Abstract
The critical impact microbiota have on health and disease make the interaction between host and microbiome increasingly important as we evaluate therapeutics. Here we highlight growing evidence that beyond disease, microbes also affect the most fundamental of host physiological phenotypes, the rate of aging itself.
As you look in the mirror you may only see yourself staring back, but in reality you are not alone; you share your body with trillions of others. Contained on and within our bodies thrives a dynamic population of microbes that form a ‘metaorganism’, comprising 10 bacterial cells for every one of our own. Despite co-evolving in the presence of this ‘microbiome’ for 500 million years (Cho and Blaser, 2012), only recently have advances in sequencing technology allowed us to appreciate the complexities of this relationship, and the manner by which genomes within metaorganisms interact and affect one another. Inter-individual variations in the microbiome impact multiple human pathologies, from metabolic syndrome to cancer (Cho and Blaser, 2012). However, new data in invertebrate systems indicate microbes extend their effects beyond host pathology, to systemic modulation of the rate of aging.
The gut microbiome comprises a complex community of bacterial species that live within intestines of coelomate animals. Historically, the influence of gut microflora on host health has focused on the two extremes of the relationship: pathogenesis and symbiosis. Canonical examples of symbiotic host/microbe relationships include the reliance of some herbivores on gut flora for efficient digestion of cellulose. However, the majority of intestinal microbes are neither pathogenic nor classically symbiotic, but instead ‘commensal’, meaning their effect is non-harmful and neutral. Advances in high-throughput sequencing have facilitated the characterization of these diverse populations of commensal bacteria, defining the metagenome - the ensemble of host and microbiota DNA, and by extension the meta-transcriptome, proteome and metabolome. With endeavors such as the Human Microbiome Project just beginning (Cho and Blaser, 2012), research into the complexity of mammalian microbiome dynamics is in its infancy. Assigning causality to effects of the microbiome in mammalian systems is expensive, technically challenging and requires microbe-free husbandry conditions, and gnotobiotic mice with defined and controlled microbiota (Cho and Blaser, 2012).
Invertebrate model systems such as the fruit fly Drosophila melanogaster and the nematode worm Caenorhabditis elegans also live in the presence of microbiota both in nature and the laboratory (Ren et al., 2007; Zhang and Hou, 2013). Due to their genetic tractability and inexpensive husbandry, invertebrate studies allow direct causality to be assigned to the presence or absence of microbiota. Because of their short lifespans, D. melanogaster and C. elegans are also powerful models for studying genetic pathways that modulate the aging process. The ultimate goal of this research is to develop novel therapeutics with beneficial impacts on overall health in old age, complementing disease-centric approaches that focus on proximal determinants of individual pathologies. This work has uncovered conserved genes that modulate healthy aging in invertebrates and mammals, and are linked to extreme longevity in humans (Kenyon, 2010), including the insulin/IGF-1 like signaling (IIS) pathway, the target of rapamycin (TOR) and AMP-activated protein kinase (AMPK). However, little attention has been paid to how the microbial environment and the microbiome itself might affect the ability of these interventions to promote longevity. Here we review examples of microbiota in invertebrates directly and indirectly influencing the host genome to affect longevity, and discuss the impact these emerging data have on the field of aging research.
Direct Interspecies Signaling and Aging
Given the proximity of microbes and host within the metaorganism, diffusible molecules originating in bacteria can directly affect cells in the host (Ryan and Dow, 2008). Recent work suggests this interspecies signaling can impact host aging rate. C. elegans are commonly grown ‘monoxenically’ in co-culture with non-pathogenic strains of E. coli ‘OP50’ bacteria. E. coli act as nutrition for the worm, providing mandatory nutrients and essential components for life that the nematode cannot synthesize de novo. Although not considered pathogenic, E. coli OP50 do proliferate inside older animals, and raising C. elegans on non-dividing bacterial lawns suppresses this infection and increases worm lifespan (Zhang and Hou, 2013). In young worms however, E. coli are mechanically broken down before entering the gut and do not live in the animal as a true microbiome. Despite this, it is becoming apparent that beyond their role as a nutrient source and potential pathogen, E. coli secrete diffusible molecules, including metabolites and small RNAs that can directly impact C. elegans aging.
