Abstract
Co-culture of multiple cell types within a single device enables the study of paracrine signaling events. However, extracting gene expression endpoints from co-culture experiments is laborious, due in part to pre-PCR processing of the sample (i.e., post-culture cell sorting, nucleic acid purification). Also, significant loss of nucleic acid may occur during these steps, especially with microfluidic cell culture where lysate volumes are small and difficult to access. Here, we describe an integrated platform for performing microfluidic cell culture and extraction of mRNA for gene expression analysis. This platform was able to recover 30-fold more mRNA than a similar, non-integrated system. Additionally, using a breast cancer / bone marrow stroma co-culture, we recapitulated stromal-dependent, estrogen-independent growth of the breast cancer cells, coincident with transcriptional changes. We anticipate that this platform will be used for streamlined analysis of paracrine signaling events as well as for screening potential drugs and/or patient samples.
INTRODUCTION
The detection and analysis of nucleic acid (NA) is a ubiquitous and critical process in many of the biosciences. While quantitative real-time PCR (qPCR) or reverse transcription qPCR (RT-qPCR) is often the endpoint of such protocols, the accuracy of the PCR readout depends not only on the PCR reaction itself, but also on an entire process originating with living cells and dependent on the quality and quantity of nucleic acid isolated. For cultured cells, this process includes the culture, cell lysis, NA extraction, NA purification, and qPCR or RT-qPCR. While much research has focused on streamlining and increasing throughput of the PCR process1,2, advances to the remaining processes, particularly NA extraction and purification, has lagged behind. In many labs, while the upstream sample preparation has become a potential bottleneck3, limiting the number of samples and replicates that can feasibly be performed by the lab.
Recently, microfluidics researchers have begun to take a holistic approach to NA analysis, designing integrated systems that link NA purification and PCR on a single chip. In these chips, nucleic acids typically are reversibly adsorbed to either functionalized surfaces4–6 or immobilized paramagnetic beads7,8, while a washing buffer rinses away other materials. However, each of these systems requires the “experiment” (culture and treatment of cells) to be performed on a separate chip or culture environment (e.g., tissue culture plate or flask). Following the experiment, whole or lysed cells are isolated and loaded into the microfluidic device. This transfer, which is often performed manually, impedes throughput and potentially decreases nucleic acid recovery due to difficulty pipetting a viscous lysate and/or operator variability, particularly when working with small cell numbers.
Common NA analyses (e.g., PCR, RT-PCR, chromatin immunoprecipitation (ChIP), sequencing, SNP analysis, epigenetic analysis) are often performed in culture systems containing a single cell line or clonal population (e.g., microtiter plates, tissue culture dishes) due to their operational ease and straightforward data analyses. However, the importance of complex multicellular environments is increasingly evident. While co-culture systems (e.g., transwells, Boyden chambers, conditioned media experiments) do not completely recapitulate the cellular microenvironment, they have enabled the study of multi-cell (paracrine) interactions. Previous work by Domenech et al. has demonstrated that by reducing the media volume per cell, the effects of soluble factor signaling are amplified. It is hypothesized that this increased signal is due to the increased concentration of soluble factors associated with reduced media volume per cell. Thus, signaling events will often produce a more measureable response (e.g., a higher fold induction of gene expression) in microchannels, relative to macroscale culture systems9. Microfluidic co-culture systems have recently been employed9–12 to study the paracrine interactions between cancer cells and other cell types present in the metastatic microenvironment in a high throughput manner. NA analyses would be of great utility in such experiments, permitting investigation of underlying transcriptional mechanisms behind phenotypic observations (e.g., differentiation, proliferation, apoptosis). Unfortunately, the complexity of the NA analysis workflow increases significantly when multiple cell types are cultured in one device. The increased number of conditions and variables typically introduced in co-culture experiments (e.g., different cell types, different cell type ratios) further compounds this problem, particularly when many different small populations of cells need to be analyzed (as with primary cultures or patient samples).
