Abstract
The vitamin K-dependent carboxylase is an integral membrane protein which is required for the post-translational modification of a variety of vitamin K-dependent proteins. Previous studies have suggested carboxylase is a glycoprotein with N-linked glycosylation sites. In the present study, we identified the N-glycosylation sites of carboxylase by mass spectrometric peptide mapping analyses combined with site-directed mutagenesis. Our mass spectrometric results show that the N-linked glycosylation in carboxylase occurs at positions N459, N550, N605, and N627. Eliminating these glycosylation sites by changing asparagine to glutamine caused the mutant carboxylase to migrate faster in SDS-PAGE gel analyses, adding further evidence that these sites are glycosylated. In addition, the mutation studies identified N525, a site not recoverable by mass spectroscopy analysis, as a glycosylation site. Furthermore, the potential glycosylation site at N570 is glycosylated only if all the five natural glycosylation sites are simultaneously mutated. Removal of the oligosaccharides by glycosidase from wild-type carboxylase or by eliminating the functional glycosylation sites by site-directed mutagenesis did not affect either the carboxylation or epoxidation activity when the small pentapeptide FLEEL was used as substrate, suggesting that N-linked glycosylation is not required for the enzymatic function of carboxylase. In contrast, when site N570 and the five natural glycosylation sites were mutated simultaneously, the resulting carboxylase protein was degraded. Our results suggest that N-linked glycosylation is not essential for carboxylase enzymatic activity but it is important for protein folding and stability.
The vitamin K-dependent carboxylase, also known as gamma-glutamyl carboxylase (GGCX)1, is an integral membrane protein of the endoplasmic reticulum. It catalyzes the post-translational modification of specific glutamic acid residues of vitamin K-dependent proteins to γ-carboxyglutamic acid residues. This post-translational modification is critical for the biological functions of the vitamin K-dependent proteins involved in blood coagulation, bone metabolism, signal transduction, and cell proliferation (1-3). In addition to its vitamin K-dependent substrate, the carboxylation reaction, which occurs in the lumen of the ER (4, 5), requires the co-substrates carbon dioxide, oxygen, and vitamin K hydroquinone. During the process of carboxylation, the γ-proton of the glutamic acid is abstracted, followed by the addition of carbon dioxide (6-9). Concomitant with carboxylation, vitamin K hydroquinone is oxidized to vitamin K epoxide which must be converted back to vitamin K by the enzyme vitamin K epoxide reductase, thus completing the vitamin K cycle (1, 10). The formation of vitamin K epoxide during the carboxylation reaction has been called an epoxidation reaction (9, 11, 12). It was suggested that an oxygenated intermediate of vitamin K produced by GGCX activates the γ-carbon of a glutamate and then yields vitamin K epoxide (6, 13).
It has been reported that GGCX binds to lectin suggesting that it is a glycoprotein (14, 15). Moreover, Endoglycosidase H treatment of GGCX purified from bovine liver indicates that it is an N-linked glycoprotein (16). N-linked glycosylation occurs at an asparagine residue in the canonical sequence NXS/T (glycosylation sequon) where X can be any residue except proline (17, 18). It is the most prevalent co-translational modification that occurs in the lumen of the ER. N-linked glycosylation plays an important role in maintaining protein stability, modulating protein-protein interactions, and protein membrane targeting (19-21). In addition, alterations of the sugar chain of glycoproteins are associated with various diseases such as cancer (22-24).
In human GGCX, there are 9 potential N-linked glycosylation sequons. A study of the membrane topology of GGCX (25) demonstrated that all of the potential glycosylation sites are located in the ER lumen where the glycosylation enzymes occur. GGCX migrates as a 94 kDa protein in SDS-PAGE (26). This is about 6 kDa greater than expected based upon its predicted amino acid sequence (87.6 kDa) (27). Results of sedimentation equilibrium proposed that glycosylation of GGCX causes a molecular weight increase of 12.8 kDa (28). These results support the notion that GGCX is a glycoprotein with multiple sites being glycosylated. However, the number of potential glycosylation sites that are actually modified, as well as the importance of glycosylation on GGCX activity and stability, are still unknown. In this study, we used mass spectrometry combined with site-directed mutagenesis to identify the glycosylation sites in human GGCX. We also examined the effect of glycosylation on carboxylation and epoxidation activity.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were reagent grade. CHAPS, CNBr, and α-cyano-4-hydroxysuccinnamic acid were obtained from Sigma (St. Louis, MO). 18O water (Normalized 96.5% atom% 18O) was from Isotec (Milwaukee, WI). Vitamin K1(20) (10 mg/mL) was from Abbott Laboratories (Chicago, IL). Vitamin K1(25) was from GLsynthesis Inc. (Worcester, MA). NaH14CO3 (specific activity, 54 mCi/mmol) was from ICN Pharmaceuticals, Inc. (Costa Mesa, CA). Aprotinin, pepstatin A, trypsin (modified, sequencing grade), chymotrypsin (sequencing grade), and endoproteinase Glu-C (sequencing grade) were purchased from Roche Molecular Biochemicals (Indianapolis, IN). Pentapeptide FLEEL and protease inhibitor H-D-Phe-Pro-Arg chloromethylketone were from Bachem (King of Prussia, PA). 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine was from Avanti (Alabaster, AL). The propeptide of proFIX and the fluorescein-labeled consensus propeptide were chemically synthesized and purified by Chiron Mimotopes (Clayton, Victoria, Australia). Restriction enzymes and PNGase F were from New England Biolab (Beverly, MA). Bac-to-Bac baculovirus expression system, Bis-Tris NuPAGE gel, protein standards, and oligonucleotides were from Invitrogen Life Technologies (Carlsbad, CA). Pfu Turbo DNA polymerase was from Stratagene (La Jolla, CA). Anti-HPC4 antibody coupled Sepharose resin was kindly provided by Dr. Charles Esmon (Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA).
Expression and Purification of GGCX in Insect Cells
Wild-type human GGCX cDNA with a HPC4 tag (EDQVDPRLIDGK) at the carboxyl terminus (29) was engineered into the baculovirus expression vector pFastBac1 by BamH I and EcoR I sites. The resulting plasmid, pFastBac1-GGCX-HPC4, was transformed into E. coli. strain DH10Bac and the recombinant bacmid was screened by blue/white selection according to the manufacturer's instructions (Invitrogen). Recombinant baculovirus was obtained by Cellfectin induced infection of Sf9 cells and screened by GGCX activity assay of the cell lysate. Expression of GGCX was performed by infection of 2 × 106/mL Sf9 cells with the recombinant virus at a multiplicity of infection of 1. Cells were collected after 48 h of infection, and the expressed GGCX was purified by affinity chromatography using anti-HPC4 antibody-coupled Sepharose resin as previously described (29).
