Abstract
Purpose
In planning for a potentially curative resection of the epileptogenic zone in patients with pediatric epilepsy, invasive monitoring with intracranial EEG is often used to localize the seizure onset zone and eloquent cortex. A precise understanding of the location of subdural strip and grid electrodes on the brain surface, and of depth electrodes in the brain in relationship to eloquent areas is expected to facilitate pre-surgical planning.
Methods
We developed a novel algorithm for the alignment of intracranial electrodes, extracted from post-operative CT, with pre-operative MRI. Our goal was to develop a method of achieving highly accurate localization of subdural and depth electrodes, in order to facilitate surgical planning. Specifically, we created a patient-specific 3D geometric model of the cortical surface from automatic segmentation of a pre-operative MRI, automatically segmented electrodes from post-operative CT, and projected each set of electrodes onto the brain surface after alignment of the CT to the MRI. Also, we produced critical visualization of anatomical landmarks, e.g. vasculature, gyri, sulci, lesions or eloquent cortical areas, which enables the epilepsy surgery team to accurately estimate the distance between the electrodes and the anatomical landmarks, which might help for better assessment of risks and benefits of surgical resection.
Results
Electrode localization accuracy was measured using knowledge of the position of placement from 2D intra-operative photographs in ten consecutive subjects who underwent intracranial EEG for pediatric epilepsy. Average spatial accuracy of localization was 1.31±0.69mm for all 385 visible electrodes in the photos.
Conclusions
In comparison to previously reported approaches, our algorithm is able to achieve more accurate alignment of strip and grid electrodes with minimal user input. Unlike manual alignment procedures, our algorithm achieves excellent alignment without time consuming and difficult judgements from an operator.
Keywords: Pediatric Epilepsy, Intracranial EEG, Electrode Localization, Epilepsy Surgery Planning
1. Introduction
The annual prevalence of epilepsy in the United States is approximately 7.1/1000 people [1], and according to Rochester Epidemiology Project, it has increased from 2.7/1,000 in 1940 to 6.8/1,000 in 1980 [2]. About 30 to 40% of children with epilepsy do not respond to the anti-epileptic drugs (AEDs) currently available [3,4]. Pediatric epilepsy is especially challenging because of the heterogeneity of focal seizure onset locations and the wide range of regions that may present as ictal onset zones. When a child’s seizure disorder is medically refractory, despite adequate trials of AEDs, other treatment options including epilepsy surgery must be explored. Early surgical resection is a promising intervention for pediatric epilepsy [5]. Surgical resection of the ictal onset zone and disruption of the epileptogenic network of the brain is designed to prevent or minimize the recurrence of seizures after surgery. Patients with a focal seizure onset zone and pharmacologically intractable seizures are frequently excellent candidates for surgery, but it is often challenging to identify the extent of the epileptogenic zone, and the location and extent of eloquent cortical areas.
The International League Against Epilepsy (ILAE) developed standards for epilepsy surgery and established criteria for the evaluation of children for epilepsy surgery [6–8]. Our institution follows a clinical workflow for patients with epilepsy, which is compatible with the ILAE established standards. In our institution, pediatric patients with focal epilepsy usually undergo an initial assessment with scalp EEG followed by additional assessment of seizure localization, including neuroimaging and nuclear medicine studies (“Phase I”). This evaluation aims to localize the region of the brain from which seizures arise. To this end, EEG electrodes are placed on the scalp in order to capture seizures and interictal EEG findings. EEG recordings are then discussed in an epilepsy surgery conference (ESC) to lateralize and localize the ictal onset areas and discuss the possible functional risks of a surgical resection. Epileptic foci are frequently difficult to localize during scalp EEG, and in cases of potential overlap with eloquent areas patients frequently undergo intracranial EEG (iEEG), referred to as a “Phase II” evaluation, in which invasive EEG electrodes are placed in or over the brain to pinpoint seizure onset region [9,10] and to determine regions of functional importance during cortical stimulation. Several types of electrodes are implanted inside the cranium, including subdural strip/grid electrodes consisting of electrodes mounted on a thin sheet of plastic and placed on the cortical surface, and depth electrodes consisting of electrodes attached to flexible wires and then inserted in the vicinity of an expected seizure onset zone, such as in a focal cortical dysplasia or gray matter heterotopia. After electrode placement, the ictal and interictal electrical activity is monitored for typically 5 to 7 days. The iEEG often demonstrates, in many instances, the irritative and ictal onset zones well [11,12].
In our routine clinical practice, an epilepsy surgery conference (ESC) is convened to discuss potential surgical candidates, to evaluate the full spectrum of data available for each patient, and to arrive at a consensus plan for optimal treatment based on the expertise of neurologists, neurosurgeons, radiologists, neuropsychologists, and nursing and social work staff. Seizures and normal function in any particular patient may be evaluated with several of a wide range of techniques, including video-EEG for characterization of seizure semiology, MEG, transcranial magnetic stimulation, PET, inter-ictal and ictal SPECT, anatomic MRI, functional MRI, diffusion MRI, and metabolic assessment with MRS. When concordance from a range of techniques indicate a focal epileptogenic zone, and that zone can be localized, surgical resection of the zone is associated with long term reduction in seizures [11,13].
One of the primary localizing modalities is iEEG. When there is a focal epileptogenic zone, selected events (such as seizures and spikes) are identified with particular electrodes, and this provides data for planning the margin of the resection [14]. Surgical planning has been largely based on clinical notes, sketches, and 2D radiographic images that depict, for example, white matter (WM) pathways and cortical gray matter (GM) landmarks to help localize the seizure onset zone or zones of the brain. However, these images simply cannot transfer onto the actual 3D geometry of the brain. This renders electrode locations ambiguous, at best, and hampers a clear understanding of the precise anatomical localization of electrodes. This hinders a clear assessment of the risks and benefits of intervention. Indeed, surgical planning of potentially curative resections is often restricted by the lack of reliable localization data. To address these limitations of existing practice, we have developed an innovative, computationally-driven approach to electrode localization that facilitates a clear understanding of the position of electrodes and readily enables communication and visualization of the localization information to all clinicians involved in each case.
It is desirable to precisely identify the position of the iEEG electrodes with respect to a pre-operatively acquired MRI. The pre-operative MRI provides a convenient reference frame for the localization of eloquent cortex with functional MRI, for white matter pathways from diffusion imaging, and for the visualization of vasculature. This MRI describes the geometry of the patient’s brain prior to placement of the electrodes. The position of the electrodes and brain anatomy may be identified with a number of imaging modalities, each of which has some advantages and some disadvantages. For example, despite having poor soft tissue contrast, post-operative computed tomography (CT) images are commonly used for electrode localization [15–19] because electrodes can be easily localized in 3D CT images, as opposed to 2D radiographic images [20]. In addition, CT shows relationships to bony landmarks (such as the edge of the craniotomy) very well. While post-operative MRI scans are also sometimes used for localization [21–23], and MRI has excellent soft tissue contrast, such MRI may suffer from inferior quality due to magnetic susceptibility artifacts at the position of electrodes that are manifested as black holes obscuring the underlying anatomy [24–26]. In addition, the short duration of a CT scan may be preferred to the longer duration of an MRI scan, because of the fragile post-operative condition of a patient with seizures. Furthermore, at our institution we frequently use an intracranial pressure (ICP) monitoring device that is not MRI compatible, and is a contra-indication for MRI.
A significant challenge in achieving accurate electrode localization with respect to the pre-operative MRI, lies in estimating soft tissue deformation that occurs post-operatively. Soft tissue deformation, also known as brain shift, occurs due to several factors that include brain swelling, drainage of cerebrospinal fluid, and surgical intervention [27]. Nabavi et al. [28] reported brain shift of up to 25mm after craniotomy. As reported in several other studies, electrode localization can be improved by accounting for brain shift [29–32]. Several algorithms have been proposed to compensate for brain shift. When both pre-operative and intra-operative or post-operative MRI are available, biomechanical properties of the brain can be characterized through a finite element model, and three-dimensional volumetric brain shift displacement can be accurately estimated by solving an optimization problem that balances measurements of displacements of image features with a biomechanical model of soft tissue deformation [33–36].