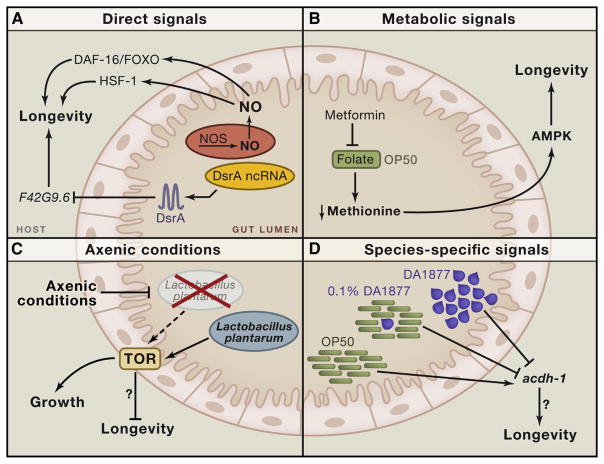
An example of a direct interspecies signal secreted by microbes that can promote longevity is nitric oxide (NO) (Gusarov et al., 2013). NO is a small, short-lived free radical that affects the activity of proteins both directly and indirectly via post-translational modifications. As a critical signaling molecule, NO has been implicated in multiple functions including neurotransmission, immunity and cardiovascular function (Gusarov et al., 2013). C. elegans are rare amongst eukaryotes as the cannot produce their own NO since they lack NO synthase. Gusarov et al. therefore speculated that worms might be utilizing NO produced from the bacteria within their microenvironment. Supporting their hypothesis, culturing C. elegans with NO-deficient Bacillus subtilis shortens their lifespan, while exogenous supplementation of NO increases it. Strikingly, an in vivo fluorescent sensor detected NO in multiple C. elegans tissues, suggesting it acts as a bona fide interspecies longevity signal. RNA-Seq analysis of worms fed NO identified specific transcriptional responses regulated by two transcription factors, DAF-16/FOXO and heat shock factor -1 (HSF-1), both known to mediate lifespan (Kenyon, 2010). Worms lacking DAF-16/FOXO or HSF-1 do not respond to the life prolonging effects of bacterial NO, meaning bacterial NO modulates C. elegans lifespan via effects on host transcription (Fig. 1A).
Figure 1. Microbe-modulation of invertebrate aging and physiology.
A. Bacterial-derived signals NO and ncRNAs regulate C. elegans longevity. B. Metformin increases C. elegans lifespan via effects on bacterial folate metabolism. C. L. plantarum drive Drosophila growth under low nutrient conditions via the longevity modulator TOR. D. Different bacterial species elicit specific transcriptional responses in C. elegans.
Not all signals from bacteria have a positive effect on worm longevity. RNA interference (RNAi), a mechanism by which short double stranded RNAs promote degradation of matching messenger RNA, was first discovered in C. elegans. Feeding E. coli expressing a double stranded C. elegans RNA sequence efficiently suppresses the worm gene (Liu et al., 2012). Interestingly, this synthetic laboratory system may be mirroring a natural communication between microbe and worm genomes. Liu et al. (2012) found that small non-coding RNAs (ncRNAs) expressed endogenously by E. coli can modulate C. elegans behavior and longevity. Feeding worms E. coli deficient in the ncRNA DsrA significantly increases their lifespan. The worm gene F42G9.6 bears sequence complementary to DsrA, and mutating this gene shortens lifespan and makes worms refractory to the effects of DsrA. Moreover, F42G9.6 expression is up-regulated in long-lived IIS mutants and over-expressing F42G9.6 slows aging (Liu et al., 2012) (Fig 1A). E. coli modulate longevity of the worm by hijacking innate RNAi machinery, as worms incapable of RNAi are resistant to the effects of DsrA. Cross-species small RNA regulation has yet to be observed between microbes and mammals. However, microRNAs derived from plants are present in human and rodent serum and tissues (Zhang et al., 2012), suggesting such interspecies communication is feasible.