Limited Trypsinization and PNGase F Digestion of GGCX
Limited trypsin digestion of GGCX in the presence of propeptide was performed as previously described (16). Cleavage was accomplished on ice in the presence of 5 μM proFIX and the reaction was stopped by the addition of H-D-Phe-Pro-Arg chloromethylketone and aprotinin to a final concentration of 1.6 μM. To examine the N-glycosylation modification, intact and/or trypsinized GGCX were incubated with 100 units/mL PNGase F in 50mM sodium phosphate (pH 7.5) at 4°C for 16 h. PNGase F treated and non-treated samples were fractionated by 10% Bis-Tris NuPAGE and visualized by silver staining.
In-gel Protease Digestion and Deglycosylation of GGCX for Mass Spectrometry Analysis
Glycoproteins often yield poor peptide maps for mass spectrometry analysis because the oligosaccharides can effectively shield proteolytic cleavage sites (30). Therefore, with the exception of the experiment using 18O labeling, freshly purified GGCX was deglycosylated before being subjected to in-gel protease digestion (31). After deglycosylation, GGCX samples were fractionated by 10% SDS-PAGE and protein bands were visualized by Coomassie staining. The GGCX band was excised, and the gel pieces were treated as described previously (31). The dry gel pieces were re-hydrated by the addition of (10 ng/μL) protease (trypsin, Endoproteinase Glu-C, or chymotrypsin) in 25 mM NH4HCO3 solution and incubated on ice for 30 minutes. After re-hydration, the protease solution was removed and the gel pieces were covered with an overlay of 20 μL of 25 mM NH4HCO3 solution. The gel pieces remained immersed throughout the digestion. The protein was digested overnight at 30°C without agitation. Digestion supernatants were directly pooled for mass spectrometry analyses.
The combined tryptic and CNBr cleavage was performed as described previously (32). Briefly, after tryptic digestion, the gel pieces were dried directly in the SpeedVac. These dried gel pieces were washed and dehydrated by acetonitrile. CNBr cleavage was started by the addition of sufficient CNBr solution in 70% TFA and the reaction was carried out overnight in the dark at room temperature. After collection of the supernatant, the peptides were extracted twice by sonication for 5 minutes in 30 μL of 60% acetonitrile with 1% TFA. The overlay and extracts were pooled and dried in a SpeedVac. The sample was dissolved in 5 μL of 40% acetonitrile with 0.1% TFA for mass spectrometry analysis.
To further confirm the glycopeptides identified by MALDI-TOF MS, the glycosylation sites of GGCX were labeled by 18O through in-gel PNGase F deglycosylation as described previously (33). Briefly, affinity purified GGCX was applied directly to SDS-PAGE without PNGase F treatment and the Coomassie blue stained GGCX band was excised and treated as described above. The dry gel pieces were swollen in 50mM sodium phosphate (pH 7.5) with 100 units/mL PNGase F and 50% H 182O. The reaction was incubated overnight at 37°C. PNGase F was removed prior to proteolysis by washing the gel pieces with 0.1% SDS in 100 mM NH4HCO3 (four changes, 1 h per change). All washings were discarded and SDS was removed from the gel pieces by incubation with methanol/water/acetic acid (50:45:5, v:v:v) for 30 minutes followed by three washes with 50% acetonitrile in 100 mM NH4HCO3. The resulting gel plugs were dried in a SpeedVac. In-gel trypsin digestion of the 18O labeled protein samples was carried out as described above.
Identification of N-glycosylation Sites of GGCX by Mass Spectrometry
Protease digested samples were applied to MALDI-TOF MS and Q-TOF MS for identifying the N-glycosylation sites of human GGCX. For MALDI-TOF MS, 0.3 μL of the above protease digested sample and 0.3 μL of 10 mg/mL 1-cyano-4-hydroxysuccinnamic acid in 50% acetonitrile containing 0.1% TFA (v/v) were deposited on the target plate and air-dried. MALDI-TOF mass spectra were recorded with an ABI 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA) MALDI-TOF/TOF mass spectrometer. A frequency-tripled Nd:YAG laser ionized samples at a pulse frequency of 200 Hz with its power adjusted between 15 and 30 μJ, depending on the sample. Laboratory air was used as the collision gas and the collision cell vacuum pressure was 7~9 × 10–7 Torr. MALDI-TOF data from tryptic digests were calibrated with auto-proteolytic peaks (internal standards) and mass errors were less than 20 ppm. The mass range for TOF MS scan functions was set to m/z 500 to 4000. TOF MS/MS scan functions were calibrated externally against the fragments of either angiotensin I or adrenocorticotropic hormone fragment 18~39 depending upon the precursor mass. The mass accuracy of CID data was typically better than 50 ppm.
ESI analyses of proteolytic fragments were conducted using nanoelectrospray LC-MS/MS on a Q-TOF API-US instrumental platform (Waters Corporation). The PepMap dC18, 5 μm, 0.075 × 100 mm column (LCPackings Corporation) was used for peptide separation. On the Q-TOF, the intensity criteria of signal to noise for MS to MSMS switch is set to 10, whereas the threshold for MSMS to MS switch is set to above 3500 Counts/s. Ions in charge states of 2+, 3+, and 4+ detected in the survey scan were selected for MSMS.
Raw data files from Q-TOF instrument are processed with Mascot Distiller (Version 1.1.1.0, Matrix Science, London) without smoothing using charge states determined from the MS scan. The resulting centroid files were searched against NCBI nr database using Protein Prospector. The mass tolerance of the precursor peptide ion was fixed at 200 ppm whereas the mass tolerance for the MS/MS fragment ions was set to 0.5 Da.
Mutation of the N-glycosylation Sites in GGCX by Site-Directed Mutagenesis
To remove the N-glycosylation sites in GGCX, the asparagine (N) residue within the canonical glycosylation sequence NXS/T was replaced with a glutamine (Q) residue by site-directed mutagenesis using sequence overlap extension PCR. The vector pFastBac1-GGCX-HPC4 was used as the template. The four N-glycosylation sites identified by mass spectrometry were removed one by one by changing asparagine to glutamine. To verify whether asparagine residues at position 389, 525, and 570 are glycosylated in vivo, we changed each of these three asparagines to glutamine individually in the GGCX mutant GGCX605 that already had the four confirmed glycosylation sites removed. To determine whether asparagine 389 or 570 is glycosylated when all the five natural glycosylation sites were removed from GGCX, these two asparagines were changed to glutamine individually in the GGCX mutant GGCX525 that had all the five natural glycosylation sites removed. The resulting GGCX mutants are shown in Table 1. All the final constructs were fully sequenced by the Genome Analysis Facility of the University of North Carolina at Chapel Hill to ensure fidelity and proper introduction of the mutations. All of the GGCX mutants were expressed and affinity purified from Sf9 cells as described above.
Table 1.