Some other recent studies project the grid electrodes onto the pre-operative cortical surface, in order to compensate for brain shift. Dykstra et al. [15] minimizes an energy function based on inter-electrode distance as well as global electrode configuration for CT-MRI registration and projection of the strips/grids on the pre-operative cortical surface, but their method needs significant time for the manual localization of the electrodes on the co-registered CT; and Yang et al. [26] minimize an energy function to project the corners of a rectangular grid of electrodes onto the cortical surface. However, they cannot localize grids that are bent or cut during implantation and cannot localize the strip electrodes. Tao et al. [18] find the electrode positions on the cortical surface by interactive manual transformation from CT to MRI images, but this might increase the subjectivity of the transformation and introduce wrong electrode projection. LaViolette et al. [17] compare the CT electrode coordinates with the intra-operative electrode coordinates measured with operating room navigational equipment at the time of resection, but due to the limitations of the navigational unit the brain shift analysis cannot be done in real time in the operating room, which makes the method of limited use. Hermes et al. [16] project each grid electrode to the point on the cortical surface in the direction of the local norm vector of the electrode grid. They also estimate the displacement of the strip electrodes after brain shift by projecting strip electrodes onto the closest points of the pre-operative MRI cortical surface. However, the closest point on the pre-operative cortical surface depends on the amount of brain shift and location of the electrodes on the post-operative cortical surface, which may substantially mislocalize the electrode positions. To the best of our knowledge, no automatic approach for the localization of the depth electrodes in the pre-operative MRI volume has been developed yet.
In contrast, we propose a novel method of projecting strip electrodes onto the pre-operative cortical surface in a direction normal to the strip plastic sheet. Furthermore, after alignment of all of the strip/grid electrodes, we estimate a volumetric displacement field based on the displacement of strip/grid electrodes, and we then adjust the position of depth electrodes accordingly. To assess the accuracy of electrode localization with our method, we utilized 2D intra-operative photographs by projecting electrodes from the 3D CT space onto the 2D photo space; and then measure the distance between the projected electrodes and the actual, corresponding electrodes that have been placed onto the 2D photo space [37,38,13]. In addition, we propose a novel approach by which we can visualize a range of anatomical structures (along with the electrodes) on a partially transparent cortical surface that will enable the epilepsy surgery team to accurately and easily estimate the distance between the electrodes and the anatomical landmarks (e.g., vasculature, gyri, sulci or lesions) which are frequently difficult at best to recognize from conventional notes and sketches (Fig 1). This will aid the surgery team in orienting to the correct seizure onset zones, eloquent cortical areas, and white matter pathways in the brain.
Fig 1.
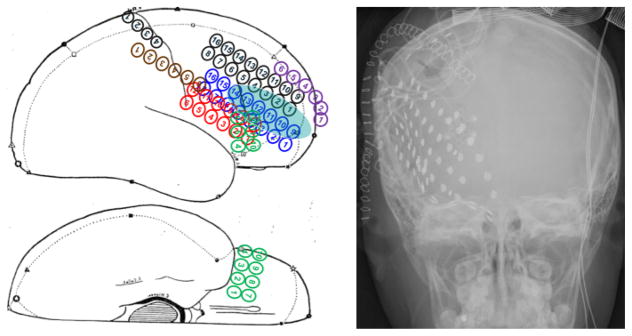
The sketch (left), radiographic image (right) of an epileptic subject presented in the epilepsy surgery conference. The blue ellipse on the sketch highlights the suspicious focal cortical dysplasia, and the high intensity solid dots in the radiographic image show the locations of the grid/strip electrodes.
Here we present a novel method for electrode localization, and describe validation experiments that assess the accuracy of our electrode localization approach (Section 2). Specifically, we describe the results of our study in ten epileptic subjects and compare our localization accuracy with that of previously described techniques (Section 3). Furthermore, we discuss the potential role of improved electrode localization in achieving better surgical outcomes for children with intractable epilepsy in the future (Section 4).
2. Materials and Methods
Ten patients (12.7±4.2 years, 5 males and 5 females) with pharmacologically intractable epilepsy participated in this study. This HIPAA-compliant study was approved by our institutional review board (IRB). Five to seven days before the iEEG was administered, structural T1-weighted images were acquired from the subjects using a 3T MR scanner (Siemens, TrioTim), with voxel size of 1.0×1.0×1.0mm3, repetition time (TR)=2.5s, echo time (TE)=2.2ms, field of view (FOV)=220mm. To determine the electrode positions with respect to the surrounding brain anatomy, CT scans were acquired shortly after craniotomy. CT of the head was carried out with voxel size of 0.5×0.5×0.625mm3, tube voltage of 125kV, and FOV=500mm (GE Medical Systems).
2.1. Electrode Characteristics
Stainless-steel or platinum-iridium alloy electrode discs (4mm dia.) are arranged in a grid (8×8, 8×4, or 8×2), strip (8×1, 6×1, or 4×1), or 8–10 cylindrically-shaped contact depth electrodes (AdTech, Racine, WI, USA). The inter-electrode spacing is 10mm in strips/grids, and 5mm in depth electrodes. After implantation, the strips and grids are sutured to the dural membrane. In addition, a head wrap bandage is used, in part to limit movement during iEEG monitoring.
2.2. Localization of Electrode Positions
This section explains our algorithm for localizing electrodes in CT and for projecting them onto the pre-operative MRI cortical surface. The alignment of the electrodes to the pre-operative MRI is achieved through a sequence of analytical steps (Fig 2).
Fig 2.
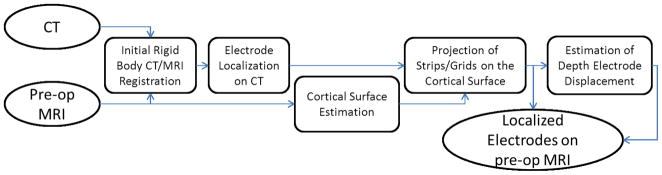
The pipeline of our electrode localization on the pre-operative MRI cortical surface.
2.2.1. Initial Rigid Body CT/MRI Registration
Initial alignment of the post-operative CT to the pre-operative MRI is carried out to bring most of the anatomy into alignment, and to provide an initialization for the search for non-rigid displacements. Following the craniotomy and placement of the electrodes, most of the skull bone and brain tissue has undergone only a rigid body transformation. This allows a rigid body transformation compensating for translation and rotation in space to be efficiently computed. This calculation needs to be robust to the presence of non-rigid displacements of the craniotomy and some of the cortical surface. We carry out rigid registration of the CT to the MRI using maximization of mutual information [39,40]. In practice, robust registration is readily achieved using a careful optimization of registration parameters using Powell’s method [41,42] together with dense sampling of intensities from the MRI and CT using Insight Toolkit (ITK) libraries in a multi-resolution framework. Our mutual information estimation is based on Maes’ technique which uses trilinear partial volume distribution interpolation to update the joint histogram for each voxel, which results in a smoother behavior of the registration criterion compared with Nearest Neighbor or Trilinear interpolations [43]. Initial rigid body alignment is computed by alignment of the geometric centers of each image. If the CT was acquired with gantry tilt, then this is eliminated by affine transformation of the CT.
2.2.2. Electrode Localization on CT
As electrodes have higher intensity than the surrounding brain tissues in the CT images, we apply a threshold to extract all the electrodes and attached wires. The appropriate threshold was determined by calibration across a retrospective set of patient CT scans. To remove any unnecessary wires, we erode the thresholded binary image with a spherical kernel of radius one voxel (Fig 3). Next, the center of mass of each connected component is calculated to determine the center of each electrode. Since depth electrodes are smaller than strip/grid electrodes, the former may be removed after the morphological filter is applied. In this scenario, we check each connected component and then restore any depth electrodes that were erroneously removed by the morphological filtering. In Table 4, we present the evaluation of the accuracy of this electrode detection and localization procedure.