Microbial Metabolites and Host Aging
Commensal bacteria can affect the nutrient landscape of the host – the majority of metabolites in human plasma are microbe-derived (Nicholson et al., 2012) - but recent work suggests bacterial-derived metabolites can affect host longevity. Metformin is the most widely prescribed treatment for metabolic disease and has gained attention as a potential pro-longevity molecule, as one of its key targets is AMPK, a conserved energy sensor implicated in lifespan extension by dietary restriction (DR) (Kenyon, 2010). Feeding worms metformin robustly increases lifespan in an AMPK-dependent manner. Surprisingly this response is indirectly mediated by the effect of the drug on bacteria, rather than the worms themselves. Metformin has no effect on worms cultured axenically (with no bacteria) or on lawns of killed bacteria (Cabreiro et al., 2013), yet extends lifespan of worms fed live but antibiotic-arrested bacteria, ruling out suppression of bacterial pathogenicity. Metformin suppresses folate metabolism specifically in E. coli, and feeding worms either the antibiotic trimethoprim, which inhibits dihydrofolate reductase in bacteria, or E. coli mutants with suppressed folate metabolism (Virk et al., 2012) also increases lifespan. Reduced folate metabolism decreases methionine content of bacteria, and this effect is causal to metformin-induced longevity in C. elegans. Although reduced dietary methionine is known to promote longevity in both mammals and Drosophila (Kenyon, 2010), these data add a new level of complexity into how pro-longevity drugs might mediate effects on an organism indirectly via affecting metabolites in associated microbiota (Fig 1B). Metformin increases lifespan in rodents (Martin-Montalvo et al., 2013), and a side effect of metformin treatment in humans is reduced folate levels (Cabreiro et al., 2013), and whether physiological effects of metformin function via microbe folate metabolism warrants investigation.
Effects of Axenic Culture on the Host
Culturing worms axenically robustly increases their lifespan (Houthoofd et al., 2002), yet since bacteria are also the food source, determining relative contributions of DR versus microbial presence on worm lifespan is challenging. An alternative model system is Drosophila, which are cultured with anti-fungal agents but not kept axenically, and host a bona fide microbiome of up to twenty different bacterial species (Ren et al., 2007). Unlike worms, the effect of associated bacteria on Drosophila lifespan is less clear, and may depend upon the precise composition of the microbiota population. Early reports suggested culturing Drosophila axenically shortened lifespan, with the presence of bacteria within the first week of adult life slowing aging (Brummel et al., 2004). However, follow up studies observed no effect of microbiota on aging (Ren et al., 2007) and such variation highlights the need to consider the holobiome when examining genetic modifiers of aging. Both groups performed lifespans using the same Canton-S strain, yet the composition of the microbiome associated with those strains differed, likely due to differences in husbandry (Ren et al., 2007). One explanation is that the Brummel study housed Drosophila with specific-symbiotic microbiota, which likely benefited them, while the Ren study had only neutral commensal microbes. The complexity of the microbiotic component therefore should not be ignored when interpreting differences in data between laboratories studying the effects of single genes on aging.
Bacteria can also affect the capacity of flies to sense nutrients (Storelli et al., 2011). Storelli et al. compared growth of Drosophila larvae on nutrient rich versus nutrient-poor diets, with and without microbiota. Bacteria had no effect on the growth of larvae replete with nutrients. However, the growth retardation of larvae on nutrient-poor food was exaggerated in the absence of bacteria. The ability of microbes to promote growth in low nutrient conditions is due to one specific species, Lactobacillus plantarum, which can promote growth to adulthood of larvae given no dietary yeast, a condition that usually arrests development. L. plantarum therefore overrides the host’s ability to match growth to nutrient availability, and does so via up-regulation of the TOR pathway. Flies overexpressing the inhibitor of TOR Complex 1 are resistant to the effects of L. plantarum on growth (Storelli et al., 2011). Given that suppression of TOR is known to promote longevity across species (Kenyon, 2010), it will be of great interest to determine if L. plantarum might regulate aging similar to its effects on growth (Fig 1C). Whether microbiome composition affects the capacity of the host to utilize nutrients has implications for the effects of diet and DR on aging. Transplanting microbiota from genetically obese ob/ob mice into germ free lean C57BL/6J mice induces obesity in the recipients compared to controls despite similar caloric intake (Turnbaugh et al., 2006), suggesting their ability to glean nutrients is affected by the microbes. How this type of interaction might affect lifespan extension via DR has yet to be explored – might transplanting a ‘dietary restricted microbiome’ into ad. libitum fed mice promote healthy aging?