GGCX mutants of asparagine to glutamine within the potential N-linked glycosylation sites
| GGCX mutants | Sites mutated |
|---|---|
| GGCX459 | N459Q |
| GGCX627 | N627Q, N459Q |
| GGCX550 | N550Q, N627Q, N459Q |
| GGCX605 | N605Q, N550Q, N627Q, N459Q |
| GGCX389 | N389Q, N605Q, N550Q, N627Q, N459Q |
| GGCX525 | N525Q, N605Q, N550Q, N627Q, N459Q |
| GGCX570 | N570Q, N605Q, N550Q, N627Q, N459Q |
| GGCX389/525 | N389Q, N525Q, N605Q, N550Q, N627Q, N459Q |
| GGCX570/525 | N570Q, N525Q, N605Q, N550Q, N627Q, N459Q |
GGCX Activity Assays
GGCX activity was determined by the incorporation of 14CO2 into the pentapeptide substrate FLEEL in the presence of propeptide. For assays of cell lysates, 10 μL of cell pellet was mixed with 105 μL of lysis buffer (final concentrations-0.5% CHAPS, 25 mM Tris-HCl (pH 7.5), 500 mM NaCl, 4 μM proFIX, and 1.25 mM FLEEL). Samples were placed on ice for 30 minutes with occasional vortexing. The carboxylation reaction was started by the addition of 10 μL of an ice-cold mix of NaH14CO3 (40 μCi/mL final concentration) and vitamin K hydroquinone (222 μM final concentration) to bring the volume to 125 μL. The reaction mix was immediately transferred to a 20 °C water bath and incubated for 30 minutes. The reactions were terminated by the addition of 1 mL 5% trichloroacetic acid, and the amount of 14CO2 incorporation was determined as described (29). The assays for purified GGCX were performed at a final concentration of 25 mM Tris-HCl (pH 7.5), 500 mM NaCl, 0.8 M (NH4)2SO4, 0.12% 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine, 0.28% CHAPS, 4 μM proFIX, and 1.25 mM FLEEL; the carboxylation reaction was carried out as above. The concentration of active GGCX was determined from the fraction of protein binding to the fluorescein-labeled consensus propeptide by fluorescence anisotropy as described previously (28).
Vitamin K Epoxidation Activity Assay
Vitamin K epoxidation activity was determined by quantitation of the vitamin K epoxide formed during the carboxylation of FLEEL as described above, except that unlabeled NaHCO3 was used. The reactions were terminated by the addition of 500μL isopropanol. The mixture was extracted with 500μL hexane containing 2.52 μM vitamin K1(25) as an internal standard. Vitamin K epoxide formation was quantitated by HPLC (34) on a C18 column (4.6×250 mm, Vydac, Hesperia, CA) with an isocratic elution. The mobile phase was acetonitrile:isopropanol:water (100:7:2, v:v:v), flow rate was 2.00 mL/min, and the UV detection wavelength was 248 nm.
RESULTS
N-glycosylation Sites of GGCX Are Located on the Carboxyl-terminal Tryptic Fragment
There are 9 potential glycosylation sites in GGCX. These sequons are at asparagine residues 159, 389, 459, 525, 550, 570, 605, 627, and 735. To re-confirm that human GGCX is a N-linked glycoprotein, affinity purified recombinant human GGCX was treated with PNGase F, an enzyme that removes carbohydrates from asparagine residues. Figure 1 demonstrates that PNGase F treated GGCX (lane 2) migrates faster than the untreated sample (lane 1) in SDS-PAGE analysis indicating that GGCX is an N-linked glycoprotein. Previous work from our laboratory showed that limited trypsin digestion of GGCX results in a disulfide-linked 30 kDa amino-terminal and 60 kDa carboxyl-terminal fragments (16) (Figure 1, lane 3). Figure 1 (lane 4) shows that only the 60 kDa carboxyl-terminal tryptic fragment is sensitive to PNGase F digestion indicating, in agreement with our previous report (16), that N-linked glycosylation occurs only on the 60 kDa carboxyl-terminal tryptic fragment of GGCX. Asparagine 159 is the only potential glycosylation site in the 30 kDa amino-terminal tryptic fragment and these results indicate that it is not glycosylated in vivo.
Figure 1. PNGase F treatment of intact and trypsin nicked GGCX.
A reducing NuPAGE silver-stained gel is depicted. Lane 1, intact wild-type GGCX; lane 2, intact wild-type GGCX digested by PNGase F; lane 3, trypsin nicked wild-type GGCX; lane 4, trypsin nicked wild-type GGCX treated by PNGase F; lane 5, PNGase F.
Identification of the N-linked Glycosylation Site in GGCX by Mass Spectrometry
Removal of the oligosaccharides from asparagines by PNGase F converts the asparagine (114 Da) to aspartate (115 Da). This 1-Da mass increase of the deglycosylated peptide relative to the corresponding nonglycosylated peptide has been widely used for identifying glycoproteins using MALDI-TOF MS (35-37). In this study, we used this 1-Da shift to identify the N-linked glycosylation sites in GGCX. Table 2 summarizes the proteolytic glycopeptides detected by mass spectrometry. Six glycopeptides containing glycosylation sites N459, N605, and N627 of human GGCX were recovered by MALDI-TOF based on the 1-Da mass shift (Table 2, top panel). Using Q-TOF MS analyses of proteolytically treated GGCX, we identified another glycosylation site at N550 (Table 2, bottom panel). Due to the hydrophobic characteristics of transmembrane peptides and the lack of appropriate protease sites to generate the appropriate size peptides for mass spectrometry analysis, we were unable to recover peptides containing the potential glycosylation sites N389, N525, and N570.
Table 2.
Peptide mass mapping of the potential N-glycosylation sites in GGCX by MALDI-TOF and Q-TOF mass spectrometry
| Measured m/z | Measured peptide mass | Expected peptide mass | Corresponding residues | Glycosylation site | Digestions method |
|---|---|---|---|---|---|
| 1821.9355a | 1820.9355 | 1819.9911 | 452-466 | N459 | Chymotrypsin |
| 2373.1978 | 2372.1978 | 2371.1562 | 458-476 | N459 | Trypsin |
| 2433.2903 | 2432.2903 | 2431.2561 | 625-647 | N627 | Trypsin, Trypsin/CNBr |
| 2617.3433 | 2616.3433 | 2615.3449 | 601-622 | N605 | Trypsin/CNBr |
| 2646.3074 | 2645.3074 | 2644.4038 | 618-641 | N627 | Chymotrypsin |
| 2690.4270 | 2689.4270 | 2688.3936 | 623-647 | N627 | Trypsin, Trypsin/CNBr |
| 694.84 (2)b | 1387.68 | 1386.68 | 547-559 | N550 | Endoproteinase Glu-C |
| 811.71 (3) | 2432.13 | 2431.26 | 625-647 | N627 | Trypsin, Trypsin/CNBr |
| 897.48 (3) | 2689.44 | 2688.39 | 623-647 | N627 | Trypsin, Trypsin/CNBr |
| 939.44 (1) | 938.44 | 937.44 | 602-609 | N605 | Chymotrypsin |
| 1217.06 (2) | 2432.12 | 2431.26 | 625-647 | N627 | Trypsin, Trypsin/CNBr |
Top panel: deglycosylated peptide detected by MALDI-TOF MS according to the 1-Da shift theory.