Fig 3.
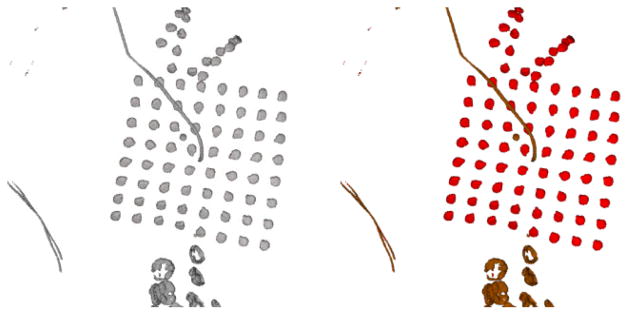
The extracted electrodes and wires after thresholding the CT image (Left). The narrower wires inside the cranium (Brown) are removed by morphological filtering, and the electrodes (Red) inside the skull are preserved (Right).
Table 4.
The accuracy of the electrode center localization in the CT image for 10 patients in this study.
| Patients | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean(mm) | 0.09 | 0.10 | 0.08 | 0.10 | 0.08 | 0.10 | 0.11 | 0.07 | 0.08 | 0.10 | 0.09 |
| SD(mm) | 0.16 | 0.20 | 0.14 | 0.18 | 0.15 | 0.18 | 0.20 | 0.12 | 0.12 | 0.17 | 0.16 |
2.2.3. Cortical Surface Estimation
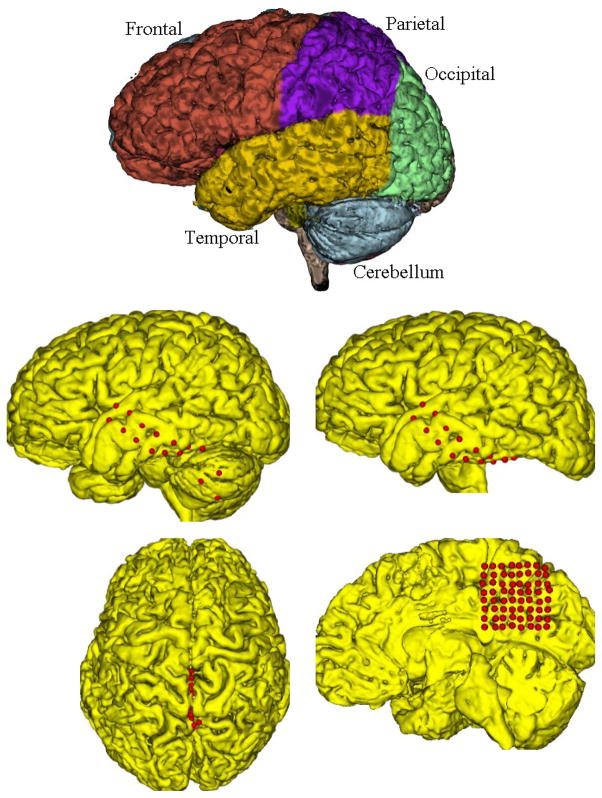
In order to localize the subdural strip/grid electrodes on the pre-operative cortical surface, we first need to estimate the pre-operative cortical surface, and then, project the electrode centers extracted from CT onto the pre-operative MRI cortical surface. It is not sufficient to project to the convex hull of the brain. In fact, it is necessary to project to the nearest surface of the hemisphere or lobe of the brain on which the electrodes are placed. The placement of electrodes in the interhemispheric fissure involves the placement of electrodes on the surface of the brain, but in a location where surrounding anatomy obscures their visualization. They are more easily visualized if the hemisphere not involved is removed from the visualization, exposing the buried surface of the remaining hemisphere for straightforward visualization. To this end, we utilize the Local MAP PSTAPLE algorithm1 with default parameters [44–46], to produce the pre-operative MRI brain parcellation, as shown Fig 4. Row1, remove the anatomical regions obscuring the electrodes after projection, e.g. left/right hemisphere or cerebellum, combine the remaining parcels into one binary image, and then smooth the binary image using a closing filter with a spherical kernel of radius three voxels. The parcellation can be completed once the pre-operative MRI image is acquired which is usually a few days before Phase II. Fig 4. Row2 illustrates an 8×2 grid pushed between cerebellum and the temporal lobe during implantation. We removed the cerebellum and projected the grids onto the temporal lobe surface so that the electrodes are correctly projected under the temporal/occipital lobe and can easily be seen. Further, Fig 4. Row3 shows an 8×8 grid inserted between two hemispheres, the left hemisphere was removed to project the grids on the smoothed right hemisphere surface.
Fig 4.
Row 1: The brain parcellation segments different anatomical regions. Row 2: A single red 8×2 grid is pushed between the cerebellum and temporal lobe. If we use the total brain cortical surface containing the cerebellum, some of the electrodes are wrongly projected on the cerebellum (left). To solve this, the cerebellum is removed to correct the projection (right). (Row 3): Since the red 8×8 grid is inserted between two hemispheres, its location is not clearly identifiable (left). To project the grid on the right hemisphere, the left hemisphere is removed so that the grid is projected on the parietal lobe of the right hemisphere (right).
2.2.4. Projection of Grids on the Cortical Surface
Grid electrodes are mounted on a thin sheet of plastic, which is placed on the cortical surface during implantation. In order to project the electrode back on the cortical surface before craniotomy, we first calculate each electrode center, as described in Section 2.2.2. Next, we estimate the pre-operative cortical surface using the brain parcellation of the T1-weighted image into a single binary image as in Section 2.2.3. Finally, we displace each electrode in a direction normal to the plastic sheet [16], and then, project it onto the smoothed cortical surface (Fig 5). Since brain shift is not an isometric deformation (i.e., distance preserving), the distance between each pair of electrodes may not be preserved after the projection is complete.
Fig 5.
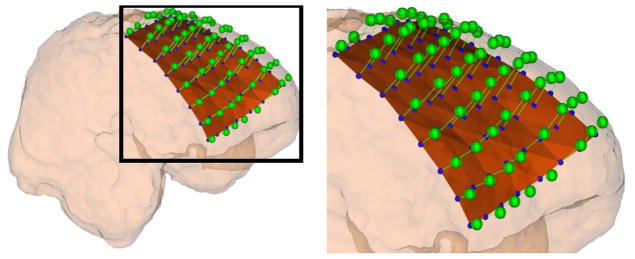
The extracted electrodes (blue spheres) in the CT image are displaced in a direction normal to the plastic sheet (brown surface) and then projected onto the smoothed cortical (pink) surface. The extracted electrodes (blue spheres) will be localized onto the MRI cortical surface (green spheres).
2.2.5. Projection of Strips on the Cortical Surface
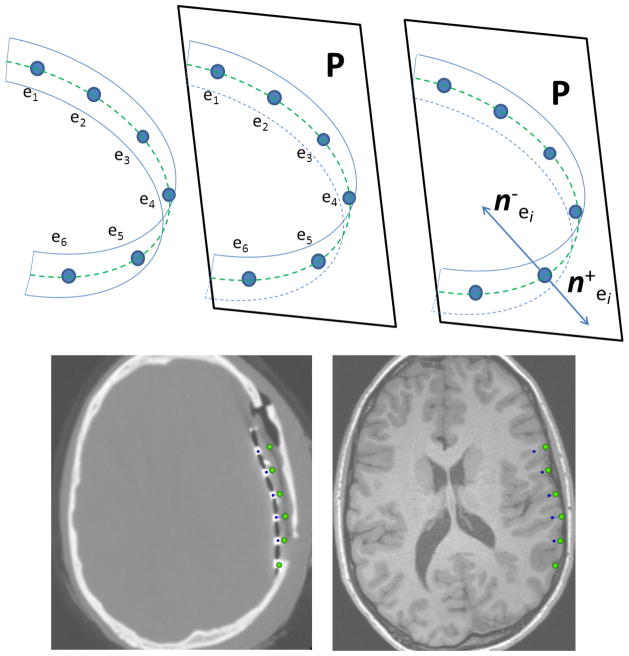
In Hermes et al. [16], the electrodes are projected onto the nearest points on the cortical surface. However, the closest point on the pre-operative cortical surface depends on the amount of brain shift and location of the electrodes on the post-operative cortical surface, which might dramatically change the electrode projection and can result in an incorrect projection. In contrast, we project the strip electrodes in a direction normal to the plastic sheet onto the smoothed cortical surface. However, the strips consist of only a single row of electrodes for which the normal vectors cannot be determined (as in a grid). Despite this, we are able to approximate the appropriate normal direction, since the narrow, rectangular-shaped plastic sheet with the electrodes is only bent in one direction and was twisted only slightly during the implantation (Fig 6). This suggests the implanted strip electrodes are roughly located in a plane, P, which is perpendicular to the rectangular plastic sheet, and passes through line l, effectively connecting the electrode centers (Fig 6). Thus, Plane P should have the minimum sum of distances from all electrodes ei(i = 1,…, L), i.e.,
Fig 6.
