Abstract
N6-methyladenosine (m6A) is the most ubiquitous mRNA base modification, but little is known about its precise location, temporal dynamics, and regulation. Here, we generated genomic maps of m6A sites in meiotic yeast transcripts at nearly single-nucleotide resolution, identifying 1,308 putatively methylated sites within 1,183 transcripts. We validated 8/8 methylation sites in different genes with direct genetic analysis, demonstrated that methylated sites are significantly conserved in a related species, and built a model that predicts methylated sites directly from sequence. Sites vary in their methylation profiles along a dense meiotic time-course, and are regulated both locally, via predictable methylatability of each site, and globally, through the core meiotic circuitry. The methyltransferase complex components localize to the yeast nucleolus, and this localization is essential for mRNA methylation. Our data illuminates a conserved, dynamically regulated methylation program in yeast meiosis, and provides an important resource for studying the function of this epitranscriptomic modification.
Introduction
DNA, RNA and proteins are all covalently modified post-synthesis, potentially impacting their function. While DNA and protein modifications have been extensively studied, our understanding of mRNA modifications is limited. The methylation of adenosine at the N6 position to form N6-methyladenosine (m6A) is among the most abundant base modifications known in eukaryotic mRNA (Desrosiers et al., 1975). Orthologs of the RNA-based N6-adenosyl methyltransferases (MTases) that catalyze this modification are present in almost all eukaryotes, and its depletion or disruption causes lethality in metazoans (Bokar, 2005; Dominissini et al., 2012; Hongay and Orr-Weaver, 2011) and severe developmental defects in plants (Zhong et al., 2008). Interestingly, the FTO protein, which is genetically associated with human obesity, acts as a specific m6A demethylase (Jia et al., 2011).
Significant technical and experimental limitations have hindered the study of m6A modifications. First, since m6A neither changes the base-pairing properties nor inhibits reverse transcription, identification of modified transcripts has depended on immunoprecipitation using antibodies against m6A (Bodi et al., 2010; Bringmann and Luhrmann, 1987; Dominissini et al., 2012; Meyer et al., 2012). Recent transcriptome-wide mappings, termed m6A-seq (Dominissini et al., 2013) or MeRIP-Seq (Meyer et al., 2012), have revealed that m6A accumulates near stop codons and atypically long exons and that methylation sites in mammals are associated with an RRACT (R=A/G) consensus sequence, consistent with earlier studies (Dimock and Stoltzfus, 1977; Schibler et al., 1977; Wei et al., 1976). However, the resolution of these maps was only ~24nt around the methylation site, as estimated from the median distance from an identified peak to the closest consensus sequence (Dominissini et al., 2012). Thus, to date, only a single methylated site has been mapped at single nucleotide resolution on eukaryotic mRNA (Horowitz et al., 1984; Narayan and Rottman, 1988). Second, experimental depletion of the methylation complex in mammals results in apoptosis (Bokar, 2005; Dominissini et al., 2012; Hongay and Orr-Weaver, 2011), rendering it difficult to dissect the functional role of methylation. Third, the mammalian methylation landscape appears to be mostly static across cell types, tissues, and stimuli (Dominissini et al., 2012; Meyer et al., 2012), limiting our ability to elucidate how methylations emerge.
By contrast, mRNA methylation in the yeast Saccharomyces cerevisiae occurs only during meiosis (Agarwala et al., 2012; Clancy et al., 2002; Hongay et al., 2006; Shah and Clancy, 1992), providing a unique opportunity to dissect its dynamics and regulation. Genetic screens in yeast have identified a core RNA methyltransferase (MIS) complex comprised of Ime4 (orthologous to mammalian methyltransferase like 3, METTL3), Mum2 (orthologous to mammalian Wilm’s tumor 1 associated protein, WTAP), and a third ancillary factor, Slz1 (Agarwala et al., 2012). The MIS complex is induced during meiosis, and defects that abrogate its mRNA methylation activity delay meiotic entry (Agarwala et al., 2012; Clancy et al., 2002; Hongay et al., 2006; Shah and Clancy, 1992). Elimination of MIS components in yeast is not lethal (Agarwala et al., 2012; Clancy et al., 2002; Hongay et al., 2006; Shah and Clancy, 1992), allowing experimental exploitation of such strains.
Here, we used a high resolution assay coupled with mutants defective in methylation to identify m6A sites, at nearly single-base resolution, in meiotic yeast transcripts. Our approach allows us to dissect ‘cis’ and ‘trans’ elements governing methylation onset and offset, and provides a broad overview on a conserved and dynamically regulated methylation program in yeast meiosis and an important resource towards addressing its function.
Results
m6A-seq defines the MIS-dependent yeast methylome
To map m6A sites in yeast, we used a highly optimized m6A-seq approach (Figure S1A). Previously published protocols (i) required substantial input material, (ii) had relatively low resolution around the actual methylated site, and, (iii) did not provide a way to directly assess false positives (Dominissini et al., 2012; Meyer et al., 2012). We optimized the protocol (Experimental Procedures) to decrease the required mRNA starting material (from 400 µg polyA+ mRNA to 5 µg), increase resolution (by decreasing fragment size and employing a ligation-based strand-specific library preparation protocol capturing both ends of the fragmented RNA, ensuring that the methylated position is within the sequenced fragment), and increase scale. Finally, to determine false positives, we used a negative control of strains with ime4Δ/Δ, which do not accumulate m6A.
We applied m6A-seq to (i) mRNA isolated from an ndt80Δ/Δ strain undergoing meiosis, which arrests during meiotic G2/prophase when bulk m6A-mRNA levels are at their peak (Agarwala et al., 2012), and (ii) as a negative control, an ime4Δ/Δ strain, which arrests at the same time-point as the ndt80Δ/Δ strain but does not accumulate m6A (Agarwala et al., 2012; Clancy et al., 2002). We aligned reads from immunoprecipated (IP) and input samples to the SK1 reference genome, and called peaks based on presence in the IP samples and absence in the input (Experimental Procedures).
Of the 3,294 sites present in at least two of three ndt80Δ/Δ biological replicates, 1,664 (50.5%) were absent from both the input samples (Experimental Procedures) and the ime4Δ/Δ samples, suggesting these are true methylated sites (‘MIS dependent’, Figure 1A). The remaining peaks were ‘MIS independent’: present in both wild-type and ime4Δ/Δ experiments, but not in the input samples. To confirm that these MIS independent sites were experimental artifacts, we applied m6A-seq to non-methylated RNA from 17 in vitro transcribed genes with MIS-independent peaks. In 13 of 17 cases, we obtained peaks in precisely the same regions as in the yeast samples (Figure S1B). These false positive sites were enriched in degenerate purine-rich sequence motifs (Figure S1C), suggesting that the antibody may be biased towards such sequences. The ime4Δ/Δ strain allows us to remove these false positive sites. In all subsequent analyses we only considered a conservative, high quality, yeast mRNA methylome of 1,308 putative m6A sites within 1,183 genes detected only in the presence of IME4 (Experimental Procedures, Table S1).