The presence of microbiota at the interface of the gut epithelium also has systemic effects on host aging. In Drosophila, maintaining balance between intestinal stem cell (ISC) quiescence and proliferation is critical for intestinal function and organismal longevity. Tipping the balance towards quiescence limits intestinal rejuvenation, while ISC over-proliferation causes epithelial dysplasia (Jasper and Jones, 2010). As flies age, hyperproliferation of ISCs leads to intestinal dysfunction, but this does not occur in animals reared axenically (Guo and Jasper, 2013). Aging induces activation of intestinal genes regulated by the single fly FOXO transcription factor. Specific FOXO inhibition in intestinal epithelial cells (ECs) prevents the effects of both age and microbiota on ISC hyperproliferation (Guo and Jasper, 2013). The peptidoglycan recognition protein ‘PGRP-SC2’ is a FOXO target that reduces with age and the beneficial effects of FOXO knockdown in ECs is not seen in flies with reduced PGRP-S2. Finally, PGRP-SC2 overexpression in Drosophila ECs significantly increases lifespan dependent upon the presence of commensal bacteria in the gut. Together, this highlights how host and bacteria interactions extend beyond intestinal homeostasis to influence systemic health and longevity.
Microbial Species-specific Effects on Host Aging
Beyond the presence or absence of bacteria, specific bacterial species can have unique effects on host physiology. Worms fed Comamonas DA1877- a bacterial species found in their natural soil habitat - have faster development time, reduced fecundity and shorter lifespans compared to those fed the standard laboratory E. coli OP50 diet. A comparison of gene expression on the two diets, led to the identification of acdh-1 as being specifically induced by OP50 but not DA1877. MacNeil et al. developed an acdh-1-promoter-based in vivo fluorescent reporter as a ‘diet’ sensor. Interestingly worms are acutely sensitive to signals emanating from DA1877; diluting OP50 with only 0.1% DA1877 still suppressed the reporter and induced similar transcriptional responses to DA1877 alone. (MacNeil et al., 2013). Watson et al. performed forward and reverse genetic screens for genes affecting the GFP sensor, identifying a complex mitochondrial metabolic and nuclear gene regulatory network. Hence DA1877 can induce whole-scale metabolic responses in the worm to modulate development and aging (Watson et al., 2013) (Fig 1D). Several enzymes identified in the screens have human orthologs that when mutated lead to inborn errors of metabolism. If gut microbes can re-model metabolic networks in humans hosts as they do in C. elegans, might pre-biotics offer candidate therapies to combat metabolic defects?
The effects of single gene mutations on lifespan can also depend upon the presence of specific bacteria. TOR is found in two complexes and in C. elegans, mutations in the TOR Complex 2 specific factor Rictor have bacterial-specific effects on longevity. Rictor mutants are short-lived when cultured on standard B-derived OP50 bacteria, yet when grown on the K12-derived E. coli HT115 they are long-lived by 76 % (Soukas et al., 2009). In contrast, worms harboring mutations in the aldehyde dehydrogenase gene alh-6, are short-lived on OP50 yet normal-lived on HT115 (Pang and Curran, 2014). Both of these effects were assigned to nutritional compositions of the bacteria, yet alternative explanations involving differential metabolites or signals produced in the alternate E. coli strains have not been fully explored. Although wildtype worms do not have markedly different lifespans on B versus K12-derived bacteria, they do have different lipid profiles, which reflect the different lipid compositions of the bacteria (Brooks et al., 2009). This effect on worm lipid composition is abolished in animals lacking the peptide transporter PEPT-1 (Brooks et al., 2009). Therefore a peptide signal might at least in part be mediating the effects of bacteria on worm physiology beyond nutritional differences. Several whole-genome RNAi screens in C. elegans have successfully identified key longevity targets, yet all have been performed using HT115. Given emerging data on worm/microbe interactions, perhaps using a single, uniform microbial background is a limited strategy, and the search for longevity genes should be extended on alternative microbial backgrounds.