In this manuscript, we integrate the “front end” of the NA analysis process flow by linking cell culture, lysis, and NA extraction / purification on a single chip for both single cell-type cultures (termed “mono-culture”) and two cell-type cultures (termed “co-culture”). First, by comparing integrated and non-integrated mono-culture/ NA extraction devices we showed that the integrated system displays operational advantages. Next, we transitioned to a co-culture version that enabled study of paracrine signaling phenomena. These devices combine two easy-to-use technologies pioneered by our lab: “passive pumping” - based microfluidics13,14 and Immiscible Phase Filtration Assisted by Surface Tension (IFAST)15–20. Passive pumping leverages surface tension to “pump” liquids through a microchannel in a passive manner that does not require any tubing, valves, or mechanical pumps – only a micropipette. IFAST provides one-step isolation of mRNA by capturing this analyte on paramagnetic particles (PMPs), and then drawing them through a barrier of oil to separate the mRNA from the bulk lysate. A previous publication by our group15 has compared the mRNA extraction performance of IFAST to “gold standard” kits, including the Ambion MagMAX Kit and the Qiagen RNeasy spin column on metrics of mRNA recovery and mRNA purity, concluding that IFAST performance was equivalent to these commercial kits. Furthermore, similar results were obtained for DNA extraction when comparing IFAST to commercially available kits (ChargeSwitch gDNA Mini Bacterial Kit, Invitrogen)17. The combination of the two technologies results is an arrayable microfluidic platform that is operated using only a pipette and a magnet. The resulting extracted mRNA can then be analyzed with multiple high-throughput, multi-gene PCR systems (e.g., RT2 Profiler PCR Array, SA Biosciences; 96.96 Dynamic Array, Fluidigm) and multiplexed PCR reactions that are already commercially available. We envision that our devices will be employed upstream of existing PCR-based technologies, alleviating the bottleneck caused by excess throughput at the PCR stage of the analysis process flow and inadequate throughput at the NA purification stage.
In our initial IFAST paper15 we analyzed the level of mRNA remaining in the sample after IFAST extraction and found that very little was left behind, suggesting a near 100% extraction. However, this experiment was evaluating the IFAST process in isolation. In this manuscript, we wanted to take a more holistic approach – looking at the performance of the entire, integrated workflow from culture to RT-qPCR. A common concern regarding gene expression analysis in microchannel culture systems is that there will not be enough mRNA to measure the expression levels of all desired genes. Therefore, we wanted to use RT-qPCR as our primary endpoint and demonstrate that we could obtain better results at a complete process level, compared to a non-integrated system.
EXPERIMENTAL
To examine the operational advantages of integrating a microfluidic culture NA system we utilized a mono-culture integrated device that consists of a cell culture microchannel attached to an IFAST component. This device was fabricated from PDMS using traditional soft lithography and bonded to a glass coverslip or cyclic olefin copolymer (COC) bottom using oxygen plasma to activate the PDMS surface. Devices were heated to 100° C for 1 hour to restore hydrophobicity, and then the culture surface was coated through exposure to 0.1% gelatin in PBS for 20 minutes to facilitate cell attachment. After removal of the excess gelatin solution, the culture channel was filled with cell culture medium devoid of steroids (charcoal-stripped, phenol-free DMEM) and then 5 μl breast cancer cell suspension (MCF-7 cells, 1 million cells per mL) was added to the channel via passive pumping, a previously-characterized13, simple method for manipulating liquids in microchannels. A microfluidic constriction between the culture region and the oil barrier region of the IFAST component prevents cell media or suspension from exiting the culture region such that culture occurs with the IFAST region of the chip empty.
Following the culture experiment, the IFAST section of the device was filled by adding 8.5 μL of mRNA elution buffer (10 mM Tris-HCl) to the output well and 8.5 μL of oil (either FC-40 oil, 3M Corp. or olive oil, Unilever Corp.) to the oil barrier well. Five μL of RIPA lysis buffer (Millipore) containing 2.5 mg/mL oligo-dT paramagnetic particles (PMPs; from Invitrogen mRNA Direct Kit) was passively pumped into the culture region and incubated for 5 minutes at room temperature to allow lysis of the cells and PMP capture of the mRNA. Next, a magnet (B444-N52, K&J Magnetics) was used to draw the PMP-captured mRNA through the oil barrier and into the output well containing mRNA elution buffer (Figure 1A). This single traverse of the oil barrier was sufficient to separate mRNA from unbound lysate components, including PCR inhibitors, without additional washing steps (see Berry, 2011a for more details of this process). Arrays of devices (Figure 1B) were operated in parallel using a multi-channel pipette and an array of magnets or a long bar magnet (such as BX041-N52, K&J Magnetics).