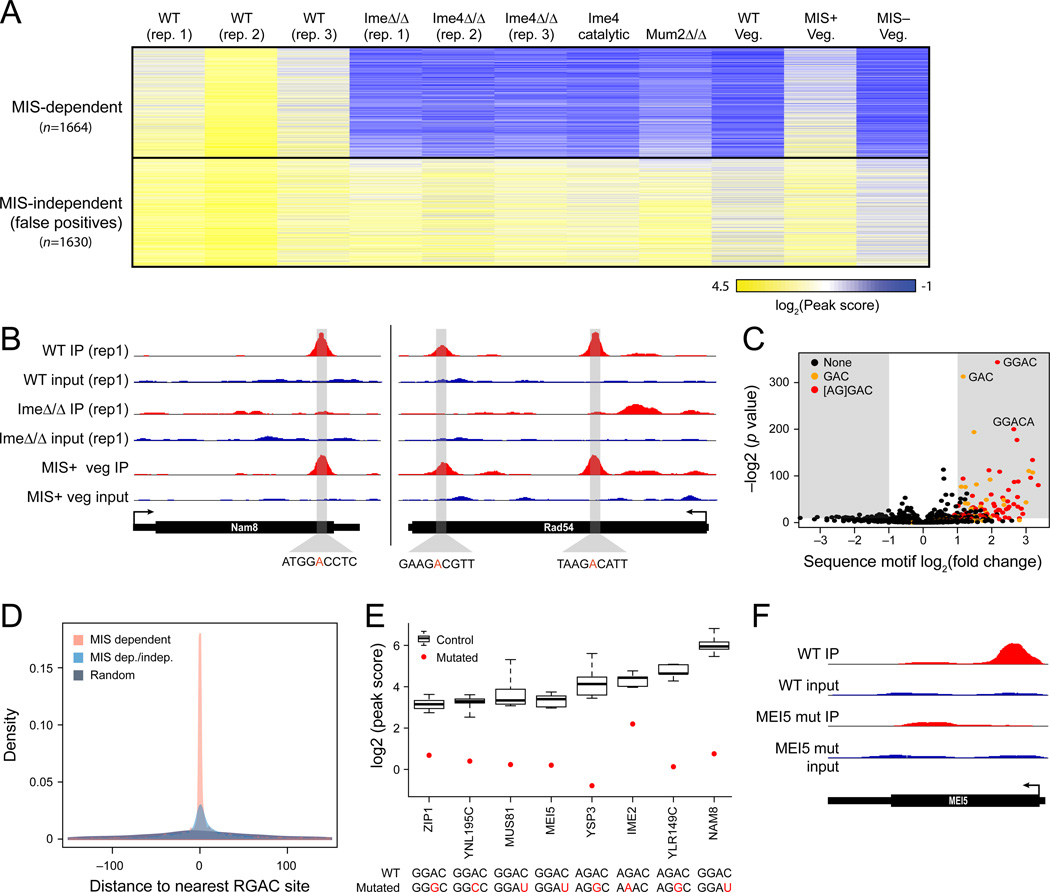
Figure 1. Genome-wide identification of MIS-dependent m6A sites with m6A-Seq.
(A) Methylation profiles. Heatmap shows the log2 transformed peak scores (fold-change of enrichment of a site over the median level of the gene; yellow: high; blue: low) for 3,294 peaks (rows) that were enriched in at least two of the three WT (ndt80Δ/Δ, SAy841) replicates across different conditions and perturbations (columns; ime4Δ/Δ ndt80Δ/Δ—SAy996, ime4-cat ndt80Δ/Δ—SAy1280, mum2Δ/Δ—SAy1310, wild-type—SAy821, MIS Induction—SAy1248). Sites are clustered using k-means clustering. MIS-dependent and -independent sites are marked on top and bottom, respectively. (B) Example methylated loci. Sequence coverage from m6A-Seq (IP) and control (input) experiments in different strains (tracks) for NAM8 (left) and RAD54 (right). Grey highlight: a 50-nt region surrounding the called peak position, with putative methylation consensus sequence (bottom). (C) Volcano plot of the enrichment (Y axis) and fold change (X axis) of all 3–6nt k-mers in a 50-nt window surrounding identified methylation sites, compared to randomly selected regions from the same genes. Shaded regions: statistically depleted (left) or enriched (right) regions (fold change >2; Bonferroni-corrected P values < 0.05). Orange: sites comprising the GAC core motif; red: sites comprising a full RGAC motif. (D) Methylated sites at near single nucleotide resolution. Density plots of the distribution of the distance between the identified peak and the most adjacent RGAC motif (X axis), for the 1,308 MIS-dependent peaks (red), all MIS-dependent and independent sites (blue), and randomly selected sites within the same genes as the MIS dependent peaks (grey). (E,F) Sequence motif is essential for methylation. (E) m6A-seq peak scores (Y axis) for 8 genes measured in strains where the methylated sequence motif was either WT (top sequence) or mutated (bottom sequence, mutation in red). The distribution of peak scores along WT strains (n=9) is indicated with boxplots (error bars: min and max); red dot: mutant peak score. (F) m6A-Seq (IP) and control (input) for the WT (two top tracks) and mutant (two bottom tracks) alleles of MEI5.
We further validated that the yeast mRNA methylome defines targets for MIS-mediated methylation. First, m6A-seq of cells either encoding a catalytically-defective allele of IME4 background (ime4-cat) or a deletion of MUM2, leads to loss of enrichment specifically at the MIS-dependent sites (Figure 1A), showing that MIS complex function is necessary for site methylation. Second, m6A-seq of cells growing under vegetative conditions but overexpressing the three components of the MIS complex (Agarwala et al., 2012) yields profiles very similar to those obtained in meiosis, suggesting that induction of the three components of the MIS complex is sufficient to faithfully recapitulate the meiotic mRNA methylation program (Figure 1A,B).
Function of methylated genes
The 1,183 methylated genes span diverse function, and are enriched in functions highly relevant to meiosis, including DNA replication (P=1.8×10−6), mismatch repair (P=1.3×10−4) and synaptonemal complex formation (P=1.5×10−3), even when using an expression matched gene set as background (data not shown). In particular, 105 of 376 curated meiosis-specific genes are methylated (Table S1, Extended Experimental Procedures). Other methylated transcripts span a wide set of functions, including signaling, maintenance and metabolism, though we cannot preclude their meiosis specific role. Notably, we did not observe m6A in the IME1 and IME4 transcripts (Bodi et al., 2010), and found a methylated region in the IME2 transcript different from that previously described (Bodi et al., 2010) (Figure S1D).
m6A occurs in a consensus motif that is necessary for methylation
The overwhelming majority of motifs enriched within a 50-bp window centered around m6A-peaks harbored an RGAC (R=A/G) consensus sequence (Figure 1C,D; Experimental Procedures), reminiscent of, yet distinct from, the RRACU consensus motif around mammalian m6A sites (Dimock and Stoltzfus, 1977; Dominissini et al., 2012; Meyer et al., 2012; Schibler et al., 1977; Wei et al., 1976). Reflecting nearly single-nucleotide resolution, the median distance between the enriched peak and the nearest RGAC consensus site is 3 nt (57 nt in an equally sized randomly selected control set; Mann-Whitney, P=6.9×10−215, Figure 1D). Notably, before filtering the MIS-independent peaks, this median distance is ~18 nt (Figure 1D), similar to those previously reported in mammals (Dominissini et al., 2012; Meyer et al., 2012).
To validate the consensus, we selected 8 peaks in distinct genes and generated diploid strains carrying single point-mutations that eliminate the methylation consensus sequence. We mutated either the methylated adenosine, or one of its two flanking positions, without altering the protein sequence. In all 8 cases, the peaks were eliminated in the mutated strains (Figure 1E, F), to the same effect when the mutation was in the methylated adenosine, or in positions +1 or −1, suggesting that all three positions are required for methylation. In none of the 8 cases did we observe compensatory methylation of adjacent non-canonical positions, as was previously reported in an in vitro point-mutation study (Narayan et al., 1994).