(Row 1): Strip electrodes e1, …,e6; (blue circles) are mounted on a narrow rectangular plastic sheet which is bent in one direction during implantation. The hypothetical dashed green line l on the plastic sheet connects the centers of the electrodes (left). Due to the rectangular shape of the plastic sheet, the electrode centers are roughly located on one plane, P, even after the strip is bent; this plane is estimated using the linear least square method (middle). The two normal vectors, and , are in plane P, and perpendicular to both line l and the plastic sheet, which show the projection directions. (Row 2): illustrates the projection of a 6×1 strip array onto the MRI cortical surface using the proposed method. First, the CT image is registered on the MRI image, and then, the electrode centers on the CT image (blue spheres) are displaced in direction normal to the plastic sheet which can be distinguished as a dark line connecting the high intensity electrodes on the CT image. As seen in the MRI image, the electrodes are accurately projected on the MRI cortical surface before craniotomy (green spheres).
| (1) |
where dis(P, ei) determines the Euclidean distance between plane P and electrode ei. We utilize the linear least square method to minimize the functional in Eq.(1) and fit a plane to the strip electrode centers, that’s,
| (2) |
where d = (d1, d2, d3)T are the parameters defining plane P, ei = (eix, eiy, eiz) are the coordinates of electrode ei, and
| (3) |
The plane parameters are d = A+b, where A+ = (ATA)−1AT is the pseudo inverse of A (Fig 6). Residuals of strip electrode distance from the plane P are demonstrated in Table 2 for all the patients participated in this study. As seen, the mean and standard deviation of the residual distance for plane P is 0.55±0.56mm, which shows the electrodes are roughly located on a plane. Now, we only consider the normal vectors in plane P, which are perpendicular to line l, and obviously normal to the plastic sheet. This yields two normal vectors in two opposite directions at each electrode location on the plastic sheet, e.g. and at electrode ei. Next, we displace each electrode in the normal directions ( and ) and find their intersection points, and , with the smoothed cortical surface. Finally, we choose the intersection point closest to the original electrode location as the projection point, i.e.,
Table 2.
The accuracy of our method for 10 study patients with various strips and grids. C1: Type and number of the implanted electrode arrays, G: Grid electrodes, S:Strip electrodes and D: Depth electrodes, the number of each array type is shown by a number after colon, and the arrays visible in the photos are indicated in brackets, C2: The number of visible electrodes in the photos, C3–C5: min, max and median of the error (mm), C6: mean±SD of the error (mm), C7: Residuals of strip electrode distance from the considered plane P (mean±SD mm) as described in Section 2.2.5.
| Patients | C1 | C2 | C3 | C4 | C5 | C6 | C7 |
|---|---|---|---|---|---|---|---|
| S1 | [G8×8:1] - S8×1:2 | 58 | 0.13 | 2.58 | 1.16 | 1.23±0.62 | 0.29±0.27 |
| S2 | [G8×4:1] - G8×2:3 - S8×1:1 - D10×1:1 | 28 | 0.12 | 2.37 | 0.85 | 0.90±0.56 | 1.10±0.88 |
| S3 | [G8×2:2- G6×2:1- S6×1:2] - G6×2:1 | 36 | 0.45 | 4.41 | 1.71 | 1.91±0.92 | 0.38±0.37 |
| S4 | [G8×8:1- G8×4:1]- G8×2:2 | 62 | 0.15 | 3.13 | 1.13 | 1.24±0.63 | - |
| S5 | [G8×4:1- G8×2:3]- G8×8:1- D10×1:1 | 23 | 0.16 | 3.05 | 0.81 | 1.06±0.68 | - |
| S6 | [G8×4:1- G8×2:2] - G8×2:1-S8×1:2 | 44 | 0.34 | 2.46 | 1.39 | 1.31±0.61 | 0.50±0.44 |
| S7 | [G8×2:3] - G8×2:1- S8×1:1 | 19 | 0.64 | 3.53 | 1.93 | 2.01±0.94 | 0.57±0.58 |
| S8 | [G8×4:2] - G8×2:2 | 48 | 0.18 | 2.77 | 0.97 | 1.08±0.59 | - |
| S9 | [G8×8:1]- G8×2:1- S8×1:2- D10×1:1 | 32 | 0.28 | 2.92 | 1.10 | 1.14±0.65 | 0.72±0.62 |
| S10 | [G8×4:2- G8×2:2]- D8×1:1 | 35 | 0.62 | 3.93 | 1.43 | 1.62±0.93 | - |
| Total | 385 | 1.31±0.69 | 0.55±0.56 |
| (4) |
The point is chosen as the projection point, if , or the point , otherwise.
2.2.6. Estimation of Depth Electrode Displacement
Depth electrodes are guided through the craniotomy and cortical surface into subcortical targets, and unlike strip/grid electrodes, the depth electrodes cannot be projected onto the smoothed cortical surface. In order to determine their displacements, we estimate a volumetric deformation field between the CT and MRI based on the displacement of strip/grid electrodes. Let E = (e1, …, eN) be the centers of the N strip/grid electrodes in the CT image, and Ē = (ē1, …, ēN) be their corresponding projections on the smoothed cortical surface as estimated in the previous steps. These two sets can be considered as two sets of landmarks selected on the two images. Our goal is to approximate the deformation field f: ℝ3 → ℝ3 such that ēi = f(ei)(i = 1, …, N). One can simply estimate the deformation field and the displacement of each point under the cortical surface using distance weighted averaging [47]. Let x be a depth electrode center in CT under the cortical surface; the assigned weight to x with respect to ei is modeled with a Gaussian kernel as,
| (5) |
where Wi(x) is the assigned weight, which depends on the Euclidean distance between x and ei, and σ2 determines the regularization of the deformation. Finally, the location of x before craniotomy, x̄, is calculated as,
| (6) |
which is the barycenter of all the strip/grid electrodes in the MRI image coordinates based on the assigned weights in Eq.(5).
2.3. Method of Assessing Strips/Grids Alignment Accuracy
The 2D intra-operative photographs depicting the strip/grid electrodes on the cortical surface are used to measure the localization accuracy [16,38,18,26]. The photographs were taken by a digital camera with a deflectable LCD screen placed perpendicular to the exposed cortex. The quality of the photographs was controlled on the LCD screen. We measure the localization accuracy by projecting the electrodes from the 3D MRI coordinate system onto the 2D intra-operative photograph coordinate system. The distance between projected electrodes and their actual, corresponding electrodes on the photograph is measured as the spatial accuracy. First, we select various anatomical landmarks including the gyri and sulci that appear on both the 2D photograph and the 3D MRI cortical surface. We can now utilize these landmarks to project electrodes onto the photo space in the following manner:
A 3×4 camera projection matrix P is calculated using the 2D photograph and 3D cortical landmarks based on the normalized Direct Linear Transformation method (DLT) [48,49].