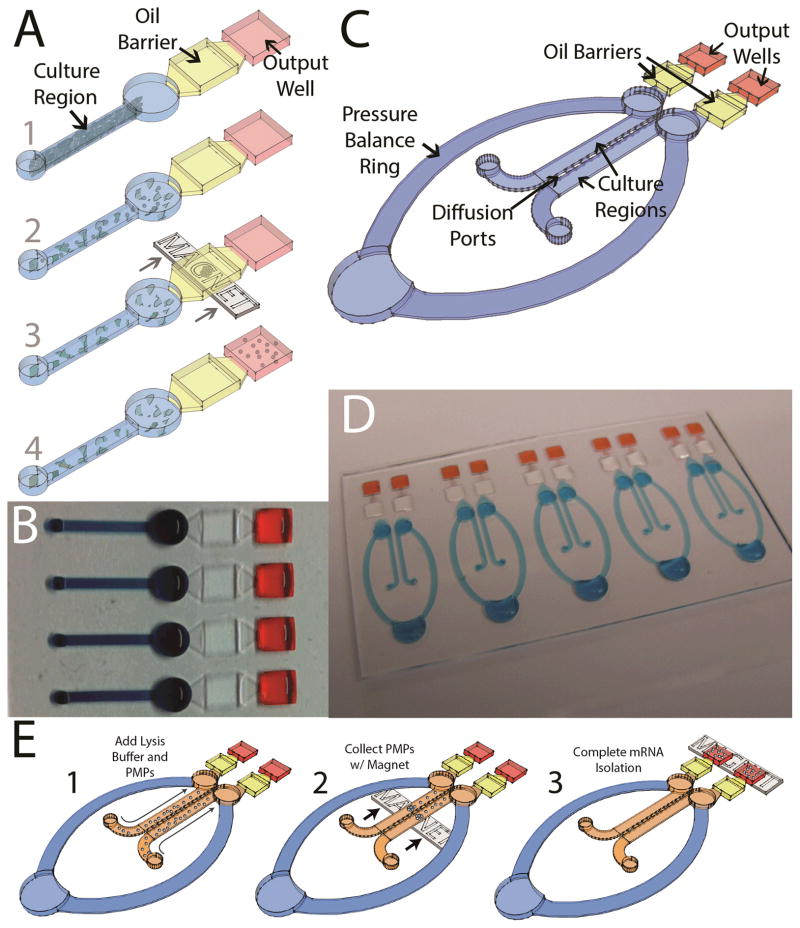
Figure 1.
A) Operation of the integrated mono-culture device. 1) Cells are loaded via passive pumping into the microchannel culture region; 2) Lysis buffer containing PMPs functionalized to capture mRNA is added to the channel via passive pumping; 3) A magnet is used to draw the PMP-captured mRNA across an oil barrier; 4) The mRNA is eluted in the output well and effectively isolated from the remainder of the lysate by the immiscible oil phase. B) Arrays of devices can be operated in parallel. C) Schematic of the integrated co-culture device. D) An array of 5 co-culture devices. E) Schematic showing operation of the co-culture device.
To compare integrated versus non-integrated systems, an equal number of cells were seeded into the mono-culture cell culture microchannel devices without an attached NA extraction component. Following culture, cell lysates were collected and transferred via pipette to a standalone IFAST device for mRNA purification. mRNA from the integrated and non-integrated systems were analyzed for the expression level of a housekeeping gene (large ribosomal protein, RPLP0) using RT-PCR (iScript cDNA Synthesis Kit was used for RT, iQ SYBR Green Supermix was used for PCR, and the reaction was performed on MyIQ Thermal Cycler, all items from Bio-Rad; PCR primer sequences and run conditions are given in Table S-1 in the Supplemental Information). In addition, to demonstrate that a known change in gene expression could be induced in cells cultured in the integrated device, estrogen-sensitive MCF-7 cells were treated with either 100 nM 17β-estradiol (E2; Steraloids, Inc.) or ethanol (EtOH) vehicle control, and expression of an E2-responsive gene21,22, estrogen receptor alpha (ERα), was measured. Gene expression levels were calculated by the ΔCT method, using RPLP0 as the control gene due to its similar level of expression compared to the other genes measured in this study and previously-reported stability in breast cancer cells23. Fold changes in expression levels were calculated using the ΔΔCT method.
The influence of the microenvironment around a cancer cell or tumor is an emergent theme in cancer research, particularly with regards to the study of metastasis. However, this interaction is difficult to study in vitro with current tools since multiple cell types must be included in the culture system. To address this problem as well as the problem of handling-induced variance (which may be compounded in a multi-cell type system), we modified our integrated mono-culture platform to include a second cell culture region. This new co-culture platform, shown in Figure 1C and 1D, consists of two parallel culture microchannels connected by twelve “diffusion ports” (500 μm long, 440 μm wide, and 10–15 μm high). These diffusion ports enable the free exchange of soluble signaling molecules (e.g., cytokines) between the cell types while maintaining sufficient separation to enable the seeding or lysis of cells in one channel without disturbing the cells in the adjacent channel. Each microchannel also has an attached independent IFAST component such that NA from each side can be collected independently without cross contamination (Figure 1E).