Bottom panel: deglycosylated peptide detected by Q-TOF MS according to the 1-Da shift theory. Numbers in parentheses indicate the charges of the corresponding peptides.
18O-labeling of the Asparagine Residue within the N-glycosylation Sequon
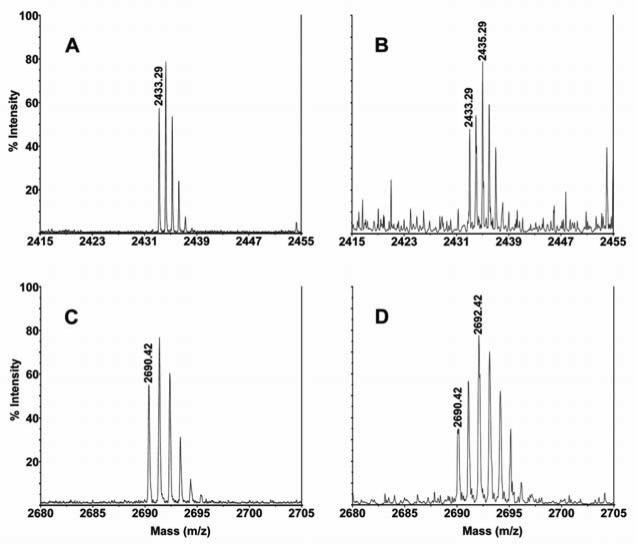
To further confirm the identity of the sites demonstrated above, we performed in-gel deglycosylation of GGCX by PNGase F in the presence of 50% 18O water as described in Experimental Procedures. When hydrolysis occurs, about 50% of the resulting aspartic acid residues will be labeled with 18O. This proteolytically derived deglycosylated peptide incorporating 18O can be recognized in MALDI-TOF mass spectrum as doublets peaks with a characteristic 2-Da spacing (33). Figure 2 shows the MALDI-TOF spectrum of 18O-labeled and non-labeled tryptic glycopeptides of GGCX at m/z 2433.29 and 2690.42 as listed in Table 2. Doublet peaks differing by 2-Da appear in the 18O-labeled sample (Figure 2 B and D) indicating that these two peptides are glycopeptides and site N627 is a functional glycosylation site in vivo. Using this 18O-labeling technology, all of the tryptic peptides previously identified as glycosylated were confirmed (Data not shown).
Figure 2. MALDI-TOF mass spectra of 18O-labeled and non-labeled deglycosylated peptides of GGCX.
Panel A, spectrum of deglycosylated tryptic peptide of GGCX at m/z 2433.29; Panel B, spectrum of 18O labeled deglycosylated tryptic peptide of GGCX at m/z 2433.29; Panel C, spectrum of deglycosylated tryptic peptide of GGCX at m/z 2690.42; Panel D, spectrum of 18O labeled deglycosylated tryptic peptide of GGCX at m/z 2690.42.
Tandem Mass Spectrometry Analysis of the Proteolytic Glycopeptides
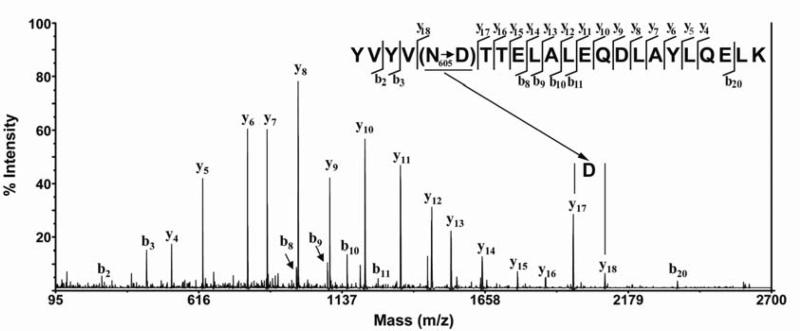
Further proof that the identified peptides are glycopeptides is provided by sequencing the precursor ions of the abundant glycopeptide by tandem mass spectrometry. We isolated precursor peptides using the timed ion selector. The selected peptides were subjected to collision-induced dissociation for amino acid sequencing. The most commonly observed product ions resulting from collision-induced dissociation belong to the y-type ion series, which results from backbone cleavage of the C–N amide linkage with the charge retained on the carboxyl-terminal fragments. The product ions are numbered according to the cleavage site from the carboxyl-terminal end. Figure 3 shows an example tandem mass spectrum of the trypsin/CNBr digested deglycosylated peptide at m/z 2617.34. This spectrum reveals a series of y-ions from the corresponding peptide YVYVNTTELALEQDLAYLQELK of wild-type GGCX. The mass difference between the y18 and y17 ions is 115 units, which corresponds to an aspartic acid residue rather than an asparagine residue (114) at position 605 in wild-type GGCX sequence, as labeled in Figure 3. This asparagine to aspartic acid conversion is due to the PNGase F deglycosylation which further supports the above assignment of the glycopeptides. In addition to the y ions, we also observed the b-ion series in the spectrum which corresponds to the same cleavage of the C–N amide linkage by collision-induced dissociation but with the charge retained on the N-terminal fragments. This result provides still further support that N605 is glycosylated. Sequencing of the abundant deglycosylated peptide at m/z 2690.42 and m/z 2433.29 also indicate that asparagine 627 is glycosylated (Data not shown). Because of the low abundance of other glycopeptides, we were unable to obtain satisfactory sequencing results that might further substantiate glycosylation of site N459.
Figure 3. Tandem mass spectrum sequencing of the deglycosylated proteolytic peptides.
Trypsin/CNBr digested deglycosylated peptide of GGCX at m/z 2617.34 was isolated using a timed ion selector and subjected to collision-induced dissociation fragmentation for amino acid sequencing. The observed y-type ions correspond to the backbone cleavage of the C-N amide linkage with the charge retained in the carboxyl-terminal fragment and the b-type ions correspond to the same cleavage with the charge retained in the amino-terminal fragment were labeled. Conversion of asparagine (N605) to aspartic acid (D) due to PNGase F deglycosylation of the glycopeptide was marked.