- The electrode centers are projected from the 3D MRI coordinates onto the 2D photo coordinates. Let ēi = (ēix, ēiy, ēiz) be the coordinates of an electrode on the smoothed cortical surface, and ti = (tix, tiy) be its projection onto the 2D photo space. This projection is,
(7) The accuracy is defined as the mean of the Euclidean distance between the actual electrodes in the 2D photograph and their projections (ti’s) from the MRI into the 2D photo space. Perfect alignment would lead to this distance being zero.
In contrast to our method that utilizes the anatomical landmarks to estimate the camera projection matrix P, previous work has utilized [20,26] all, or a subset, of the visible electrode locations to estimate matrix P. This estimation is error-prone, however, as the accuracy of the electrode locations (Ē) on the smoothed cortical surface is unknown, and cannot be used to determine P. If the electrode location on the cortical surface is not accurate, this leads to an inaccurate camera projection matrix, which, in turn, produces an inaccurate error estimate.
3. Results
Ten patients (12.7±4.2 years, 5 males and 5 females) with pharmacologically intractable epilepsy participated in this study (Table 1). T1-weighted images were acquired from each subject before craniotomy, along with CT scans acquired within 48h of implantation. We extracted the cortical surface, brain parcellation, and suspected lesions from the MRI images. Next, we applied our method of extracting the electrodes in CT and projected them on the MRI cortical surface. Last, we validated the spatial accuracy of our method based on the intra-operative 2D photo, as described in Section 2.3.
Table 1.
Characterization of ten patients with pharmacologically intractable epilepsy participated in this study.
| Patients | Gender | Age at Surgery (Year) | Type of Surgery | Pathology |
|---|---|---|---|---|
| S1 | M | 10 | Right Frontal | Reactive changes |
| S2 | M | 10 | Left Paramedial Frontal | Parenchyma with MCDa |
| S3 | F | 6 | Right Frontal | Parenchyma with MCDa |
| S4 | F | 20 | Right Parietal | Reactive changes |
| S5 | F | 10 | Right Frontal | Glioma |
| S6 | M | 11 | Medial Frontal | Focal cortical dysplasia |
| S7 | M | 17 | Right Frontal | Parenchyma with MCDa |
| S8 | F | 16 | Right Occipital | Reactive changes |
| S9 | F | 16 | Left anterior temporal | Focal cortical dysplasia |
| S10 | M | 11 | Left lateral temporal | Focal cortical dysplasia |
MCD: Malformation of Cortical Development
3.1. Spatial Accuracy
We selected between 12 and 16 anatomical landmarks on the gyri or sulci of each subject, and then determined the camera projection matrix based on landmarks identified. Table 2 shows how our method creates a unique electrode localization profile (including degree of accuracy) for each study subject.
In total, 385 electrodes were visible in the intra-operative photographs. The average electrode localization accuracy was 1.31±0.69mm, which is sufficiently accurate for surgical planning. This accuracy is superior to those obtained with state of the art techniques in the literature (Table 3). The average spatial accuracy for subjects, S3, S5, S6, S7 and S10 with 8×2 grids, was 1.56±0.80mm, yet for the subjects with only 8×8 or 8×4 grids, was 1.14±0.61mm. This shows that the 8×2 grids have moved after implantation, as they are not sutured in place, as opposed to the 8×8 or 8×4 grids, which are sutured in place and did not move (Fig 7).
Table 3.
The accuracy of the electrode localization studies based on the intra-operative photographs.
Fig 7.
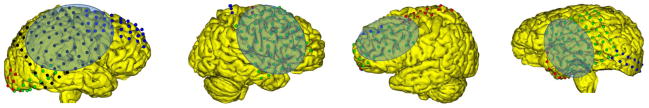
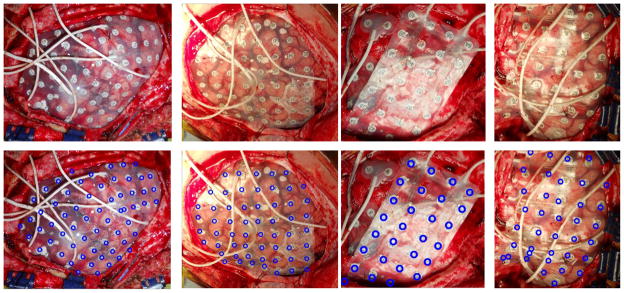
(Row 1) The electrode localization of four subjects on the MRI cortical surfaces are illustrated with craniotomy regions highlighted in blue. (Row 2) The intra-operative photos of the same subjects showing the electrode locations, (Row 3) the projection of the electrodes to the photos shown by blue rings.
We carried out an experiment to evaluate the accuracy of the electrode localization procedure in Section 2.2.2 for identifying electrode centers. We applied the algorithm described above in Section 2.2.2 to segment each electrode and to estimate the center of mass of each electrode. We then interactively identified the center of each electrode in the CT scan, and measured the distance between the center of the electrode identified manually and by our algorithm. The accuracy of localization of 1002 electrodes in 10 subjects is presented in Table 4.
3.2. Execution Time
Table 5 shows a breakdown of each task in the pipeline and the estimated amount of time needed to complete each step. The localization of electrodes on the cortical surface is very straightforward and does not exceed 13min. However, the depth electrodes might sometimes be removed by the morphological filter, which needs the user to add them back manually, which usually takes less than 5min depending on the number of the eliminated electrodes.
Table 5.
Execution of tasks: Time (in minutes) required for each step in the pipeline.
| Pipeline Steps | Estimated time to completion: ≤ 13 min |
|---|---|
| CT/MRI Registration | 5 to 7 min |
| Electrode localization in CT | 5 min |
| Projection of the electrodes | ≤ 1 min |
3.3. Illustrative Case
In order to qualitatively measure the effectiveness of our electrode localization method, we compared our localization results with the preliminary sketches provided during electrode implantation in the OR. However, due to inaccuracies in ratio of brain vs. grids, or the movement of strips/grids after implantation [13], identification of the relation in location of landmarks vs. electrodes is difficult. Fig 8 was presented as the sketch illustrating the electrode locations after implantation. As shown in the sketch, the red and green 8×2 grids are placed on the inferior and middle temporal gyri. However, based on the results of intra-operative photos, the red and green grids should be shifted upward such that the superior temporal gyrus would be covered. Our method localized the red and green grids on an area that encompassed the both inferior and middle temporal gyri as well as the superior temporal gyrus. Also note the tip of the red grid is located on the supramarginal gyrus (Fig 8). These results indicate that our approach provides improved grid localization and supplementary information to confirm that the sketch is correct.
Fig 8.
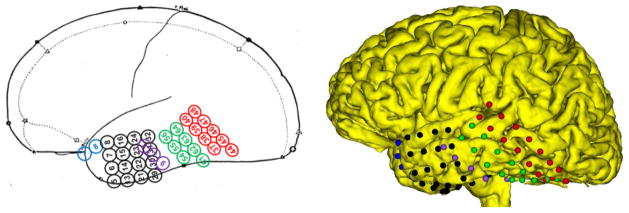
The primary sketch presented by the neurophysiologists shows the red and green grids covering only the inferior and middle temporal gyri (left). By contrast, our localization method localizes these areas of the brain as well as the superior temporal gyrus (right).
3.4. The Vasculature Visualization
Neurophysiologists most often use sulci/gyri landmarks to orient to the correct strip/grid electrode locations. However, because the sulci/gyri can be difficult to distinguish on the cortical surface, especially in case of repeat surgeries, developmental abnormalities, and with a small size of craniotomy, accurate electrode localization can be challenging, if no additional information is available. Under these circumstances, surface vasculature, mostly venous, are used in epilepsy planning as alternative anatomical landmarks for electrode localization, as surface vasculature is easier to distinguish than other landmarks. Furthermore, blood vessels coursing under the cortical surface can serve as important fiducials that help ensure accurate depth electrode localization in some of the deepest recesses of the brain where seizure activity may be taking place. Specifically, we extracted and enhanced the vasculature from magnetic resonance angiography (MRA) images, and then projected it onto the cortical surface along with the electrodes, which takes less than 5 minutes. In this way, neurophysiologists can compare the utility of the vasculature landmarks to the sulci/gyri in terms of electrode localization (Fig 9).
