999). High-titer recombinant adeno-associated virus production utilizing a recombinant herpes simplex virus type I vector expressing AAV-2 rep and cap. Gene Ther. 6, 986–993 [DOI] [PubMed] [Google Scholar]
- Excoffon K.J., Koerber J.T., Dickey D.D., et al. (2009). Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc. Natl. Acad. Sci. USA 106, 3865–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert L., and Merten O.W. (2011). Latest developments in the large-scale production of adeno-associated virus vectors in insect cells toward the treatment of neuromuscular diseases. J. Invertebr. Pathol. 107Suppl, S80–S93 [DOI] [PubMed] [Google Scholar]
- Gao G.P., Alvira M.R., Wang L., et al. (2002). Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99, 11854–11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Alvira M.R., Somanathan S., et al. (2003). Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. USA 100, 6081–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Kern A., Rittner K., et al. (1998). Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 9, 2745–2760 [DOI] [PubMed] [Google Scholar]
- Grimm D., Kay M.A., and Kleinschmidt J.A. (2003). Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther. 7, 839–850 [DOI] [PubMed] [Google Scholar]
- Heilbronn R., Engstler M., Weger S., et al. (2003). ssDNA-dependent colocalization of adeno-associated virus Rep and herpes simplex virus ICP8 in nuclear replication domains. Nucleic Acids Res. 31, 6206–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüser D., Weger S., and Heilbronn R. (2003). Packaging of human chromosome 19-specific adeno-associated virus (AAV) integration sites in AAV virions during AAV wild-type and recombinant AAV vector production. J. Virol. 77, 4881–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W., Wang L., Harrell H., et al. (2008). An efficient rHSV-based complementation system for the production of multiple rAAV vector serotypes. Gene Ther. 16, 229–239 [DOI] [PubMed] [Google Scholar]
- Kastelein J.J., Ross C.J., and Hayden M.R. (2013). From mutation identification to therapy: discovery and origins of the first approved gene therapy in the Western world. Hum. Gene Ther. 24, 472–478 [DOI] [PubMed] [Google Scholar]
- Kohlbrenner E., Aslanidi G., Nash K., et al. (2005). Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol. Ther. 12, 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck D., Kern A., and Kleinschmidt J.A. (2007). Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J. Virol. Methods 140, 17–24 [DOI] [PubMed] [Google Scholar]
- Lock M., Alvira M., Vandenberghe L.H., et al. (2010). Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum. Gene Ther. 21, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Frederick A., Luo Y., et al. (2013). Generation and characterization of adeno-associated virus producer cell lines for research and preclinical vector production. Hum. Gene Ther. Methods 24, 253–269 [DOI] [PubMed] [Google Scholar]
- Mingozzi F., and High K.A. (2011). Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 12, 341–355 [DOI] [PubMed] [Google Scholar]
- Mitchell M., Nam H.J., Carter A., et al. (2009). Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 9. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 715–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Wang L., Takeuchi T., et al. (2004). Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology 330, 375–383 [DOI] [PubMed] [Google Scholar]
- Nathwani A.C., Tuddenham E.G., Rangarajan S., et al. (2011). Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 365, 2357–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nony P., Chadeuf G., Tessier J., et al. (2003). Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: a model for generation of rep-positive AAV particles. J. Virol. 77, 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Voutetakis A., Afione S., et al. (2008). Adeno-associated virus type 12 (AAV12): a novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J. Virol. 82, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.H., Levy J.R., and Kotin R.M. (2009). A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol. Ther. 17, 1888–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F., Bleker S., Leuchs B., et al. (2006). Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 80, 11040–11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F., Schmidt K., and Kleinschmidt J.A. (2010). A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. USA 107, 10220–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F., Kother K., Schmidt K., et al. (2011). The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 85, 12686–12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.L., Wang L., Niamke J., et al. (2009). Scalable recombinant adeno-associated virus production using recombinant herpes simplex virus type 1 coinfection of suspension-adapted mammalian cells. Hum. Gene Ther. 20, 861–870 [DOI] [PubMed] [Google Scholar]
- Urabe M., Ding C., and Kotin R.M. (2002). Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 13, 1935–1943 [DOI] [PubMed] [Google Scholar]
- Urabe M., Nakakura T., Xin K.Q., et al. (2006). Scalable generation of high-titer recombinant adeno-associated virus type 5 in insect cells. J. Virol. 80, 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag T., Cecchini S., and Kotin R.M. (2009). Producing recombinant adeno-associated virus in foster cells: overcoming production limitations using a baculovirus-insect cell expression strategy. Hum. Gene Ther. 20, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K., von Kietzell K., Heilbronn R., et al. (2012). Roles of E4orf6 and VA I RNA in adenovirus-mediated stimulation of human parvovirus B19 DNA replication and structural gene expression. J. Virol. 86, 5099–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba A., Weger S., Kern A., et al. (1995). Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69, 5311–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba A., Kern A., Weger S., et al. (1997). Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71, 1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Li J., and Samulski R.J. (1998). Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72, 2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin S. (2005). Production of recombinant adeno-associated virus vectors. Hum. Gene Ther. 16, 551–557 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.