Removal of the N-glycosylation Sites of GGCX by Site-Directed Mutagenesis
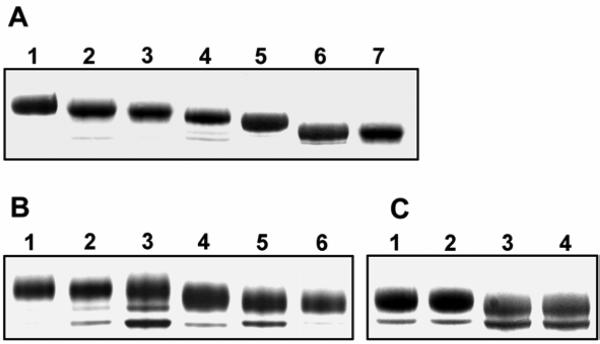
Together, the above results demonstrate that four of the glycosylation sites in GGCX, N459, N550, N605, and N627 are glycosylated during co-translational modification. However, we were unable to recover peptides covering the remaining sites. Since N-glycosylation occurs at asparagine residues within the canonical sequence of NX(S/T), changing asparagine to glutamine (or another residue) will abolish glycosylation at the mutated site. The elimination of one glycosylation site may reduce the protein molecular weight by as much as 3 kDa (19). Therefore, if the glycosylation sites are modified, we expect the mutated proteins to migrate differently in SDS-PAGE. Figure 4A shows SDS-PAGE analysis of the affinity purified GGCX mutants. Changing N459, N550, N605, or N627 (lane 2 to lane 5) to glutamine alters the migration of GGCX on reducing SDS-PAGE. This result supports the mass spectrometry results and indicates that eliminating the glycosylation sequon results in a measurable change in migration of GGCX on reducing SDS-PAGE gels. However, treatment of GGCX605 (with all 4 identified glycosylation sites removed) with PNGase F causes a further increase of its mobility (Figure 4A, lane 6), suggesting that these four glycosylation sites are not the only functional glycosylated sites in GGCX.
Figure 4. Gel-shift analyses of GGCXs with mutations at the potential glycosylation sites.
Reducing NuPAGE silver-stained gels are depicted. Panel A, Lane 1, wild-type GGCX; lane 2, GGCX459 mutant GGCX; lane 3, GGCX627 mutant GGCX; lane 4, GGCX550 mutant GGCX; lane 5, GGCX605 mutant GGCX; lane 6, GGCX605 mutant GGCX treated with PNGase F; lane 7, wild-type GGCX treated with PNGase F. Panel B, lane 1, GGCX605 mutant GGCX; lane 2, GGCX389 mutant GGCX; lane 3, GGCX570 mutant GGCX; lane 4, GGCX525 mutant GGCX; lane 5, GGCX525 mutant GGCX treated with PNGase F; lane 6, GGCX605 mutant GGCX treated with PNGase F. Panel C, lane 1, GGCX525 mutant GGCX; lane 2, GGCX389/525 mutant GGCX; lane 3, GGCX525 mutant GGCX treated with PNGase F; lane 4, GGCX389/525 mutant GGCX treated with PNGase F.
Identification of N-glycosylation Sites in GGCX by Site-Directed Mutagenesis and Gel-shift Analysis
Results shown in Figure 4A (lane 6) suggest there is, in addition to the four sites identified by MS, at least one additional glycosylation site in GGCX. It also shows that removal of a genuine glycosylation site in GGCX causes a visible decrease in size detected by SDS-PAGE. To further examine this question, we started with a GGCX molecule (GGCX605) with the four confirmed glycosylation sites mutated to glutamine. To this mutated molecule (GGCX605) were added mutations of N389Q, N525Q, and N570Q individually. Figure 4B compares the migration difference among these affinity purified GGCX mutants by SDS-PAGE analysis. As can be seen, GGCX389 (Figure 4B lane 2) and GGCX570 (Figure 4B lane 3) mutants have the same migration distance as GGCX605 (Figure 4B lane 1) suggesting that sites N389 and N570 are not glycosylated in vivo. In contrast, the GGCX525 (Figure 4B lane 4) mutant migrates faster than GGCX605 indicating that N525 is utilized for the addition of carbohydrate during co-translational modification. These results, together with the results of mass spectrometry, suggest that there are five functional N-glycosylation sites in GGCX at residues N459, N525, N550, N605, and N627.
Figure 4B (lane 5) also shows that the GGCX525 mutant, which has simultaneously removed all five glycosylation sites, is still sensitive to PNGase F digestion. This result raises the possibility that when all the natural glycosylation sites in GGCX are removed, an alternative glycosylation site functions. To verify this, we changed asparagine to glutamine individually at the two candidate sites, N389 and N570 in mutant GGCX525 yielding two mutants: GGCX389/525 and GGCX570/525. As shown in Figure 4C, GGCX389/525 (lane 2) and GGCX525 (lane 1) have the same migration distance and both of them migrate faster after PNGase F digestion (lane 3 and 4). This result suggests that GGCX389/525 and GGCX525 have the same glycosylation status, and that site N389 did not serve as the proposed alternative glycosylation site. Since GGCX389/525 mutant is sensitive to PNGase F digestion and N570 is the only candidate glycosylation site in this mutant, we propose that site N570 is the alternative glycosylation site. In addition, we were unable to properly express and purify the GGCX570/525 mutant due to a protein degradation problem. These results together suggest that site N570 may serve as an alternative glycosylation site. When site N570 and all the five natural glycosylation sites are removed (mutant GGCX570/525), GGCX apparently can not fold correctly.
N-linked Glycosylation Is not Required for the Enzymatic Function of GGCX
To investigate whether the removal of glycosylation sites affects the enzymatic function of GGCX, the specific carboxylation activity and epoxidation activity of GGCX variants were examined. Results are summarized in Table 3. As can be seen, removal of the sugar moieties from wild-type GGCX by PNGase F digestion only has a minor effect on either the FLEEL carboxylation (91.5%) or vitamin K epoxidation (86%) activities compared with that of the wild-type enzyme. GGCX mutants that have 1 to 4 glycosylation sites removed (GGCX459, GGCX627, GGCX550, and GGCX605) have normal or even higher carboxylation and epoxidation activity. In addition, simultaneous removal of all five natural glycosylation sites (GGCX525) causes only a 30% reduction in carboxylation and epoxidation activity relative to that of the wild-type enzyme. Table 3 also shows that the ratios of vitamin K epoxidation and CO2 incorporation of all these GGCX variants are between 1.4 and 2.4, which agrees with the results of Sugiura et al (38). These results suggest that N-linked glycosylation is not required for either the carboxylation or epoxidation activity of GGCX.
Table 3.