Fig 9.
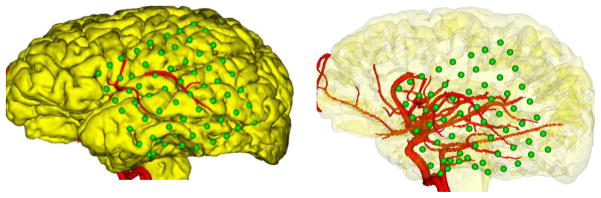
Clinicians can orient to the correct electrode locations (green) using the vasculature (red), reducing the potential for surgical injury to blood vessels during the implantation of electrodes.
In addition, during electrode implantation, the surgeon’s ability to visualize the vasculature reduces the likelihood of surgical injury to blood vessels that are underneath the skull, and thus hidden from view. This visual information is especially critical as the surgeon endeavors to cover more cortical area by pushing the strips/grids on the cortical surface under the skull and through to the edges of the craniotomy. The ability to visualize both seen and unseen blood vessels also has clear implications on the surgeon’s role in minimizing the risk of hemorrhage and stroke during surgery and post-operatively.
3.5. Lesion Visualization
Brain lesions are most often found underneath the cortical surface, which makes it hard to estimate their position and extent with respect the electrodes. In addition, in patients with a previous surgery cavity, location of the electrodes in relation to the cavity margins is very important to estimate the extent of resection needed to achieve therapeutic goals; especially, freedom from seizures, with limited adverse neurological side effects, and improved quality of life. However, the location of the electrodes in relation to the lesions and the cavity margins cannot be easily estimated from the sketches or radiographic images. To this end, we segment the lesions or the previous surgery cavity from structural MR images [50] before craniotomy, and then render them with a partially transparent cortical surface along with the electrodes. Then, the surgical team can assess the various electrode positions relative to the lesions or the cavity margins, and last, estimate the extent of resection (Fig 10). The lesion detection takes about 2h, and can be completed once the T1-weighted image is acquired which is usually few days before the electrode implantation (Phase II).
Fig 10.
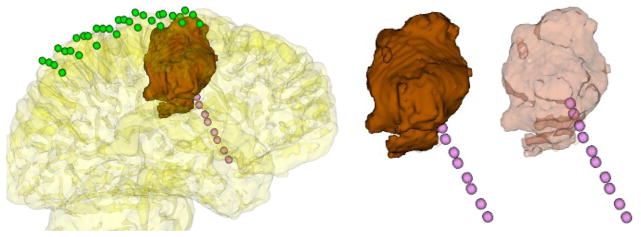
An 8×4 grid (green) and a 10×1 depth electrode array (pink) are illustrated along with a lesion (brown) extracted and visualized under a partially transparent cortical surface. The extent of the lesion and its position relative to the electrodes can be analyzed more easily on a partially transparent cortical surface (left). In addition, depth electrodes and the extracted lesion (middle) are likewise illustrated on a partially transparent cortical surface (right). In this way, the epilepsy surgery team can identify the electrode positions relative to the lesion. As seen in the figure to the right, the last two depth electrodes on the strip are actually inside the tumor, which is not evident when the lesion is shown opaquely, as in the middle figure.
3.6. Visualization of Functional/Structural Regions
Visualization of brain parcellation along with the electrodes allows neurophysiologists to visually recognize the regions where the electrodes are placed. Fig 11 (left) illustrates two 8×4 and 8×2 grids along with parcellation of the cortical surface, which provides useful structural and functional information that guides surgical planning. We can map functionally critical cortical areas that are localized using functional MRI on the cortical surface (Fig 11-right) or on white matter pathways to investigate the functional and structural changes in the brain due to lesions or ongoing seizures. Fig 11 (middle) shows the optic radiations nearby a focal cortical dysplasia (red).
Fig 11.
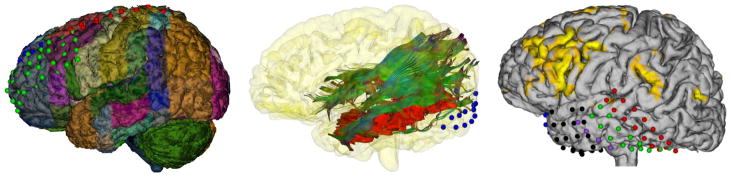
The brain parcellation helps neurophysiologists recognize the functional/structural areas where the electrodes are placed (Left). The optic radiation (OR) tractography is illustrated along with the focal cortical dysplasia in red and an 8×2 grid in blue (Middle). The electrode localization and the activation areas in a fMRI experiment on the cortical surface (Right). The functional activation areas and the tractography fibers on the cortical surface may facilitate judgement of the desired surgical resection margin.
4. Discussion
Our method of electrode localization offers precise and reliable information about the areas of the brain where the electrodes are placed during implantation, in conjunction with an improved visualization of the location of normal function. The successful application of the approach we have described may ultimately enable more accurate surgical planning, and could thus significantly improve patient outcomes by facilitating a reduction in the number and frequency of seizures as well as fewer neurological deficits post-operatively.
In our proposed method, after alignment of the post-operative CT with the pre-operative MRI, the grid electrodes are projected onto the cortical surface in a direction normal to the plastic sheet at the position of each electrode. For the strip electrode projection, we define a unique normal by determining a plane through minimizing the sum of the squares of distances between the strip electrodes and the plane. Next, we displace each electrode in this plane in a direction normal to the plastic sheet. However, this method cannot be employed for the depth electrodes since they are not projected onto the cortical surface, as are the strips/grids. Therefore, we estimate the volumetric brain shift based on the displacement of the strips/grids and then calculate the displacement of the depth electrodes. Previous studies, however, do not estimate the depth electrode displacement after brain shift [20,26]. Further, during implantation, the strips/grids are usually bent while being pushed underneath the skull, or occasionally, the surgeon cuts them into smaller grids. Nonetheless, our method accurately localizes the electrodes, despite the strip/grid electrodes being cut or bent during implantation.
We validated the localization accuracy of our method on ten epileptic patients using intra-operative photographs. We report accuracy of 1.31±0.69mm for all 385 visible electrodes, which is superior to localization errors reported in previous studies (Table 3). Further, our validation shows that the spatial accuracy of subjects with grids (e.g., 8×8) sutured in place is greater than the spatial accuracy of subjects with strips/grids not sutured in place, which are also more likely to move after implantation. However, our method has two primary limitations: 1) certain depth electrodes might be erroneously removed after the morphological filter step; and 2) overlapping electrodes might be connected and considered as a single electrode after the thresholding step. In cases such as these, electrode centers must be manually selected on the CT image, a process that takes a few minutes for each subject.
Accurate localization of iEEG electrodes in the brain is crucial for planning of a resection in pediatric epilepsy. Further, visualization of critical anatomical landmarks, functionally eloquent cortex and white matter pathways, enables clinicians to rapidly integrate the diverse information that is crucial for the assessment of a surgical resection plan. This information is not readily integrated through sketches and radiographic images that are routinely used. Indeed, without this critical anatomical information and precise localization of electrodes, the functional consequence of a planned resection margin is challenging to assess [51], and thus limits the utility of pre-operatively images for surgical planning [52,53]. Such assessments are crucial in meeting key treatment goals, namely, to reduce or eliminate the frequency and severity of seizures, and to minimize the likelihood of post-surgical neurological deficits. The techniques we described2 enable us to visualize lesions, vasculature, and electrodes on a partially transparent cortical surface along with a brain parcellation which provides useful structural and functional information that may also guide surgical planning. This information combination and image fusion enables the neurologists and neurosurgeons involved in each case to recognize critical structural and functional landmarks of the brain, which might facilitate surgical planning.