Outlook
Collectively these studies represent the beginning of the exploration into how genomes within metaorganisms interact to influence host longevity. Although it is too soon to know how invertebrate results will translate to mammalian aging, they highlight a need to consider the ‘holobiome’ when thinking about the impact of single gene manipulations on longevity – context is key. Anecdotal evidence for this perhaps already exists for mammalian aging. Snell Dwarf Mice, deficient in thyroid hormone, are long-lived by up to 50%, yet decades earlier, these mice were touted as a model of rapid aging and progeria (Flurkey et al., 2002). The difference did not lie in the mice, but in their environment. As husbandry facilities improved, a once progeric mouse became a long-lived hope. Although husbandry factors other than microbes may have mediated this transition, it is a striking reminder of how environmental context can drastically alter the effect of a single mutation on aging.
Aging research aims to find single interventions with beneficial effects on multiple age-related disorders, by impacting the plasticity of the aging process. As humans age the composition of our microbiome changes (Yatsunenko et al., 2012). If microbiome dynamics severely alter the effects of single gene interventions, might we be targeting the wrong genome in the metaorganism? Perhaps alongside focusing on the genetic regulation of healthy aging, we should pay equal attention to pre-biotic regulation of longevity as an alternate means to the same end.
Acknowledgments
We apologize to colleagues whose work was not cited due to space limitations. W.M is funded by the Ellison Medical Foundation and the NIH/NIA 1R01AG044346.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brooks KK, Liang B, Watts JL. PLoS ONE. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummel T, Ching A, Seroude L, Simon AF, Benzer S. Proc Natl Acad Sci USA. 2004;101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cochemé HM, Noori T, Weinkove D, Schuster E, Greene NDE, Gems D. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Harrison DE. Mech Ageing Dev. 2002;123:121–130. doi: 10.1016/s0047-6374(01)00339-6. [DOI] [PubMed] [Google Scholar]
- Guo L, Jasper H. Cell. 2013 In Press. [Google Scholar]
- Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E. Cell. 2013;152:818–830. doi: 10.1016/j.cell.2012.12.043. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. Exp Gerontol. 2002;37:1371–1378. doi: 10.1016/s0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Jasper H, Jones DL. Cell Metab. 2010;12:561–565. doi: 10.1016/j.cmet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang X, Wang HD, Wu J, Ren J, Meng L, Wu Q, Dong H, Wu J, Kao TY, et al. Nat Commun. 2012;3:1073. doi: 10.1038/ncomms2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil LT, Watson E, Arda HE, Zhu LJ, Walhout AJM. Cell. 2013;153:240–252. doi: 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, et al. Nat Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Pang S, Curran SP. Cell Metab. 2014 doi: 10.1016/j.cmet.2013.12.005. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Webster P, Finkel SE, Tower J. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Dow JM. Microbiology (Reading, Engl) 2008;154:1845–1858. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Virk B, Correia G, Dixon DP, Feyst I, Jia J, Oberleitner N, Briggs Z, Hodge E, Edwards R, Ward J, et al. BMC Biol. 2012;10:67. doi: 10.1186/1741-7007-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Arda HE, Zhu LJ, Walhout AJM. Cell. 2013;153:253–266. doi: 10.1016/j.cell.2013.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, et al. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Hou A. ISRN Microbiol. 2013;2013:356451. doi: 10.1155/2013/356451. [DOI] [PMC free article] [PubMed] [Google Scholar]