Comparison of the specific carboxylation and epoxidation activity of GGCX variants
| GGCX | Specific activity for carboxylation |
Specific activity for epoxidation |
Epoxidation/Carboxylation | ||
|---|---|---|---|---|---|
| mole 14CO2/min/mole enzyme * | Percentage activity | mole KO/min/mole enzyme * | Percentage activity | ||
| WT | 22.3±0.3 | 100 | 32.3±0.4 | 100 | 1.4 |
| WT+PNGase F | 20.4±0.5 | 92 | 27.9±0.5 | 86 | 1.4 |
| GGCX459 | 24.7±0.2 | 111 | 49.7±2.0 | 154 | 2.0 |
| GGCX627 | 22.2±0.4 | 100 | 42.0±0.2 | 130 | 1.9 |
| GGCX550 | 26.2±0.2 | 117 | 63.4±2.3 | 196 | 2.4 |
| GGCX605 | 35.9±0.1 | 161 | 60.7±2.4 | 188 | 1.7 |
| GGCX525 | 17.4±0.2 | 78 | 23.9±0.3 | 74 | 1.4 |
Specific activities were determined by fluorescence titration of the fraction of the active enzyme according to Ref. 28 and the value listed are the average±SD, n=3
DISCUSSION
The goal of this study was to identify the N-linked glycosylation sites in GGCX and determine the importance of this post-translational modification to GGCX structure and/or function. Evidence that GGCX is a glycoprotein was published about 20 years ago (14). However, the details of which GGCX glycosylation sequons are modified and how this modification affects GGCX maturation and enzymatic function is still unknown.
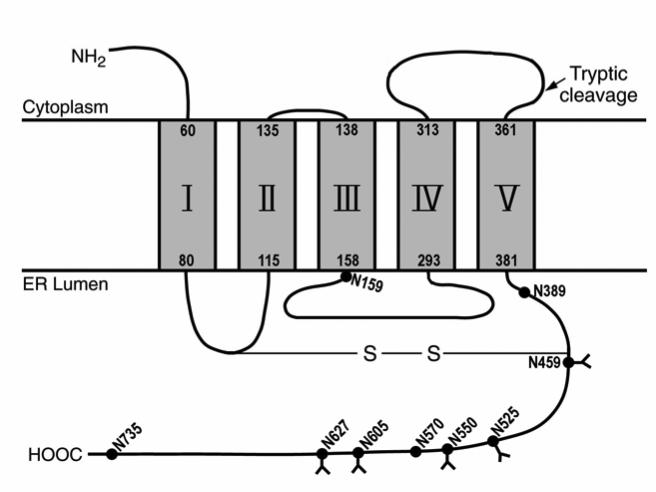
According to our results, N159, the only potential glycosylation site in the 30 kDa tryptic peptide, is not modified (Figure 1). This result is not surprising since according to our membrane topology model, N159 is located immediately after the third transmembrane domain (138-158) (Figure 5) (25). According to the “minimum glycosylation distance” rule (39, 40), for efficient glycosylation, the minimum number of residues between the glycosylation acceptor Asn and the lumenal end of a transmembrane segment is between 10 and 14 amino acids. Therefore, N159 would not be expected to be modified. This result then lends further support to our topology model.
Figure 5. Membrane topology of vitamin K-dependent GGCX.
Five transmembrane domains of GGCX are labeled by Roman numerals and the boundary residues noted. The single disulfide bond is between cysteines 99 and 450. Potential glycosylation sites are marked by filled dots (•) and the residue number of the asparagines is shown. Functional glycosylation sites identified in this paper are designated (Y).
Our mass spectrometric studies identified 4 glycosylation sites in human GGCX: N459, N550, N605, and N627. However, peptides containing the potential sites at N525, 389 and 570 were not recovered. To investigate the status of these sites, to confirm the sites identified by MS, and to allow evaluation of the functional importance of the various sites, we created a series of mutant GGCXs. The four sites identified by MS were confirmed by our mutational studies (Figure 4A). In addition site N525 was glycosylated under normal circumstances (Figure 4B). Therefore, there are five functional glycosylation sites in GGCX.
The results (Figure 4B) with all the five glycosylation sites, N459, 525, 550, 605 and 627, changed to Q indicated that there was at least one additional site in GGCX. But this site is only glycosylated when the five sites were mutated simultaneously. The only possibility for this additional glycosylation site is residues 389 or 570. We have already shown that neither of these sites is glycosylated in the mutant GGCX605 (Figure 4B). Results of SDS-PAGE gel-shift experiments (Figure 4C) show that site N570 is modified when all five of the natural glycosylation sequons are removed. This interpretation is derived from the result with the GGCX mutant GGCX389/525, since the GGCX with 5 mutations plus N570Q (GGCX570/525) is degraded. We do not know whether GGCX570/525 is targeted for degradation in the cell or is degraded during purification. That N389 is not glycosylated in addition to the results with N159, adds further support to our membrane topology model since it too falls under the “minimum glycosylation distance” rule. N389 is 8 residues away from the last transmembrane domain (361-381) (Figure 5) (25).
Thus, glycosylation of GGCX is essential for proper, stable structure formation. It is generally accepted that one of the key roles of N-linked glycosylation is to promote proper protein folding of glycoproteins. When glycosylation is blocked, many glycoproteins do not fold correctly (19, 41). Proteins in non-native conformations fail to pass ER quality control and are targeted for degradation (42). Our results (Table 3) show that when five of the natural glycosylation sites are removed, even one alternative glycosylation site (N570) is sufficient for GGCX to pass ER quality control and to form an active enzyme (GGCX525); however, the GGCX with N570 mutated, as well as the 5 natural sites, is degraded. This suggests that a minimum of one glycosylation site is necessary either for folding of GGCX or for its stability once folded. This effect does not appear to be a local effect on structure, since the exact glycosylaion site seems unimportant. This indicates the global importance of glycosylation for the overall structure of GGCX. Therefore, glycosylation is important for proper folding or for GGCX to pass quality control.
Enzymatic removal of the glycan moiety from purified GGCX by PNGase F has little effect on the carboxylation (91.5%) and epoxidation (86%) activity. So while glycans are essential for protein stability and/or folding, no glycosylation is necessary for expression of enzymatic activity. Interestingly, the mutant enzymes with one to four sites mutated, retained similar or even higher enzymatic activities compared to those of the wild-type enzyme. The GGCXs with 3 and 4 mutated glycosylation sites had almost two-fold higher epoxidase activity than that of wild type GGCX.
The enzymatic removal of glycans yields an aspartic acid residue in place of asparagine. This changes the charge character of the enzyme. In this case the polar non-charged sugars are replaced by a negative charge. On the other hand, mutation to Q simply replaces the non-charged polar sugars with a smaller non-charged polar glutamine. Thus, in the latter case the change is steric in nature. The hypothesis of how the carboxylation reaction occurs suggests that oxidation of vitamin K hydroquinone to vitamin K epoxide activates the removal of the γ-proton from the glutamic acid of the substrate to form a carbanion intermediate (6, 13). Then, a CO2 molecule attacks this carbanion intermediate to form the final product of γ-carboxyglutamic acid. One interpretation of our results is that replacing the glycan with the smaller Q side-chain may allow easier access to vitamin K by removing steric hindrance to this hydrophobic substrate's binding. In contrast, the smaller substrate CO2 is not as significantly affected by this change. This may explain why the epoxidation activity increases proportionately more than carboxylation in the mutant GGCX relative to that of wild type GGCX.