Methylation sites can be predicted from sequence and structural features
We next leveraged the enhanced resolution and quality of our assay to examine which sequence and structural features are associated with bona fide methylated sites. First, we determined a broader sequence motif from a conservatively selected subset of 711 sites in which the peak was within 5 nt of an RGAC site. Position +4 was a uridine in 73% of the cases, and position −4 was an adenosine in 63% of the cases (Figure 2A), resulting in a full yeast consensus sequence ANRG-m6A-CNNT. Second, methylated sites were strongly biased towards the 3’ end of transcripts (Figure 2B, Mann-Whitney, P=5.9×10−51), with the 3’ bias increasing with peak strength (Figure S2). Finally, consistent with previous hypotheses from small scale studies (Bokar, 2005), methylated sites were significantly less structured in comparison to randomly selected counterparts from the same genes (Mann-Whitney, P=1.7×10−11, Figure 2C), or when controlling for 3’ bias of these regions (Mann-Whitney, P=1×10−9), possibly since these are more exposed to the methylation machinery.
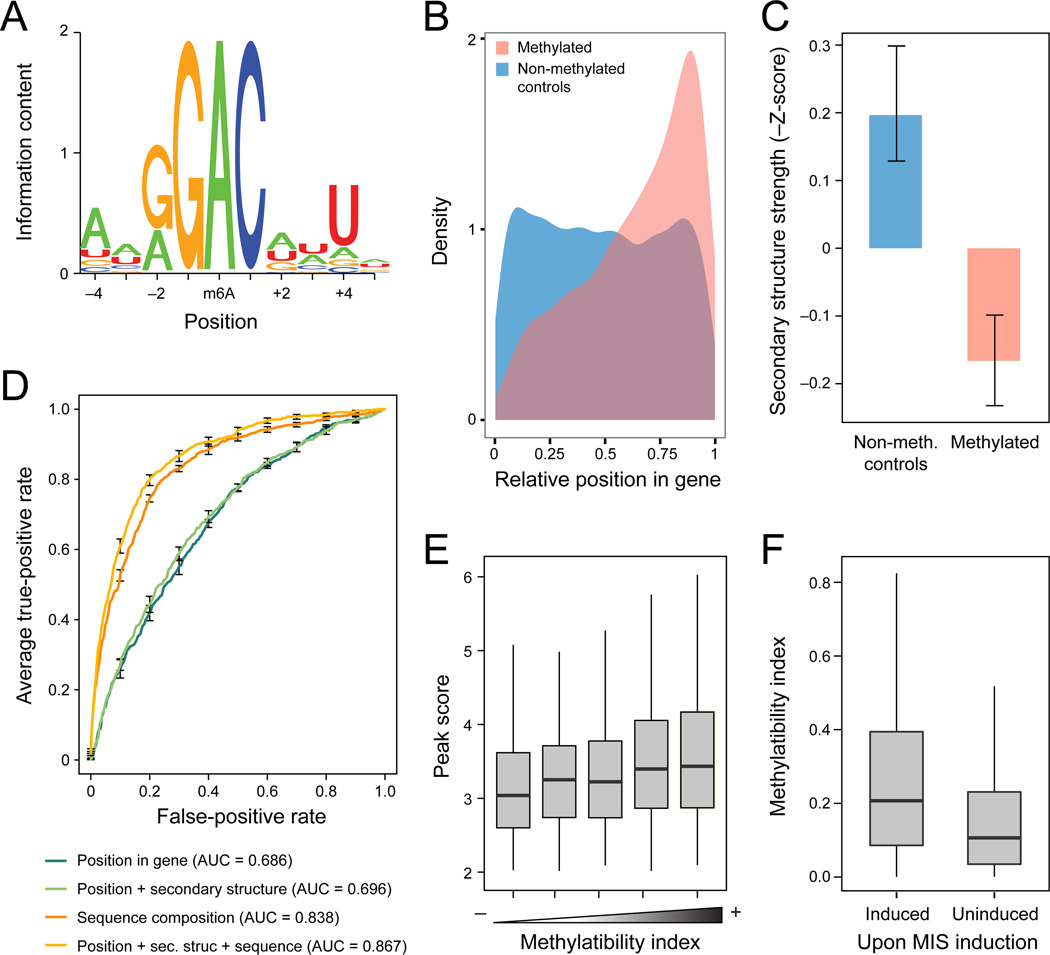
Figure 2. A methylatability model accurately predicts methylated sites solely from sequence, structure and relative position.
(A) Methylation motif. Sequence logo of the methylation consensus sequence, based on 711 conservative sites where the peak was within 5 nt of an RGAC site. (B) 3’ end bias of methylated. Distributions of the relative position within the transcript (X axis, 0: 5’ end; 1: 3’ end) for methylated sites (pink) and for sites randomly selected within the same genes (blue). (C) Methylated sites are less structured. Z scores for stability of local secondary structure (Y axis) in a 50 nt window surrounding the methylated position (pink, right) and in random controls (blue, left). Z scores calculated as the minimal free energy by RNAfold, normalized against randomly shuffled sequences of the same length and nucleotide composition. Error bars: SEM. (D) Methylatability model. Receiver-operator curves (ROC) depicting the performance of different logistic regression classifiers in predicting a site’s methylation state based on different sets of features. A model using position, secondary structure and sequence motif information (orange) performs best, with the sequence motif contributing the most (red). (E) Methylatability Index. Boxplots (boxes: lower quartile, median, and upper quartile; whiskers extend to most extreme point no more than 1.5 fold interquartile range) depicting the distributions of the experimentally measured peak score (y-axis) as a function of the computationally assigned Methylatability Index (x-axis). (F) Sites methylated upon MIS activation have higher Methylatability Indexes. Boxplots depicting the distributions of the experimentally measured peak score (y-axis) across sites that underwent methylations upon MIS induction (SAy1248, Induced), or failed to become methylated under these conditions (Uninduced).
Combining these features, we built a high-quality logistic regression classifier to predict methylated sites from these extended sequence, transcript position, and structure features alone. We trained the classifier to distinguish between a ‘negative set’ of ~10,000 non-methylated sequences surrounding an RGAC sites and a stringent ‘positive set’ of 832 methylated sites centered around an RGAC consensus (Extended Experimental Procedures), and assessed its performance by 10-fold cross validation. A classifier using all three feature types performs best (area under the curve (AUC) = 0.87, Figure 2D), with the largest contribution from the extended sequence motif features. There is a positive correlation between the model-assigned probability of methylation (‘Methylatability Index’) and the experimental peak score (a feature on which the model was not trained, P=2.4×10−11, Figure 2E), suggesting that the model quantitatively recapitulates the same features perceived by the methylating machinery. Furthermore, sites that were not as robustly methylated following ectopic induction of the MIS complex in non-meiotic cells also had lower Methylatability Indices (Figure 2F), suggesting that they are poorer substrates for methylation, and more sensitive to the suboptimal vegetative conditions.