A large ring connecting the two culture regions was added to prevent passive pumping of liquid (cell suspension or lysis buffer) through the diffusion ports, by balancing the pressure between the two microchannels (Figure 2A). If the pressure in one side is disturbed by the addition or removal of liquid, flow will occur around the ring to balance the pressure rather than through the diffusion ports due to the higher fluidic resistance of the small diffusion ports. In accordance with the physics of passive pumping13,14, flow will progress toward the largest droplet in the system, which is located at the base of the pressure balance ring, thus ensuring that liquid from one side of the device does not flow around the ring to contaminate the other side. The pressure balance ring serves a secondary purpose as a large volume reservoir for media exchange. 20 μL of fresh media can be added to the ring with minimal disturbance to any cytokine concentrations or gradients established in the culture region (Figure 2B). Nutrients from this new media can gently diffuse into the culture regions while waste products diffuse into the ring (Figure 2C). Because only approximately half of the media is replaced at each exchange, this process is performed daily to ensure sufficient media availability and to prevent excessive buildup of waste. As with the integrated mono-culture device, arrays of the integrated co-culture device can be fabricated (Figure 1D).
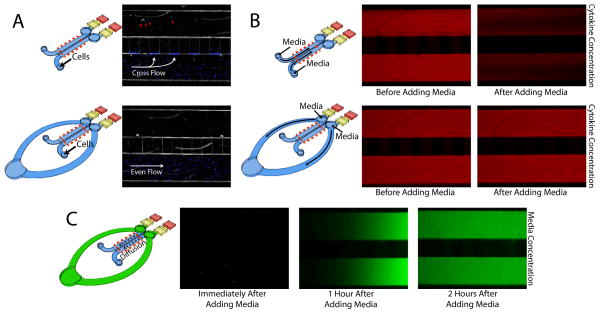
Figure 2.
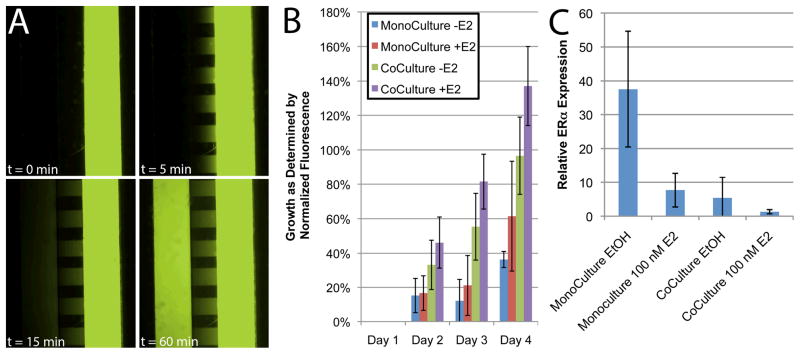
A) Cells added to a device with no ring will be drawn toward the diffusion ports due to an imbalance of pressure between the two cell culture channels. Many cells will become lodged in the shallow diffusion ports while others make it through the ports to contaminate the other side of the device (shown with red arrows). B) Without a ring, media must be added directly to the culture regions, thus immediately displacing any signaling molecules (modeled by a red dye). With the ring, media can be added to the system without disturbing the secreted factors. Both “after” pictures were taken within 1 minute of the media addition. C) Media added to the ring (modeled as a green dye) will diffuse across the culture region within a few hours of media change. Red dotted lines mark the section of each device that was imaged.
To demonstrate that cells from one culture region could be selectively lysed and purified without disturbing the connected culture region, MCF-7 cells constitutively expressing enhanced green fluorescent protein (eGFP) were loaded into one side of the device while bone marrow stromal cells (HS-5) were loaded into the other side. The co-culture of a breast cancer cell line (MCF-7) with bone marrow stroma (HS-5) has significant biological and clinical implications, as bone is one of the most common metastatic sites of breast cancer. As with the mono-culture device, culture regions were coated with gelatin and the device was filled with media prior to seeding. Following a 24-hour incubation for cell adhesion, each side was lysed with RIPA buffer and the mRNA was purified on-chip as described with the mono-culture system. A fluorescent microscope (Olympus IX-70) was used to directly observe the lysis event. Each side of the co-culture was lysed and extracted individually, enabling us to visibly confirm that no PMPs traveled through the diffusion ports during this step. RT-PCR was used to quantify markers for both cell types (eGFP for the fluorescent MCF-7 cells and vimentin, a stromal-expressed gene, for the HS-5 cells) from mRNA purified from both sides. It was confirmed that the MCF-7 cells did not express a measureable level of vimentin and the HS-5 cell did not express eGFP.