While our results suggest that N-linked glycosylation is not required for the enzymatic function of GGCX, it is based on the carboxylation of the small pentapeptide substrate FLEEL in the presence of a separate factor IX propeptide in the in vitro assay system. In vivo, the natural substrates for GGCX are macromolecules of vitamin K-dependent proteins. These proteins, especially the coagulation factors are also glycoproteins. It has been reported that the glycans in the coagulation factors are important for protein-protein interaction (43-45), and that the glycosylation modification occurs before post-translational carboxylation modification (46). Therefore, it is possible that the glycans in GGCX play a role in the carbohydrate interaction between GGCX and its natural glycoprotein substrates.
The principal binding site for coagulation factors to GGCX is their propeptide (47). It has been reported that the propeptide binding site in GGCX is located in the region of residues 495-513 (48). As shown in Figure 5, three out of the five functional glycosylation sites (N459, N525, N550) are close to this region in the primary structure of GGCX sequence. Binding of the propeptide, which causes a conformational change in GGCX, may bring the glycans of the substrate and enzyme into proximity and facilitate substrate binding, and thus, the carboxylation reaction.
In conclusion, we have identified the N-linked glycosylation sites of GGCX by mass spectrometry and site-directed mutagenesis. Our results suggest that there are five glycosylation sites in GGCX at positions N459, N525, N550, N605, and N627. The potential glycosylation site at N570 is not glycosylated under normal conditions, but it serves as an alternative site for glycosylation when all the natural glycosylation sites are eliminated. Even this one site ensures that the protein folds correctly. While glycosylation is important for GGCX folding and to prevent degradation, it is not required for GGCX enzymatic function.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Kevin L. Carrick for mass spectrometry data acquisition.
The abbreviations used are
- GGCX
gamma-glutamyl carboxylase
- ER
endoplasmic reticulum
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight
- Q-TOF MS
quadrupole time of flight mass spectrometry
- CHAPS
(3-[(3-Cholamidopropyl)-dimethylammonio]-1-propanesulfonate
- CNBr
cyanogen bromide
- Vitamin K1(20)
2-methyl-3-phytyl-1,4-naphthoquinone
- Vitamin K1 (25)
2-methyl-3-(3,7,11,15,19-pentamethyl-2-eicosenyl)-1,4-naphthalenedionel
- PNGase F
Peptide:N-Glycosidase F
- proFIX
19 amino acid peptide comprising residues TVFLDHENANKILNRPKRY of human factor IX
- TFA
Trifluoroacetic acid
- SDS- PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
Footnotes
This work was supported by National Institutes of Health Grant HL48318 (to D. W. S.)
REFERENCE
- 1.Stafford DW. The vitamin K cycle. J Thromb Haemost. 2005;3:1873–1878. doi: 10.1111/j.1538-7836.2005.01419.x. [DOI] [PubMed] [Google Scholar]
- 2.Presnell SR, Stafford DW. The vitamin K-dependent carboxylase. Thromb Haemost. 2002;87:937–946. [PubMed] [Google Scholar]
- 3.Berkner KL. The vitamin K-dependent carboxylase. Annu Rev Nutr. 2005;25:127–149. doi: 10.1146/annurev.nutr.25.050304.092713. [DOI] [PubMed] [Google Scholar]
- 4.Carlisle TL, Suttie JW. Vitamin K dependent carboxylase: subcellular location of the carboxylase and enzymes involved in vitamin K metabolism in rat liver. Biochemistry. 1980;19:1161–1167. doi: 10.1021/bi00547a019. [DOI] [PubMed] [Google Scholar]
- 5.Bristol JA, Ratcliffe JV, Roth DA, Jacobs MA, Furie BC, Furie B. Biosynthesis of prothrombin: intracellular localization of the vitamin K-dependent carboxylase and the sites of gamma-carboxylation. Blood. 1996;88:2585–2593. [PubMed] [Google Scholar]
- 6.Larson AE, Friedman PA, Suttie JW. Vitamin K-dependent carboxylase. Stoichiometry of carboxylation and vitamin K 2,3-epoxide formation. J Biol Chem. 1981;256:11032–11035. [PubMed] [Google Scholar]
- 7.Dubois J, Gaudry M, Bory S, Azerad R, Marquet A. Vitamin K-dependent carboxylation. Study of the hydrogen abstraction stereochemistry with gamma-fluoroglutamic acid-containing peptides. J Biol Chem. 1983;258:7897–7899. [PubMed] [Google Scholar]
- 8.Suttie JW. Mechanism of action of vitamin K: synthesis of gamma-carboxyglutamic acid. CRC Crit Rev Biochem. 1980;8:191–223. doi: 10.3109/10409238009105469. [DOI] [PubMed] [Google Scholar]
- 9.Suttie JW. The metabolic role of vitamin K. Fed Proc. 1980;39:2730–2735. [PubMed] [Google Scholar]
- 10.Willingham AK, Matschiner JT. Changes in phylloquinone epoxidase activity related to prothrombin synthesis and microsomal clotting activity in the rat. Biochem J. 1974;140:435–441. doi: 10.1042/bj1400435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matschiner JT, Willingham AK. Influence of sex hormones on vitamin K deficiency and epoxidation of vitamin K in the rat. J Nutr. 1974;104:660–665. doi: 10.1093/jn/104.6.660. [DOI] [PubMed] [Google Scholar]
- 12.Suttie JW, Geweke LO, Martin SL, Willingham AK. Vitamin K epoxidase: dependence of epoxidase activity on substrates of the vitamin K-dependent carboxylation reaction. FEBS Lett. 1980;109:267–270. doi: 10.1016/0014-5793(80)81102-1. [DOI] [PubMed] [Google Scholar]
- 13.Dowd P, Hershline R, Ham SW, Naganathan S. Vitamin K and energy transduction: a base strength amplification mechanism. Science. 1995;269:1684–1691. doi: 10.1126/science.7569894. [DOI] [PubMed] [Google Scholar]
- 14.Brody T, Suttie JW. Evidence for the glycoprotein nature of vitamin K-dependent carboxylase from rat liver. Biochim Biophys Acta. 1987;923:1–7. doi: 10.1016/0304-4165(87)90118-8. [DOI] [PubMed] [Google Scholar]
- 15.Berkner KL, Harbeck M, Lingenfelter S, Bailey C, Sanders-Hinck CM, Suttie JW. Purification and identification of bovine liver gamma-carboxylase. Proc Natl Acad Sci U S A. 1992;89:6242–6246. doi: 10.1073/pnas.89.14.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu SM, Mutucumarana VP, Geromanos S, Stafford DW. The propeptide binding site of the bovine gamma-glutamyl carboxylase. J Biol Chem. 1997;272:11718–11722. doi: 10.1074/jbc.272.18.11718. [DOI] [PubMed] [Google Scholar]
- 17.Gavel Y, von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Dor S, Esterman N, Rubin E, Sharon N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14:95–101. doi: 10.1093/glycob/cwh004. [DOI] [PubMed] [Google Scholar]