Methylation sites are evolutionarily conserved across yeast species at levels comparable to transcription factor binding sites
We next evaluated the evolutionary conservation of individual methylation sites. We applied m6A-seq to Saccharomyces mikatae, which sporulates efficiently under lab conditions. We generated an ndt80Δ/Δ S. mikatae strain and a negative control ndt80Δ/Δ ime4Δ/Δ strain, and applied m6A-seq to each strain under meiotic conditions, as well as to a wild-type strain under vegetative growth, as an additional control. Global analysis of methylated sites revealed very similar patterns to the ones observed in S. cerevisiae, with both MIS-dependent and MIS-independent sites (Figure 3A). The 635 S. mikatae IME4 dependent peaks (Table S2) were strongly enriched for a consensus very similar to the S. cerevisiae one (Figure 3B), were similarly close to the consensus (median: 4 nt), and were had similar 3’ bias (Figure S3).
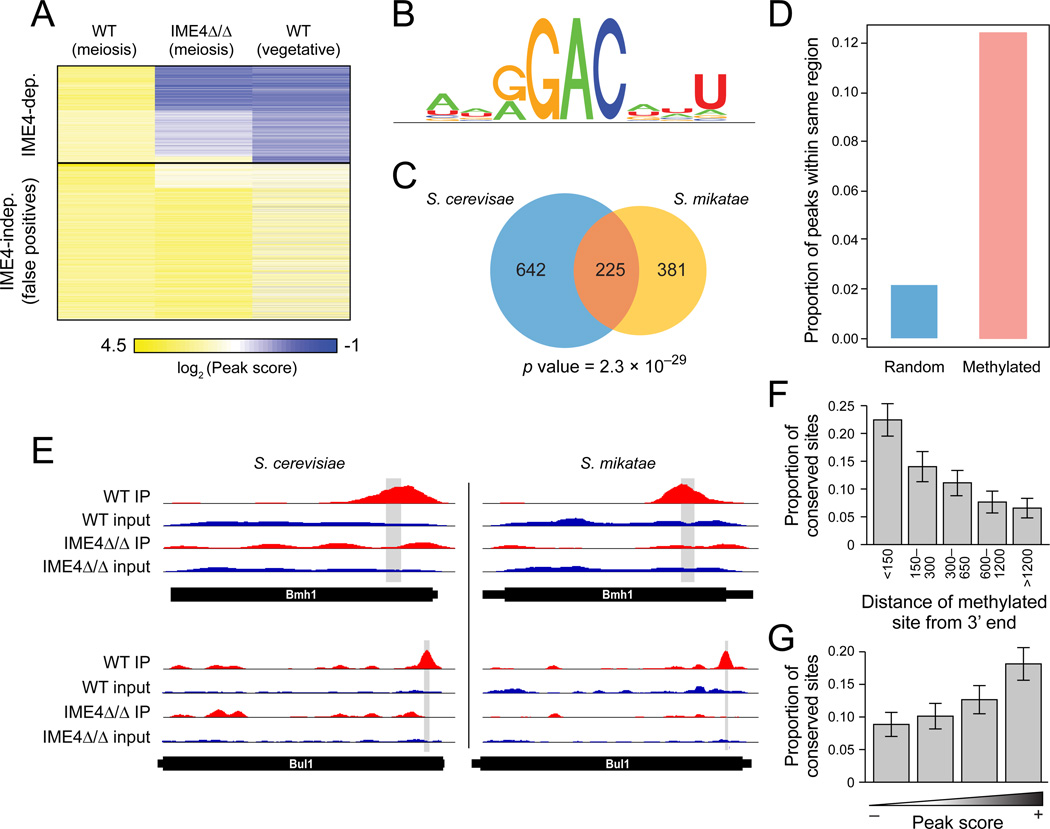
Figure 3. Evolutionary conservation of methylation between S. cerevisiae and S. mikatae.
(A) m6A-Seq of S. mikatae. Heatmap shows the peak scores (as in Figure 1) for 3,345 peaks (rows) that were enriched (peak score>2) in S. mikatae WT strain under prophase arrest conditions (columns; wild-type meiosis, ndt80Δ/Δ—SAy1428, ime4Δ/Δ ndt80Δ/Δ—SAy1429, wild-type vegetative—SAy1426). Sites are clustered using k-means clustering. MIS-dependent (top) and independent (bottom) sites are denoted. (B) S. mikatate methylation consensus motif. (C) Significant conservation of methylated genes. Venn-diagram depicting the overlap between genes methylated in S. cerevisiae (blue), S. mikatae (orange) and both (pink), and the associated hypergeometric P value. (D) Significant conservation of methylated sites. The proportion of sites detected in S. cerevisiae that are also detected within the orthologous 100-nt region in S. mikatae (pink bar). Blue bar: proportions for a random set of controls. (E) m6A-seq profiles for two example meiosis genes with orthologous methylated positions. (F,G) Stronger and more 3’ sites are more conserved. Proportion of conserved sites (Y axis), as a function of distance from the 3’ end of the transcript (F), or of peak score in S. cerevisiae (G). Error bars: SEM.
There is a highly statistically significant overlap in methylated genes between the two species (229/610 methylated genes in S. mikatae are also methylated in S. cerevisiae, P=4.6×10−27; Figure 3C). This extent of conservation is similar to, albeit slightly lower than, that previously observed for transcription factor binding events (Borneman et al., 2007). In 54 cases, the methylated sites were at precisely orthologous (conserved) positions (e.g., Figure 3D,E), 64 were within a 100-nt window (possibly reflecting positional bias), and the remainder were more distant. The extent of conservation was significantly higher than expected by chance (5 and 16, respectively; Experimental Procedures), and increased with 3’ proximity (Figure 3F) and peak strength (Figure 3G), suggesting that conserved sites are more likely to be functional. Thus, our results suggest that mRNA methylation is conserved at the gene level, although there is substantial turnover of both sites and targeted transcripts, consistent with the evolutionary patterns of cis-regulatory sequences.
Dynamic changes in methylation during yeast meiosis
The dynamic nature of methylations in yeast offers a unique opportunity to explore their onset and offset. We measured the RNA methylome with m6A-Seq at six time points along a 3-hour time-course from induction of sporulation until meiotic prophase, and five timepoints along a 105 minute time-course after release from prophase arrest (Figure 4A,B).