In future work, we will address the infrequent need to manually segment certain depth electrodes. The use of a local adaptive threshold filter will be investigated for depth electrode segmentation, in order to reduce the effect of the intensity variations in CT images around strips/grids and the depth electrodes, and prevent incorrect depth electrode removal. Further, a morphological filter with adaptive kernel size will be utilized rather than a constant kernel size, which more likely preserves the depth electrodes. In addition, the prior information about the number of each electrode type implanted for each subject may be incorporated in the electrode segmentation process.
Acknowledgments
This research was supported in part by NIH grants R01 RR021885, R01 EB008015, R03 EB008680, R01 LM010033 and R01 EB013248.
Footnotes
This software will be available online (http://crl.med.harvard.edu/software/STAPLE/)
The currently distributed software is available from http://www.crl.med.harvard.edu/software
Conflict of Interest
Vahid Taimouri, Alireza Akhondi-Asl, Xavier Tomas-Fernandez, Jurriaan M. Peters, Sanjay P. Prabhu, Annapurna Poduri, Masanori Takeoka, Tobias Loddenkemper, Ann Marie R. Bergin, Chellamani Harini, Joseph R. Madsen and Simon K. Warfield declare that they have no conflict of interest.
Ethical Standards Statement
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5).
Statement of Informed Consent
Informed consent was obtained from all patients for being included in the study.
References
- 1.Hirtz D, Thurman D, Gwinn-Hardy K, Mohamed M, Chaudhuri A, Zalutsky R. How common are the “common” neurologic disorders? Neurology. 2007;68 (5):326–337. doi: 10.1212/01.wnl.0000252807.38124.a3. [DOI] [PubMed] [Google Scholar]
- 2.Hauser WA, Annegers JF, Kurland LT. Prevalence of epilepsy in Rochester, Minnesota: 1940–1980. Epilepsia. 2007;32 (4):429–445. doi: 10.1111/j.1528-1157.1991.tb04675.x. [DOI] [PubMed] [Google Scholar]
- 3.Kwan P, Brodie MJ. Effectiveness of first antiepileptic drug. Epilepsia. 2001;42 (10):1255–1260. doi: 10.1046/j.1528-1157.2001.04501.x. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt D. Efficacy of new antiepileptic drugs. Epilepsy Currents. 2011;11 (1):9–11. doi: 10.5698/1535-7511-11.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engel J, Jr, McDermott MP, Wiebe S, Langfitt JT, Stern JM, Dewar S, Sperling MR, Gardiner I, Erba G, Fried I. Early Surgical Therapy for Drug-Resistant Temporal Lobe Epilepsy A Randomized Trial. JAMA: The Journal of the American Medical Association. 2012;307 (9):922–930. doi: 10.1001/jama.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binnie CD, Polkey CE. Commission on Neurosurgery of the International League Against Epilepsy (ILAE) 1993–1997: recommended standards. Epilepsia. 2000;41 (10):1346–1349. doi: 10.1111/j.1528-1157.2000.tb04616.x. [DOI] [PubMed] [Google Scholar]
- 7.Eksioglu Y, Riviello JJ., Jr Intractable epilepsy in children and selection of surgical candidates. Pediatric Epilepsy Surgery: Preoperative Assessment and Surgical Treatment 2010 [Google Scholar]
- 8.Harvey AS, Cross JH, Shinnar S, Mathern GW. Defining the spectrum of international practice in pediatric epilepsy surgery patients. Epilepsia. 2008;49 (1):146–155. doi: 10.1111/j.1528-1167.2007.01421.x. [DOI] [PubMed] [Google Scholar]
- 9.Behrens E, Zentner J, Van Roost D, Hufnagel A, Elger C, Schramm J. Subdural and depth electrodes in the presurgical evaluation of epilepsy. Acta neurochirurgica. 1994;128 (1):84–87. doi: 10.1007/BF01400656. [DOI] [PubMed] [Google Scholar]
- 10.Engel AK, Moll CKE, Fried I, Ojemann GA. Invasive recordings from the human brain: clinical insights and beyond. Nature Reviews Neuroscience. 2005;6 (1):35–47. doi: 10.1038/nrn1585. [DOI] [PubMed] [Google Scholar]
- 11.Cataltepe O, Jallo G. Pediatric Epilepsy Surgery: Preoperative Assessment and Surgical Treatment. Thieme; 2010. [Google Scholar]
- 12.Rosenow F, Lüders H. Presurgical evaluation of epilepsy. Brain. 2001;124 (9):1683–1700. doi: 10.1093/brain/124.9.1683. [DOI] [PubMed] [Google Scholar]
- 13.Wellmer J, Von Oertzen J, Schaller C, Urbach H, König R, Widman G, Van Roost D, Elger CE. Digital Photography and 3D MRI–based Multimodal Imaging for Individualized Planning of Resective Neocortical Epilepsy Surgery. Epilepsia. 2002;43 (12):1543–1550. doi: 10.1046/j.1528-1157.2002.30002.x. [DOI] [PubMed] [Google Scholar]
- 14.Datta A, Loddenkemper T. Wyllie’s Treatment of Epilepsy. In: Wyllie ECG, Gidal B, Goodkin H, editors. Principles & Practice. 5. Lippincott, Williams & Wilkins; Philadelphia: 2011. pp. 818–827. [Google Scholar]
- 15.Dykstra AR, Chan AM, Quinn BT, Zepeda R, Keller CJ, Cormier J, Madsen JR, Eskandar EN, Cash SS. Individualized localization and cortical surface-based registration of intracranial electrodes. NeuroImage. 2011 doi: 10.1016/j.neuroimage.2011.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermes D, Miller KJ, Noordmans HJ, Vansteensel MJ, Ramsey NF. Automated electrocorticographic electrode localization on individually rendered brain surfaces. Journal of neuroscience methods. 2010;185 (2):293–298. doi: 10.1016/j.jneumeth.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 17.LaViolette PS, Rand SD, Ellingson BM, Raghavan M, Lew SM, Schmainda KM, Mueller W. 3D visualization of subdural electrode shift as measured at craniotomy reopening. Epilepsy research. 2011;94 (1):102–109. doi: 10.1016/j.eplepsyres.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao JX, Hawes-Ebersole S, Baldwin M, Shah S, Erickson RK, Ebersole JS. The accuracy and reliability of 3D CT/MRI co-registration in planning epilepsy surgery. Clinical Neurophysiology. 2009;120 (4):748–753. doi: 10.1016/j.clinph.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Agarwal R, Nguyen D, Domocos V, Gotman J. Intracranial electrode visualization in invasive pre-surgical evaluation for epilepsy. Engineering in Medicine and Biology Society, 2005. IEEE-EMBS 2005; 27th Annual International Conference of the; IEEE; 2006. pp. 952–955. [DOI] [PubMed] [Google Scholar]
- 20.Dalal SS, Edwards E, Kirsch HE, Barbaro NM, Knight RT, Nagarajan SS. Localization of neurosurgically implanted electrodes via photograph–MRI–radiograph coregistration. Journal of neuroscience methods. 2008;174 (1):106–115. doi: 10.1016/j.jneumeth.2008.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreas HJ, Huppertz HJ, Comeau RM, Honegger JB, Spreer JM, Zentner JK. Visualization of subdural strip and grid electrodes using curvilinear reformatting of 3D MR imaging data sets. American journal of neuroradiology. 2002;23 (3):400–403. [PMC free article] [PubMed] [Google Scholar]
- 22.Kovalev D, Spreer J, Honegger J, Zentner J, Schulze-Bonhage A, Huppertz HJ. Rapid and fully automated visualization of subdural electrodes in the presurgical evaluation of epilepsy patients. American journal of neuroradiology. 2005;26 (5):1078–1083. [PMC free article] [PubMed] [Google Scholar]
- 23.Morris K, O’brien TJ, Cook MJ, Murphy M, Bowden SC. A computer-generated stereotactic “Virtual Subdural Grid” to guide resective epilepsy surgery. American journal of neuroradiology. 2004;25 (1):77–83. [PMC free article] [PubMed] [Google Scholar]