To investigate the phenotypic effects of culturing breast cancer cells in the context of bone marrow stromal microenvironment, MCF-7 cells and HS-5 cells were co-cultured in the integrated device for 4 days. Imaging of the co-culture 1 day after seeding (termed “Day 1”) indicated that cells were evenly distributed in the cell culture regions of the device without entering the diffusion ports. MCF-7 cells were cultured under four conditions: with and without HS-5 stroma (i.e., MCF-7 cells in one microchannel and HS-5 cell in the other vs. MCF-7 cells in both culture microchannels) and with and without daily treatment with 100 nM E2 (provided through the pressure balance ring). Experiments without E2 were treated with EtOH as a vehicle control.
To quantify the phenotypic changes (e.g., morphology, proliferation) of the MCF-7 cells in the presence of HS-5 cells, images of the cultured cells were analyzed using ImageJ software. We have previously reported20 that MCF-7 cell number correlates with eGFP signal intensity for MCF-7 cells transfected to stably express eGFP. Thus, to quantify MCF-7 cell proliferation, eGFP signal intensity was measured each day. Measurements included both the MCF-7 side of the co-culture (or one side of the devices with MCF-7 cells in both sides) and the diffusion port region to ensure cells that migrated out of the channel region were counted. Proliferation data was normalized to the signal on Day 1 to account for any loading variations. Measurements from both gene expression data and image analysis were compared using a two-tailed Student’s t-test.
RESULTS AND DISCUSSION
RT-PCR measurements of a housekeeping gene (RPLP0) were significantly increased in the integrated mono-culture device relative to the non-integrated system (p < 0.01, Figure 3A) as indicated by earlier amplification of the NA from the integrated system. Specifically, the threshold cycle decreased from 31.3 ± 2.7 (non-integrated) to 26.3 ± 1.7 (integrated), corresponding to a 32-fold increase in gene expression signal and a reduction in threshold cycle coefficient of variation from 8.6% (non-integrated) to 6.5% (integrated). This data suggests that substantial loss of mRNA occurs in the non-integrated system, presumably during transfer of the viscous lysate between the culture platform and the NA purification device. Importantly, by integrating the two components, we were able to limit this loss and recover substantially more mRNA. As expected22, treating MCF-7 cells in the integrated device with E2 resulted in a significant decrease (p < 0.01) in the expression of ERα (Figure 3B), demonstrating activation of the estrogen receptor within microchannel-cultured cells.
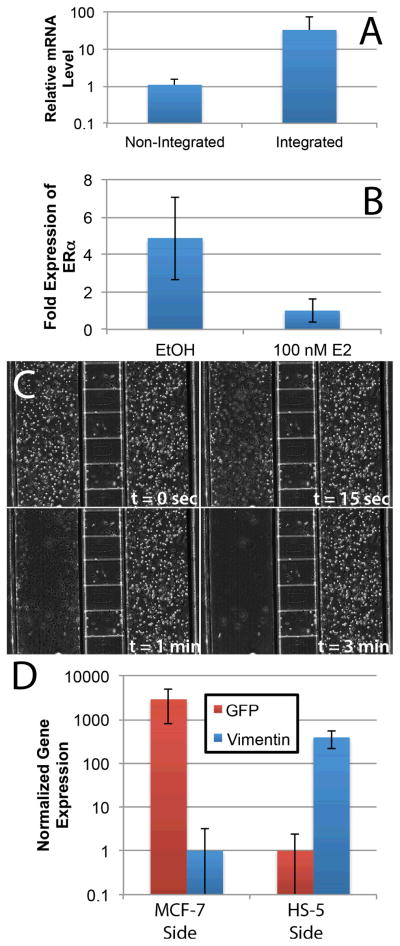
Figure 3.
A) RT-PCR of RPLP0 for mRNA isolated using non-integrated (microchannels and IFAST on two separate chips) and integrated (microchannels with IFAST integration, as in Figure 1A) configurations shows increased recovery of mRNA with the integrated device (n = 25). B) Down-regulation of ERα is observed in the presence of 100 nM E2 in the integrated device. This response is well known and demonstrates that expected transcriptional changes are induced within the integrated culture platform. C) The selective lysis of HS-5 cells from one side of the device with minimal effect on the other side. Note – diffusion ports can be seen as the horizontal conduits connecting the two culture regions. D) Quantification of mRNA collected from each side of integrated co-culture devices with MCF-7 cells on one side and HS-5 cells on the other. The data indicates that each side can be independently lysed with minimal cross-contamination as indicated by the low levels of vimentin detection in the MCF-7 side and GFP in the HS-5 side. Error bars represent standard deviation in all graphs.