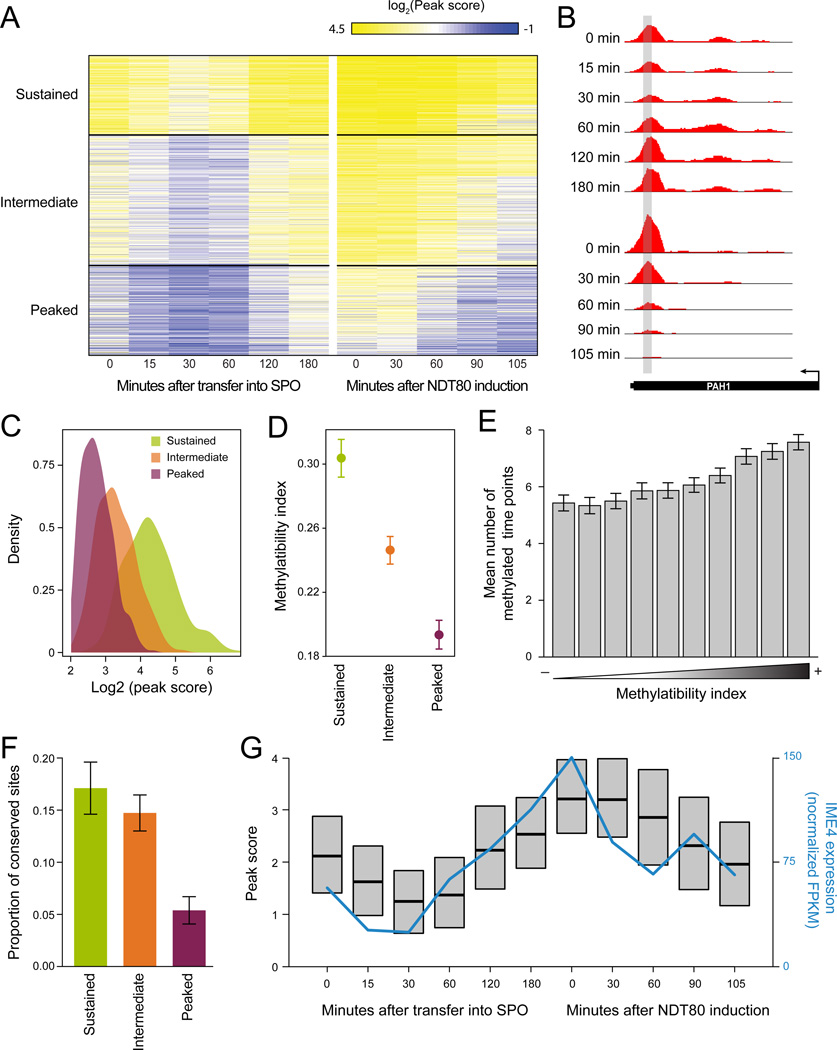
Figure 4. Dynamic changes in methylation across meiosis reflect inherent methylatability.
(A) Sustained, intermediate and peaked methylation profiles across a meiosis time course. Peak scores (as in Figure 1) for 1,308 peaks (rows) at 6 time points up to prophase arrest, and five time points following NDT80 induction and release from arrest (SAy995) (columns). Sites clustered using k-means. (B) m6A-seq at the ‘sustained’ PAH1 transcript. (C) The temporal window of methylation is consistent with the peak score. Density plots of the distributions of peak-score at prophase arrest (Figure 1A) in the sustained (green), intermediate (orange) and peaked (purple) clusters. (D,E) The temporal window of methylation is longer for genes with higher methylatability index. (D) Mean methylatability index (Y axis; error bars: SEM) for transcripts in the sustained (green), intermediate (orange) and peaked (purple) clusters. (E) Barplots of the average span of methylations (number of timepoints throughout methylation in which peak scores were greater than 2) at 10 quantiles of methylatability index (X axis). Error bars: SEM. (F) Sustained and intermediate methylated sites are more likely to be conserved. Proportion of conserved sites between S. cerevisiae and S. mikatae (Y axis) in each of the three temporal clusters (X axis). (G) IME4 expression correlates with average methylation. Box plots of the distributions (interquartile range and medians) of peak scores across the timecourse, and IME4 expression levels by RNA-Seq across the time points.
The temporal methylation profiles were partitioned to three clusters: A ‘sustained’ cluster of sites methylated throughout the time course (26% of sites); a ‘peaked’ cluster of sites methylated only during a narrow time-window in meiosis prophase (43%), and an ‘intermediate’ cluster of sites methylated during a broader window (31%). The extent of the methylated window correlated with the peak score (Figure 4C), the Methylatability Index (Figure 4D–E), the presence of an ‘A’ at position −4 of the consensus (P=0.03; Figure S4A), 3’ bias of the site (P=5.4×10−68, Figure S4B), and the extent of conserved methylation in S. mikatae (P=2.6×10−5, Figure 4F). Thus, a site’s Methylability Index and its conservation may reflect both the extent of methylation and its temporal span.
Methylation scores peaked at meiotic prophase across all clusters (Figures 4A,G), with onset correlating with increased accumulation of all MIS complex components (Fig. 4F, S4C), but offset correlating only with decreased expression of IME4 (Figure 4G, Figure S4C). Collectively, these results suggest a model where the methylation profiles across yeast meiosis are determined locally (‘in cis’) via the inherent methylatability of the transcript, and globally (‘in trans’) via the interplay of different components of the methylation machinery.
m6A methylation is induced by IME1-mediated induction of SLZ1
To dissect the global regulation imposed on the meiotic methylation machinery, we examined the role of IME1, the master regulator of meiosis in yeast. We found that IME1 is required for mRNA methylation by the MIS complex in the meiotic cell cycle. Diploid cells lacking IME1 (ime1Δ/Δ) failed to accumulate m6A-mRNA during meiosis (Figure 5A). This was not merely due to defects in progression through the meiotic cell cycle, as deletion of IME2, which arrests cells at the same point as ime1Δ/Δ, did not affect m6A-mRNA levels.
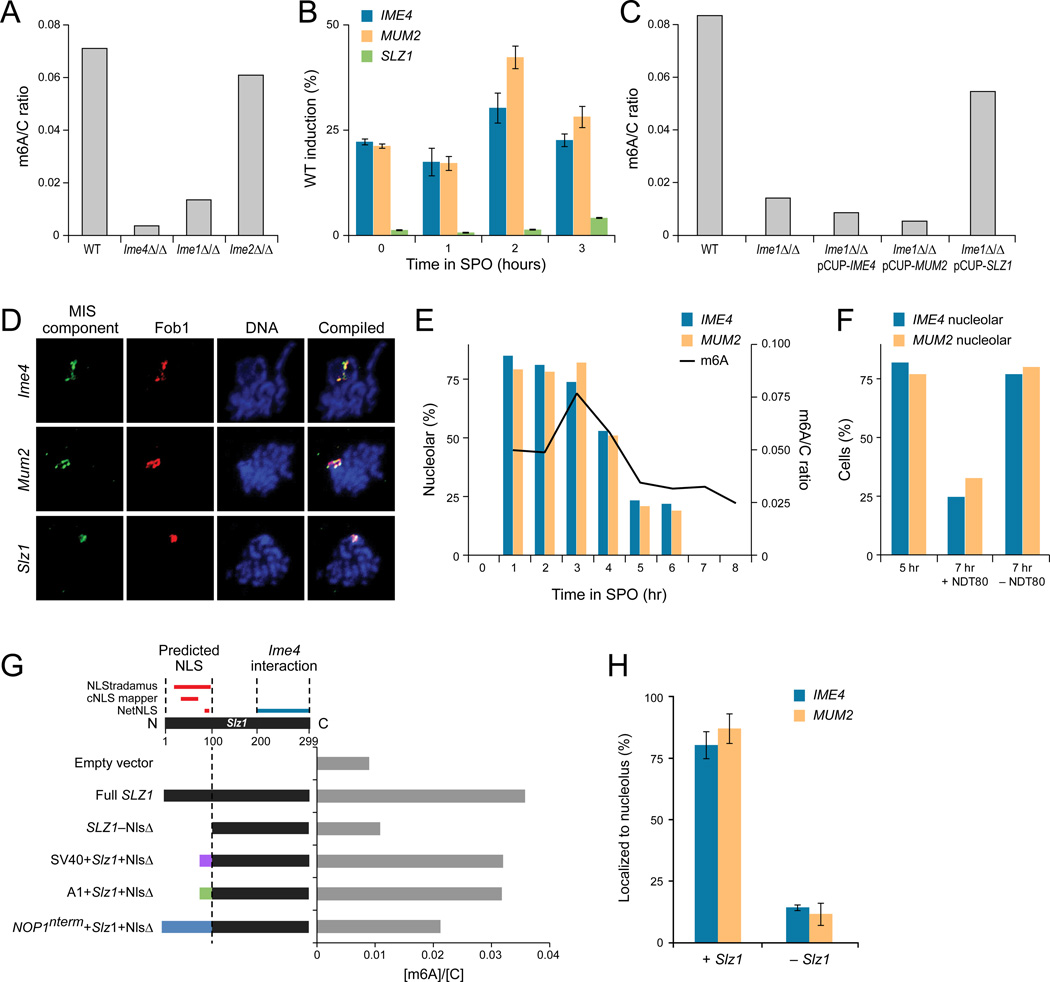
Figure 5. IME1 regulates MIS complex induction and nucleolar localization.