- 24.Carmichael DW, Thornton JS, Rodionov R, Thornton R, McEvoy A, Allen PJ, Lemieux L. Safety of localizing epilepsy monitoring intracranial electroencephalograph electrodes using MRI: Radiofrequency-induced heating. Journal of Magnetic Resonance Imaging. 2008;28 (5):1233–1244. doi: 10.1002/jmri.21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis LM, Spencer DD, Spencer SS, Bronen RA. MR imaging of implanted depth and subdural electrodes: is it safe? Epilepsy research. 1999;35 (2):95–98. doi: 10.1016/s0920-1211(99)00007-8. [DOI] [PubMed] [Google Scholar]
- 26.Yang AI, Wang X, Doyle W, Halgren E, Carlson C, Belcher TL, Cash SS, Devinsky O, Thesen T. Localization of dense intracranial electrode arrays using magnetic resonance imaging. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daga P, Modat M, Micallef C, Mancini L, White M, Cardoso MJ, Kitchen N, McEvoy A, Thornton J, Yousry T. Near real time brain shift estimation for interventional MRI suite. High-Performance MICCAI 2010 [Google Scholar]
- 28.Nabavi A, Black PML, Gering DT, Westin CF, Mehta V, Pergolizzi RS, Jr, Ferrant M, Warfield SK, Hata N, Schwartz RB. Serial intraoperative magnetic resonance imaging of brain shift. Neurosurgery. 2001;48 (4):787–798. doi: 10.1097/00006123-200104000-00019. [DOI] [PubMed] [Google Scholar]
- 29.Darcey TM, Roberts DW. Technique for the localization of intracranially implanted electrodes. Journal of neurosurgery. 2010;113 (6):1182–1185. doi: 10.3171/2009.12.JNS091678. [DOI] [PubMed] [Google Scholar]
- 30.Immonen A, Jutila L, Könönen M, Mervaala E, Partanen J, Puranen M, Rinne J, Ylinen A, Vapalahti M. 3-D reconstructed magnetic resonance imaging in localization of subdural EEG electrodes: Case illustration. Epilepsy research. 2003;54 (1):59–62. doi: 10.1016/s0920-1211(03)00038-x. [DOI] [PubMed] [Google Scholar]
- 31.Kamida T, Anan M, Shimotaka K, Abe T, Fujiki M, Kobayashi H. Visualization of subdural electrodes with fusion CT scan/MRI during neuronavigation-guided epilepsy surgery. Journal of Clinical Neuroscience. 2010;17 (4):511–513. doi: 10.1016/j.jocn.2009.06.038. [DOI] [PubMed] [Google Scholar]
- 32.Murphy MA, O’Brien TJ, Morris K, Cook MJ. Multimodality image-guided surgery for the treatment of medically refractory epilepsy. Journal of neurosurgery. 2004;100 (3):452–462. doi: 10.3171/jns.2004.100.3.0452. [DOI] [PubMed] [Google Scholar]
- 33.Archip N, Jolesz FA, Warfield SK. A validation framework for brain tumor segmentation. Academic radiology. 2007;14 (10):1242–1251. doi: 10.1016/j.acra.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 34.Clatz O, Delingette H, Talos IF, Golby AJ, Kikinis R, Jolesz FA, Ayache N, Warfield SK. Robust nonrigid registration to capture brain shift from intraoperative MRI. Medical Imaging, IEEE Transactions on. 2005;24 (11):1417–1427. doi: 10.1109/TMI.2005.856734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrant M, Nabavi A, Macq B, Black PML, Jolesz FA, Kikinis R, Warfield SK. Serial registration of intraoperative MR images of the brain. Medical Image Analysis. 2002;6 (4):337–359. doi: 10.1016/s1361-8415(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 36.Ferrant M, Nabavi A, Macq B, Jolesz FA, Kikinis R, Warfield SK. Registration of 3-D intraoperative MR images of the brain using a finite-element biomechanical model. Medical Imaging, IEEE Transactions on. 2001;20 (12):1384–1397. doi: 10.1109/42.974933. [DOI] [PubMed] [Google Scholar]
- 37.Mahvash M, König R, Wellmer J, Urbach H, Meyer B, Schaller K. Coregistration of digital photography of the human cortex and cranial magnetic resonance imaging for visualization of subdural electrodes in epilepsy surgery. Neurosurgery. 2007;61 (5):340–345. doi: 10.1227/01.neu.0000303992.87987.17. [DOI] [PubMed] [Google Scholar]
- 38.Sebastiano F, Di Gennaro G, Esposito V, Picardi A, Morace R, Sparano A, Mascia A, Colonnese C, Cantore G, Quarato P. A rapid and reliable procedure to localize subdural electrodes in presurgical evaluation of patients with drug-resistant focal epilepsy. Clinical neurophysiology. 2006;117 (2):341–347. doi: 10.1016/j.clinph.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Akselrod-Ballin A, Bock D, Reid RC, Warfield SK. Accelerating Image Registration With the Johnson–Lindenstrauss Lemma: Application to Imaging 3-D Neural Ultrastructure With Electron Microscopy. Medical Imaging, IEEE Transactions on. 2011;30 (7):1427–1438. doi: 10.1109/TMI.2011.2125797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells WM, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. Medical image analysis. 1996;1 (1):35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 41.Powell MJ. An efficient method for finding the minimum of a function of several variables without calculating derivatives. The computer journal. 1964;7 (2):155–162. [Google Scholar]
- 42.Warfield SK, Jolesz FA, Kikinis R. A high performance computing approach to the registration of medical imaging data. Parallel Computing. 1998;24 (9):1345–1368. [Google Scholar]
- 43.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. Medical Imaging, IEEE Transactions on. 1997;16 (2):187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 44.Akhondi-Asl A, Warfield SK. Simultaneous Truth and Performance Level Estimation through Fusion of Probabilistic Segmentations. IEEE Transactions on Medical Imaging. 2013 doi: 10.1109/TMI.2013.2266258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Commowick O, Akhondi-Asl A, Warfield SK. Estimating A Reference Standard Segmentation with Spatially Varying Performance Parameters: Local MAP STAPLE. Medical Imaging, IEEE Transactions on. 2012;31 (8):1593–1606. doi: 10.1109/TMI.2012.2197406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. Medical Imaging, IEEE Transactions on. 2004;23 (7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atkeson CG, Moore AW, Schaal S. Locally weighted learning. Artificial intelligence review. 1997;11 (1):11–73. [Google Scholar]
- 48.Hartley R, Zisserman A. Multiple view geometry in computer vision. Vol. 2. Cambridge Univ Press; 2000. [Google Scholar]
- 49.Trucco E, Verri A. Introductory techniques for 3-D computer vision. Vol. 93. Prentice Hall; Upper Saddle River^ eNJ NJ: 1998. [Google Scholar]
- 50.Tomas-Fernandez X, Warfield S. Population intensity outliers or a new model for brain WM abnormalities. Biomedical Imaging (ISBI); 9th IEEE International Symposium on; 2012; IEEE; 2012. pp. 1543–1546. [Google Scholar]
- 51.Beyer J, Hadwiger M, Wolfsberger S, Buhler K. High-quality multimodal volume rendering for preoperative planning of neurosurgical interventions. Visualization and Computer Graphics, IEEE Transactions on. 2007;13 (6):1696–1703. doi: 10.1109/TVCG.2007.70560. [DOI] [PubMed] [Google Scholar]
- 52.Dimaio SP, Archip N, Hata N, Talos IF, Warfield SK, Majumdar A, McDannold N, Hynynen K, Morrison PR, Wells W. Image-guided neurosurgery at Brigham and Women’s Hospital. Engineering in Medicine and Biology Magazine, IEEE. 2006;25 (5):67–73. doi: 10.1109/memb.2006.1705749. [DOI] [PubMed] [Google Scholar]
- 53.Schiffbauer H, Ferrari P, Rowley HA, Berger MS, Roberts TPL. Functional activity within brain tumors: a magnetic source imaging study. Neurosurgery. 2001;49 (6):1313–1321. doi: 10.1097/00006123-200112000-00005. [DOI] [PubMed] [Google Scholar]