Upon transitioning to the MCF-7 / HS-5 co-culture system, microscopy (Figure 3C) suggested that one side could be completely lysed with minimal effect on the other. This result was corroborated by RT-PCR data collected after 1 day of co-culture, which demonstrated that the mRNA from the MCF-7 channel of the device contained an average of 3,000-fold more eGFP than vimentin while the HS-5 channel of the device contained 400-fold more vimentin than eGFP (Figure 3D). This result confirms that mRNA can be independently isolated from each side of the reaction with minimal cross-contamination. However, in some studies, this level of cross-contamination (~0.1%) may be significant (e.g., detection of a expression change in a cell type that expresses a low level of the target gene when the other cell type is a high expressor of the same gene). Therefore, we recommend that users measure expression of a target gene in both sides of the device to confirm that a perceived up-regulation during co-culture is not the result of cross-contamination from a very high expressor of the target gene in the other culture region.
Next, MCF-7 cells were co-cultured with HS-5 cells in the absence and presence of E2. Co-culture devices loaded with MCF-7 cells in both sides (termed MCF-7 mono-culture) were used as controls. After four days of culture, differences were observed in each of the four culture conditions (−/+ E2, mono-culture and co-culture). When HS-5 cells were added to one side of the co-culture device, MCF-7 cells were observed in the diffusion port regions on Days 3 and 4 (Figure 4). Based on this observation, mRNA extraction was always performed on Day 2 to prevent cross-contamination. MCF-7 cells in the absence of both stroma and E2 proliferated very slowly. The addition of 100 nM E2 to the control MCF-7 cultures resulted in increased cell proliferation. MCF-7 cells in devices seeded with both cell types in the absence of E2 showed significantly increased proliferation relative to the condition without stroma or E2 (p < 0.05 at Day 4). When E2 was added to co-culture cells, MCF-7 proliferation increased even further and was significantly higher than both control conditions (p < 0.05) (Figure 5B).
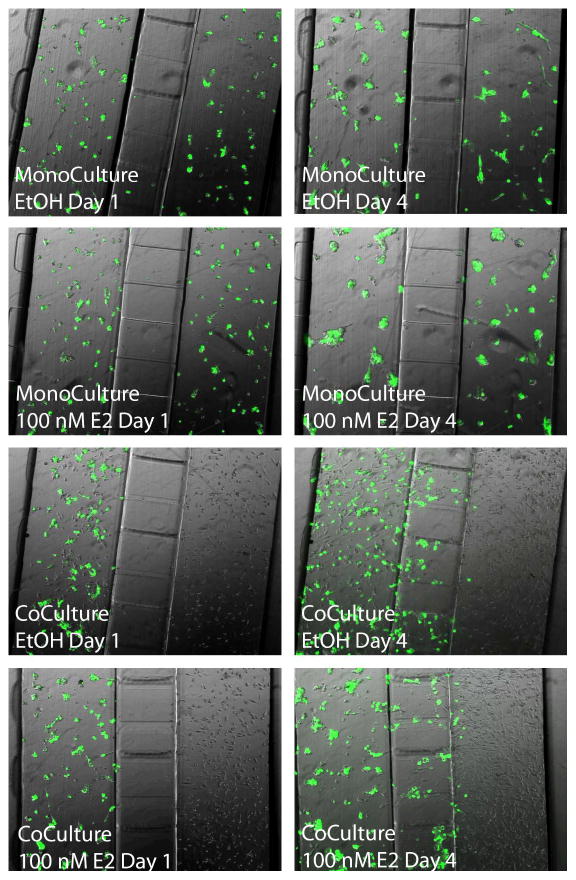
Figure 4.
Images of culture experiments in the integrated co-culture device at Day 1 and Day 4. Four experimental conditions were tested: (Top) MCF-7 cells in both sides of the device in the absence of E2 (EtOH vehicle only); (Second from top) MCF-7 cells in both sides with the daily addition of 100 nM E2; (Second from bottom) MCF-7 cells co-cultured with HS-5 cells in the absence of E2 (EtOH vehicle only); (Bottom) MCF-7 cells co-cultured with HS-5 cells in the presence of 100 nM E2. The experiment was repeated with both wild-type MCF-7 and MCF-7 cells stably transfected to express eGFP to aid in the distinction between cell types. No morphological differences were observed between these two MCF-7 cell lines.