(A) Ime1 and Ime4 are essential for m6A methylation in meiosis. TLC-based quantification of mRNA m6A relative to cytosine 3 hours after meiotic starvation, when m6A accumulation is maximal in WT cells (SAy821). m6A levels are reduced in mutants in early meiosis genes ime1Δ/Δ (SAy834) and ime4Δ/Δ (SAy1196) but not ime2Δ/Δ (SAy771). (B) Ime1 regulates MIS complex gene expression. Induction of IME4 (blue), MUM2 (red), and SLZ1 (green) transcripts in ime1Δ/Δ background relative to wild-type (by qPCR). Error bars: standard deviation of four replicates. (C) SLZ1 – but not IME4 or MUM2 – overexpression rescues the ime1Δ/Δ methylation defect. Quantification of mRNA m6A relative to cytosine 3 hours after meiotic starvation. Wild-type (SAy821) levels are reduced in ime1Δ/Δ (SAy834) background. Conditional expression of IME4 or MUM2 from the CUP1 promoter in ime1Δ/Δ (SAy1383, SAy1384, respectively) does not overcome this defect, whereas expression of SLZ1 does (SAy1385). (D) The MIS complex localizes to the nucleolus during meiosis. Representative images of immunofluorescence of spread meiotic nuclei, showing colocalization of epitope-tagged Ime4 (SAy914), Mum2 (SAy1235) or Slz1 (SAy1254) (first column) with the nucleolar marker Fob1 (second column). Blue: DNA (DAPI—third column). Compilation (fourth) column: DNA: blue, MIS component: green, Fob1: red. (E) Nucleolar entry and exit of the MIS complex correspond to onset and offset of m6A methylation during meiosis. Quantification of the percentage of cells (Y axis) that show nucleolar co-localization of either Ime4 (blue bars) or Mum2 (red bars) upon induction into sporulation medium. Nucleolar co-localization was determined by immuno-fluorescence of nuclear spreads (n=100 Fob1 foci/time point); percent co-localization at 7 and 8 hours was not quantified as the majority of cells were spores at this time-point. Black curve: m6A abundance relative to cytosine throughout meiosis (right axis). All data were collected from a single meiosis in strain SAy1232. (F) Nucleolar exit dependent on NDT80. % cells showing nucleolar co-localization of Ime4 (blue bars) and Mum2 (orange bars) after treatment with 1µM estradiol (+NDT80) or with solvent (−NDT80) (strain SAy1469). Cells were treated at 5 hours and assayed for nucleolar co-localization after two hours of development in the respective medium. Nucleolar co-localization was determined by immunofluorescence of nuclear spreads (n=100 Fob1 foci/time point). (G, top) Schematic of domains of Slz1, showing predicted nucleolar localization sequence. (G, bottom) Levels of m6A normalized by cytosine (X axis, blue bars) in IME1 deletion strains with either an empty expression vector (empty plasmid--SAy1432), or various alleles of SLZ1 (SAy1422, 1434, 1438, 1441 and 1439, respectively). Samples were taken three hours after strains were induced into meiosis. (H) Quantification of nucleolar colocalization events of either epitope-tagged Mum2 (orange) or Ime4 (blue) with Fob1 in an ime1Δ/Δ PCUP1-SLZ1 strain (SAy1385) with either induction (+SLZ1, left) or no induction (−SLZ1, right). Error bars: standard deviation of three time-points during meiotic prophase, n=100 cells per time-point.
The mRNA methylation defect of ime1Δ/Δ strains likely results from a failure to express SLZ1, a MIS complex component. First, in an ime1Δ/Δ mutant, SLZ1 transcription is less than 5% of that of wild-type (Figure 5B), whereas that of the other MIS components, IME4 and MUM2, is less substantially reduced. The previously-characterized dependence of SLZ1 activation on Ume6 (the DNA-binding component of the Ime1 transcriptional activation complex) and the presence of a Ume6 DNA-binding motif in the SLZ1 promoter (Williams et al., 2002) further suggest that Ime1 directly activates SLZ1 transcription. Second, overexpression of SLZ1 from the inducible CUP1 promoter in an ime1Δ/Δ mutant restores accumulation of meiotic m6A mRNA, whereas overexpression of either IME4 or MUM2 in the ime1Δ/Δ mutant cannot bypass the m6A accumulation defect (Figure 5C).
mRNA methylation requires nucleolar localization of the MIS complex
To determine the cellular compartment in which m6A methylation occurs, we next used immunofluorescence (IF) of each of the three components of the MIS complex at meiotic prophase, when m6A-mRNA levels are maximal. IF of nuclear spreads showed that MIS components were largely excluded from the meiotic chromosomes, but concentrated in a small compartment of the chromosomes that co-stained with Fob1, a marker of the nucleolus (Figure 5D). In addition, whole-cell IF revealed these components are also found in the cytoplasm (Figure S5A).
Localization of the MIS complex along a sporulation time course (Figure 5E) shows that the complex localizes to the nucleolus only during the induction of m6A mRNA methylation; nucleolar co-localization is subsequently lost at later phases, corresponding to return to basal levels of m6A mRNA. Both Mum2 and Ime4 co-localized with the nucleolus during the period of m6A mRNA accumulation (between pre-meiotic S phase and induction of the meiotic divisions MI and MII, ~3 hours in SPO medium) (Agarwala et al., 2012), after IME1 induction and before NDT80 induction (Figure S5B). Conversely, as m6A mRNA levels returned to basal levels upon NDT80 induction and the meiotic divisions (Figure 5E, S9B, 4–7h in SPO), Ime4 and Mum2 lost their nucleolar co-localization (Figure 5E), although both remained in cytoplasmic puncta throughout the meiotic time-course (Figure S5C).
Loss of nucleolar localization and offset of m6A methylation in later phases of meiosis (Figure 5E) depend on Ndt80 activation. Cells that do not express NDT80 at the end of meiotic prophase retain high levels of m6A mRNA (Agarwala et al., 2012) and nucleolar co-localization of both Ime4 and Mum2 (Figure 5F). Induction of NDT80 at this time point, however, leads to the down-regulation of mRNA methylation (Agarwala et al., 2012), and loss of the nucleolar localization of Ime4 and Mum2 (Figure 5F).