- 19.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 20.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 21.Jones J, Krag SS, Betenbaugh MJ. Controlling N-linked glycan site occupancy. Biochim Biophys Acta. 2005;1726:121–137. doi: 10.1016/j.bbagen.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim Biophys Acta. 1999;1473:21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 23.Dennis JW, Granovsky M, Warren CE. Protein glycosylation in development and disease. Bioessays. 1999;21:412–421. doi: 10.1002/(SICI)1521-1878(199905)21:5<412::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunol Cell Biol. 2005;83:429–439. doi: 10.1111/j.1440-1711.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 25.Tie J, Wu SM, Jin D, Nicchitta CV, Stafford DW. A topological study of the human gamma-glutamyl carboxylase. Blood. 2000;96:973–978. [PubMed] [Google Scholar]
- 26.Wu SM, Morris DP, Stafford DW. Identification and purification to near homogeneity of the vitamin K-dependent carboxylase. Proc Natl Acad Sci U S A. 1991;88:2236–2240. doi: 10.1073/pnas.88.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu SM, Cheung WF, Frazier D, Stafford DW. Cloning and expression of the cDNA for human gamma-glutamyl carboxylase. Science. 1991;254:1634–1636. doi: 10.1126/science.1749935. [DOI] [PubMed] [Google Scholar]
- 28.Presnell SR, Tripathy A, Lentz BR, Jin DY, Stafford DW. A novel fluorescence assay to study propeptide interaction with gamma-glutamyl carboxylase. Biochemistry. 2001;40:11723–11733. doi: 10.1021/bi010332w. [DOI] [PubMed] [Google Scholar]
- 29.Stanley TB, Jin DY, Lin PJ, Stafford DW. The propeptides of the vitamin K-dependent proteins possess different affinities for the vitamin K-dependent carboxylase. J Biol Chem. 1999;274:16940–16944. doi: 10.1074/jbc.274.24.16940. [DOI] [PubMed] [Google Scholar]
- 30.Rudd PM, Joao HC, Coghill E, Fiten P, Saunders MR, Opdenakker G, Dwek RA. Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry. 1994;33:17–22. doi: 10.1021/bi00167a003. [DOI] [PubMed] [Google Scholar]
- 31.Tie JK, Mutucumarana VP, Straight DL, Carrick KL, Pope RM, Stafford DW. Determination of disulfide bond assignment of human vitamin K-dependent gamma-glutamyl carboxylase by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Biol Chem. 2003;278:45468–45475. doi: 10.1074/jbc.M309164200. [DOI] [PubMed] [Google Scholar]
- 32.van Montfort BA, Doeven MK, Canas B, Veenhoff LM, Poolman B, Robillard GT. Combined in-gel tryptic digestion and CNBr cleavage for the generation of peptide maps of an integral membrane protein with MALDI-TOF mass spectrometry. Biochim Biophys Acta. 2002;1555:111–115. doi: 10.1016/s0005-2728(02)00264-5. [DOI] [PubMed] [Google Scholar]
- 33.Kuster B, Mann M. 18O-labeling of N-glycosylation sites to improve the identification of gel-separated glycoproteins using peptide mass mapping and database searching. Anal Chem. 1999;71:1431–1440. doi: 10.1021/ac981012u. [DOI] [PubMed] [Google Scholar]
- 34.Thijssen HH, Soute BA, Vervoort LM, Claessens JG. Paracetamol (acetaminophen) warfarin interaction: NAPQI, the toxic metabolite of paracetamol, is an inhibitor of enzymes in the vitamin K cycle. Thromb Haemost. 2004;92:797–802. doi: 10.1160/TH04-02-0109. [DOI] [PubMed] [Google Scholar]
- 35.Zhen Y, Caprioli RM, Staros JV. Characterization of glycosylation sites of the epidermal growth factor receptor. Biochemistry. 2003;42:5478–5492. doi: 10.1021/bi027101p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kvaratskhelia M, Clark PK, Hess S, Melder DC, Federspiel MJ, Hughes SH. Identification of glycosylation sites in the SU component of the Avian Sarcoma/Leukosis virus Envelope Glycoprotein (Subgroup A) by mass spectrometry. Virology. 2004;326:171–181. doi: 10.1016/j.virol.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Fan X, She YM, Bagshaw RD, Callahan JW, Schachter H, Mahuran DJ. A method for proteomic identification of membrane-bound proteins containing Asn-linked oligosaccharides. Anal Biochem. 2004;332:178–186. doi: 10.1016/j.ab.2004.05.038. [DOI] [PubMed] [Google Scholar]
- 38.Sugiura I, Furie B, Walsh CT, Furie BC. Profactor IX propeptide and glutamate substrate binding sites on the vitamin K-dependent carboxylase identified by site-directed mutagenesis. J Biol Chem. 1996;271:17837–17844. doi: 10.1074/jbc.271.30.17837. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson I, Saaf A, Whitley P, Gafvelin G, Waller C, von Heijne G. Proline-induced disruption of a transmembrane alpha-helix in its natural environment. J Mol Biol. 1998;284:1165–1175. doi: 10.1006/jmbi.1998.2217. [DOI] [PubMed] [Google Scholar]
- 40.Nilsson IM, von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- 41.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 42.Klausner RD, Sitia R. Protein degradation in the endoplasmic reticulum. Cell. 1990;62:611–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- 43.Silveira JR, Kalafatis M, Tracy PB. Carbohydrate moieties on the procofactor factor V, but not the derived cofactor factor Va, regulate its inactivation by activated protein C. Biochemistry. 2002;41:1672–1680. doi: 10.1021/bi011304g. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez JA, Hackeng TM, Kojima K, Griffin JH. The carbohydrate moiety of factor V modulates inactivation by activated protein C. Blood. 1997;89:4348–4354. [PubMed] [Google Scholar]
- 45.Sinha U, Wolf DL. Carbohydrate residues modulate the activation of coagulation factor X. J Biol Chem. 1993;268:3048–3051. [PubMed] [Google Scholar]
- 46.Hallgren KW, Hommema EL, McNally BA, Berkner KL. Carboxylase overexpression effects full carboxylation but poor release and secretion of factor IX: implications for the release of vitamin K-dependent proteins. Biochemistry. 2002;41:15045–15055. doi: 10.1021/bi026016e. [DOI] [PubMed] [Google Scholar]
- 47.Furie B, Bouchard BA, Furie BC. Vitamin K-dependent biosynthesis of gamma-carboxyglutamic acid. Blood. 1999;93:1798–1808. [PubMed] [Google Scholar]
- 48.Lin PJ, Jin DY, Tie JK, Presnell SR, Straight DL, Stafford DW. The putative vitamin K-dependent gamma-glutamyl carboxylase internal propeptide appears to be the propeptide binding site. J Biol Chem. 2002;277:28584–28591. doi: 10.1074/jbc.M202292200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.