Figure 5.
A) Fluorescein dye was loaded into one side of the integrated co-culture device to visualize diffusion through the diffusion ports. B) Growth of MCF-7 cells in the co-culture device as measured by fluorescence. While the addition of E2 slightly increased growth, the inclusion of HS-5 cells in the other co-culture channel increased proliferation significantly. C) Treatment with 100 nM E2 or co-culture with HS-5 cells caused significant down-regulation of ERα in MCF-7 cells. This down-regulation was more pronounced when HS-5 co-culture was combined with E2 treatment. The mRNA from this experiment was lysed on-chip and extracted using the integrated IFAST component. Error bars represent standard deviation in all graphs.
We hypothesize that the proliferative changes in the MCF-7 cells are the result of exchange of signaling molecules with the HS-5 cells. Such secreted factors will diffuse through the diffusion ports and induce transcriptional changes in the MCF-7cells. This diffusive effect is visualized by the spread of fluorescein dye (1 mg/mL) from one side of the co-culture device to the other over the course of one hour (Figure 5A). While the particular cytokine(s) remain unknown at this point, it is likely to diffuse at a somewhat slower rate due to a smaller concentration gradient and a larger molecular weight (in the case of a protein) in accordance with Fick’s laws of diffusion.
Using the IFAST component of the integrated co-culture device, mRNA was isolated from the MCF-7 cells cultured with and without HS-5 cells. Expression of eGFP and vimentin was measured from the extracted mRNA to confirm purity as in Figure 3D. In addition, expression of ERα was determined in each culture condition (−/+ E2, mono-culture and co-culture). As expected, the addition of E2 to the control MCF-7 cells caused a significant (p < 0.01) reduction in ERα expression level, a response consistent with mechanism of E2 action in this cell line. However, a similar decline in ERα level was observed in the absence of E2 when the MCF-7 cells were co-cultured with HS-5 cells. ERα declined even further with the combination of E2 and HS-5 co-culture (Figure 5C). This E2-independent modulation of the ERα gene by stromal cells has been previously reported10,24. The mechanism behind this change is not completely understood, but it is likely multifactorial as many extracellular factors have been identified that activate ERα25.
Taken together with the co-culture growth observations, the ERα expression data suggests that co-culture of MCF-7 cells with HS-5 cells induces a response similar to the pro-growth behavior induced by treatment with E2. This finding has clinical implications since many breast cancer therapies target the ERα pathway. Furthermore, many drugs that are effective against the primary tumor show limited effectiveness in metastatic tumor sites, including the bone marrow, potentially due to micro-environmental effects. The data presented in this manuscript corroborates this observation since the presence of a bone marrow stromal cell line enables estrogen-independent growth of the MCF-7 cell line. While further study is needed to clarify the mechanism of E2-independent growth of breast cancer cells in the presence of stroma, the data supports the conclusion that E2-independent growth can be recapitulated in vitro in the integrated co-culture device. The inclusion of HS-5 cells into the microenvironment of the MCF-7 cells has clear effects on their proliferative, migratory and transcriptional behavior. Some of these effects (proliferation and ERα transcription) seem to parallel responses seen with estrogen treatment, while others (migration and morphology) may be unrelated. However, the ability to recapitulate a stroma-dependent, E2-independent growth response in vitro in breast cancer cells facilitates future studies of this mechanism and other micro-environmental signaling phenomena as well as novel screens involving patient samples and potential therapeutics.
CONCLUSION
The characterization of the two platforms (the integrated mono-culture systems shown in Figure 1A and the integrated co-culture system in Figure 1C) demonstrates that mRNA can be extracted and purified directly from cultured cells on a single chip. Furthermore, the data in Figure 3B suggests that this process is more repeatable than conventional non-integrated culture platforms. In co-culture scenarios, mRNA can be extracted from on cell type on either side (or both) with minimal (~0.1%) cross-contamination. Also, due to the integrated, facile, and arrayable nature of this technology, it can be utilized as a “front end” for existing high-throughput qPCR or RT-PCR technologies, potentially alleviating bottlenecks in the NA process flow. Using this method, we have demonstrated a model system for breast cancer metastasis to bone, where bone marrow stromal cells induce loss of ERα expression and E2-independent growth in breast cancer epithelial cells. We anticipate that this platform will facilitate studies of a variety of other co-culture systems, thus enabling researchers to easily and rapidly measure gene expression changes caused by inter-cellular signaling via soluble factors.