Nucleolar entry of the MIS complex is mediated by Slz1
MIS complex entry to the nucleolus is regulated by Slz1 and is necessary for full levels of m6A mRNA accumulation in meiosis. Sequence analysis indicates that Slz1, but not Mum2 and Ime4, has a nuclear localization signal (NLS) peptide encoded in its N-terminus (Figure 5G). Neither Ime4 nor Mum2 localize to the nucleolus during meiosis in the ime1Δ/Δ strain, in which Slz1 is not expressed and there is a defect in mRNA methylation. Expression of Slz1 in this strain restored mRNA methylation and nucleolar localization of both proteins (Figure 5H). Expression of a SLZ1 allele lacking its NLS failed to overcome the m6A defect. However, replacing the NLS with either the SV40 NLS, the non-canonical A1 NLS, or the n-terminal signal sequence of the nucleolar structural protein Nop1 mitigated this defect (Figure 5G). These data suggest that Slz1 is responsible for the nucleolar localization of Ime4 and Mum2, and that this localization is necessary for full levels of m6A mRNA accumulation in meiosis.
Evolutionary analysis reveals co-evolving m6A ‘writers’ and ‘readers’ from yeast to mammals
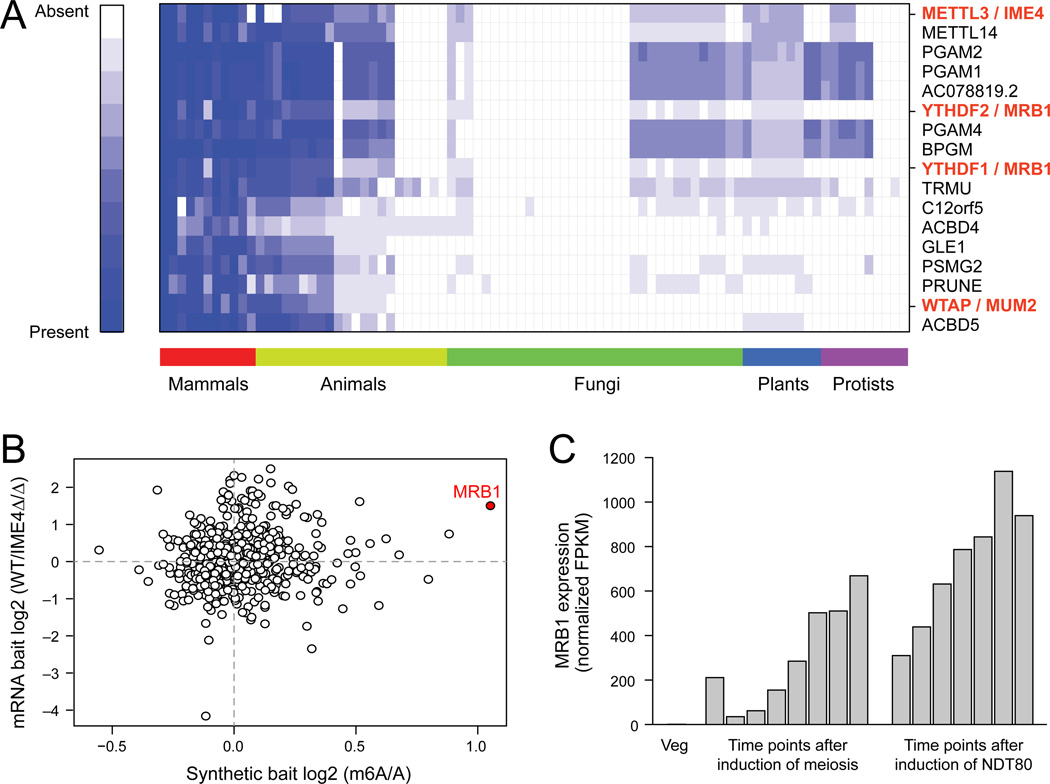
To identify additional components of the methylation program, we searched for proteins sharing a similar evolutionary trajectory with METTL3, the mammalian ortholog of IME4, across 86 eukaryotic species, spanning mammals, other animals, fungi, plants and protists (Tabach et al., 2013a; Tabach et al., 2013b) (Figure 6A). The top 20 co-evolving genes included Wilms Tumor 1 Associated Protein (WTAP), the mammalian ortholog of MUM2, and FIP37, a plant ortholog (Zhong et al., 2008), and the proteins from the YTH family, two members of which (YTHDF2 and YTHDF3) we have previously found as m6A ‘readers’ that selectively bind m6A in mammals (Dominissini et al., 2012). The yeast homolog identified in this analysis is ydr374c, henceforth referred to as Methylated RNA Binding Protein 1 (MRB1).
Figure 6. Co-evolution of m6A ‘writers’ and ‘readers’ across eukaryotes.
(A) Co-evolution of m6A readers and writers. Phylogenetic profiles for the top 20 proteins co-evolving with human METTL3 across 85 other eukaryotic genomes. For each query protein, the normalized ratio of the BLAST score for the top-scoring protein sequence similarity is indicated in the cell corresponding to each genome. White: 0, no similarity; Blue: 1, 100% similarity. (B) MRB1 binds methylated RNA. Mass-spectrometry based quantification of the levels of proteins pulled down using methylated and non-methylated RNA baits (X axis – in-vitro transcribed, Y axis – poly(A) selected RNA from WT or IME4Δ/Δ strains). Red: MRB1. (C) MRB1 expression is induced during meiosis. RNA-seq derived expression levels (TMM-normalized FPKM values) of MRB1 across a meiotic time course.
To validate MRB1 as an m6A ‘reader’, we used affinity chromatography proteomics to identify proteins that selectively bind methylated RNA, compared to non-methylated counterparts. We exposed protein lysate isolated from meiotic yeast cultures to either methylated or non-methylated baits, and analyzed the bound proteins by quantitative mass-spectrometry (Experimental Procedures). Strikingly, Mrb1 was the top candidate showing reproducible preferential association with methylated baits (Figure 6B) and the only one with a >2 fold enrichment in both experiments. MRB1 is expressed in a meiosis-specific manner, consistent with the meiosis restricted methylation (Figure 6C), and its deletion led to defects in meiotic progression, albeit less severe than in the ime4Δ/Δ strain (Figure S6). These data support a role for Mrb1 as an m6A ‘reader’, conserved from yeast to mammals, highlighting the overall conservation of this ancient cellular function.
Discussion
High-resolution methylation maps
We used our dynamic m6A maps during yeast meiosis to identify high confidence methylated sites at nearly single nucleotide resolution. We achieved this resolution by optimizing the pull-down protocol and eliminating many false positives with methylation-defficient control strains. This is a substantial improvement over mammalian m6A-seq studies (Dominissini et al., 2012; Meyer et al., 2012), and dramatically expands over the single methylated site previously validated (Narayan and Rottman, 1988). We found that ~50% of identified peaks are false positives, highlighting the importance of our controls and filtering criteria, using (1) strains/conditions in which methylation is globally absent, and (2) in vitro transcribed controls. The latter may be particularly useful in mammals, where depletion of the methyltransferase complex leads to apoptosis (Bokar, 2005; Dominissini et al., 2012). Such false positives may impact other nucleotide affinity assays (e.g., meDIP). Nonetheless, it is possible that our stringent filtering results in false negatives, and that some residual false positives remain.