Supplementary Material
Acknowledgments
Funded by a grant from the Bill & Melinda Gates Foundation through the Grand Challenges in Global Health initiative, a DOD PCRP Idea grant (W81XWH-09-1-0192), and an NIH NCI R33 grant (CA137673).
Footnotes
Conflict of Interest
S. M. Berry and D. J. Beebe are co-founders of Salus Discovery
References
- 1.Marcus JS, Anderson WF, Quake SR. Anal Chem. 2006;78:956–958. doi: 10.1021/ac0513865. [DOI] [PubMed] [Google Scholar]
- 2.Lounsbury JA, Karlsson A, Miranian DC, Cronk SM, Nelson DA, Li J, Haverstick DM, Kinnon P, Saul DJ, Landers JP. Lab Chip. 2013;13:1384–1393. doi: 10.1039/c3lc41326h. [DOI] [PubMed] [Google Scholar]
- 3.Zhang C, Xu J, Ma W, Zheng W. Biotechnol Adv. 2006;24:243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Nathaniel C, Cady SS. Sensors and Actuators B: Chemical. 2005:332–341. [Google Scholar]
- 5.Anderson RC, Su X, Bogdan GJ, Fenton J. Nucl Acids Res. 2000;28:e60–e60. doi: 10.1093/nar/28.12.e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witek MA, Hupert ML, Park DSW, Fears K, Murphy MC, Soper SA. Anal Chem. 2008;80:3483–3491. doi: 10.1021/ac8002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu RH, Yang J, Lenigk R, Bonanno J, Grodzinski P. Anal Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 8.Marcus JS, Anderson WF, Quake SR. Anal Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 9.Domenech M, Yu H, Warrick J, Badders NM, Meyvantsson I, Alexander CM, Beebe DJ. Integr Biol (Camb) 2009;1:267–274. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang JD, Berry SM, Powers GL, Beebe DJ, Alarid ET. Integrative Biology. doi: 10.1039/c3ib20265h. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su G, Sung KE, Beebe DJ, Friedl A. PLoS ONE. 2012;7:e46685. doi: 10.1371/journal.pone.0046685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung KE, Su X, Berthier E, Pehlke C, Friedl A, Beebe DJ. PLoS ONE. 2013;8:e76373. doi: 10.1371/journal.pone.0076373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker GM, Beebe DJ. Lab Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 14.Berthier E, Beebe DJ. Lab Chip. 2007;7:1475–1478. doi: 10.1039/b707637a. [DOI] [PubMed] [Google Scholar]
- 15.Berry SM, Alarid ET, Beebe DJ. Lab Chip. 2011;11:1747–1753. doi: 10.1039/c1lc00004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry SM, Regehr KJ, Casavant BP, Beebe DJ. J Lab Autom. 2013;18:206–211. doi: 10.1177/2211068212462023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strotman LN, Lin G, Berry SM, Johnson EA, Beebe DJ. Analyst. 2012;137:4023–4028. doi: 10.1039/c2an35506j. [DOI] [PubMed] [Google Scholar]
- 18.Berry SM, Maccoux LJ, Beebe DJ. Anal Chem. 2012;84:5518–5523. doi: 10.1021/ac300085m. [DOI] [PubMed] [Google Scholar]
- 19.Strotman L, O’Connell R, Casavant BP, Berry SM, Sperger JM, Lang JM, Beebe DJ. Anal Chem. 2013 doi: 10.1021/ac402162r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry SM, Strotman LN, Kueck JD, Alarid ET, Beebe DJ. Biomed Microdevices. 2011;13:1033–1042. doi: 10.1007/s10544-011-9573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Read LD, Greene GL, Katzenellenbogen BS. Mol Endocrinol. 1989;3:295–304. doi: 10.1210/mend-3-2-295. [DOI] [PubMed] [Google Scholar]
- 22.Ellison-Zelski SJ, Solodin NM, Alarid ET. Mol Cell Biol. 2009;29:4949–4958. doi: 10.1128/MCB.00383-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyng MB, Lænkholm AV, Pallisgaard N, Ditzel HJ. BMC Cancer. 2008;8:20. doi: 10.1186/1471-2407-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stossi F, Madak-Erdoğan Z, Katzenellenbogen BS. Oncogene. 2012;31:1825–1834. doi: 10.1038/onc.2011.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunone G, Briand PA, Miksicek RJ, Picard D. EMBO J. 1996;15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.