Conservation of methylation machinery and topology
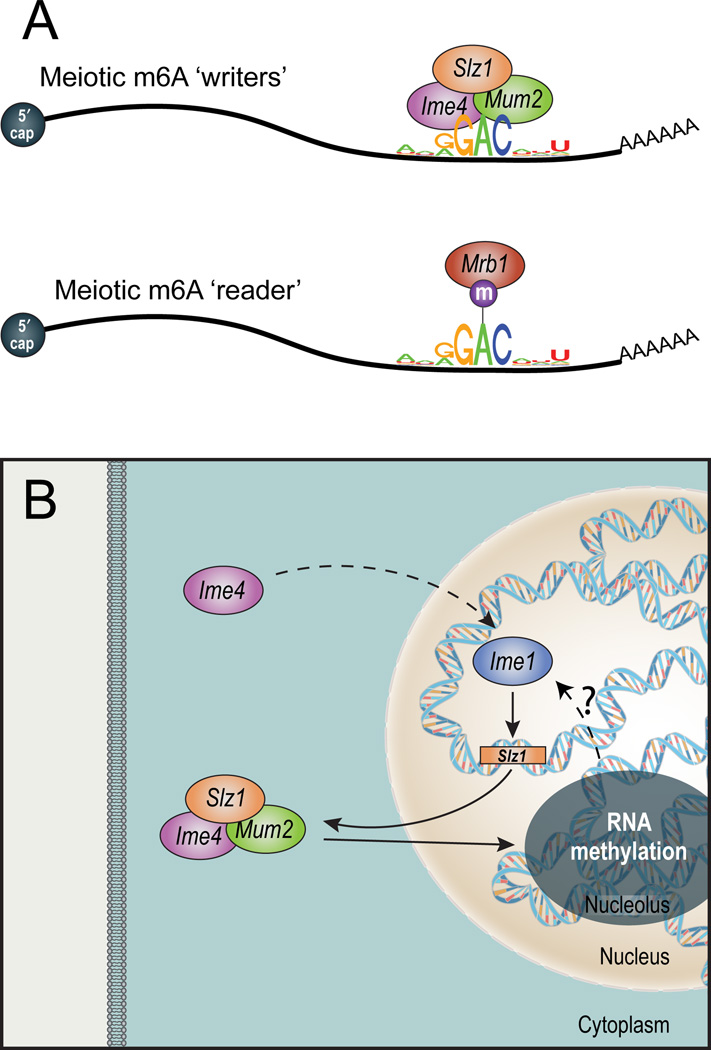
We uncovered several striking similarities between mRNA methylation in yeast and mammals, suggesting that yeast is a good general model for m6A methylation. In both yeast and mammals methylations occur at an RGAC core consensus (Dimock and Stoltzfus, 1977; Dominissini et al., 2012; Meyer et al., 2012; Narayan et al., 1994; Schibler et al., 1977; Wei et al., 1976), albeit with divergence of the broader consensus. Mammalian methylations are highly enriched near stop codons (Dominissini et al., 2012; Meyer et al., 2012) and in yeast, they are 3’ biased and tightly correlated with the stop codon (only 4.7% of the identified peaks occur in the 3’ UTR). m6A ‘writers’ (mammalian METTL3/WTAP, yeast IME4/MUM2) and ‘readers’ (mammalian YTHDFs, yeast MRB1) have co-evolved. The YTH RNA binding domain is present across all eukaryotes (Zhang et al., 2010). Notably, the Schizosaccharomyces pombe MRB1 ortholog, mmi1, is essential for eliminating meiosis-specific transcripts during vegetative growth (Harigaya et al., 2006), suggesting a common meiosis-related role of methylation in both species.
A key difference between mammal and yeast is in the dynamic nature of the methylation program. In mammals, methylation profiles are similar across examined conditions and ‘readers’ and ‘writers’ are broadly expressed, whereas in yeast, both methylation protein expression and the methylation program are confined to meiosis. While demethylation is controlled by an active process in human through the RNA demethylases FTO (Jia et al., 2011) and Alkbh5 (Zheng et al., 2013), their orthologs are not detectable in yeast, where methylation offset is coupled to MIS complex down regulation and likely proceeds passively through RNA degradation Indeed, a longer temporal span of methylation (sustained, intermediate or peaked, Figure 4) is associated – on average – with longer half-lives (under vegetative conditions (Miller et al., 2011)), supporting a passive model (Figure S7A). However, as degradation may both affect and be affected by methylations fully resolving this interplay will require quantitative monitoring and formal modeling.
Cis and trans regulation of methylation
The extent and temporal span of methylations are explained via a combination of cis and trans elements. At the cis level, the extended sequence motif, 3’-position, and lack local secondary structure all increase methylation level and span, with the latter two helping explain why only 1 in ~40 RGAC sites and only 1 in 10 extended ANNRGACNNT sites are methylated (Extended Experimental Procedures). The lack of secondary structure is consistent with findings that duplex RNA structures are incapable of m6A methylation (Narayan et al., 1994), possibly since structured sites are inaccessible to the MIS complex. A yeast cell may use differential methylation in a temporally ordered way across transcripts, analogous to ordered temporal regulation across a regulon (Kalir et al., 2001).
At the trans level, the global methylation peak at meiotic prophase is regulated by IME1 (onset) and NDT80 (offset). Onset is mediated by Ime1-dependent induction of SLZ1, leading to nucleolar localization of the complex. Notably, IME4 is necessary for IME1 accumulation (Shah, 1994, Hongay 2006, van Werven 2012), and we find that MIS complex activity is necessary for full IME1 induction (Figure S7B–D), suggesting a putative positive feedback loop between RNA methylation and IME1 expression (Figure 7B). Offset of the methylation program is triggered by NDT80 induction, leading to exit of the MIS complex from the nucleolus, down-regulation of IME4 and termination of the methylation program.
Figure 7. Key elements in the yeast meiotic methylation program.
(A) The core methylation machinery. The MIS complex (top), active during meiotic prophase, methylates within a sequence motif that is typically 3’ biased and unstructured. The ‘reader’ MRB1 (bottom) binds the methylated motif. (B) Meiotic regulation of mRNA methylation. IME4 induction leads to IME1 accumulation, which induces SLZ1 expression. SLZ1 forms the MIS complex with MUM2 and IME4, and shuttles them into the nucleolus, where mRNA methylation occurs. mRNA methylation may be required for IME1 activation (dashed arrow, ?), thereby possibly closing a positive feedback loop.
Nucleolar localization of the MIS complex
Although the nucleolus is primarily associated with ribosome biogenesis, recent studies have implicated it in mitosis regulation (reviewed in (Boisvert et al., 2007)). Intriguingly, Misu/Nsun2, an m5C RNA methyltransferase of both tRNA and mRNA (Hussain et al., 2013; Khoddami and Cairns, 2013; Squires et al., 2012), localizes to the nucleolus during interphase (Hussain et al., 2009). Thus, nucleolar localization of the MIS complex raises speculations about nucleolar regulation of both m5C and m6A RNA methyltransferases in mitosis and meiosis, as well as the possibility of coordination between mRNA methylation and ribosome biogenesis (we found no evidence for methylated rRNA in our data; data not shown). Notably, mammalian RNA methylation and demethylation occur at nuclear speckles (Bokar et al., 1997; Zheng et al., 2013), which do not appear to have a parallel in yeast (Potashkin et al., 1990).
Possible functions of methylation
IME4 deletion delays meiotic progression, as reflected by delays in the induction of NDT80 and of the two meiotic divisions (Agarwala et al., 2012), and earlier delays in the formation of the synaptonemal complex and double strand breaks (Figure S7E–G). However, the molecular function of m6A-methylation remains unknown. We observed only modest changes (up to 40% difference) in steady state expression levels in 5 of the 8 strains in which we eliminated methylation by point mutating the consensus sequence. Three genes had increased levels of expression, and two decreased (Figure S7H). Furthermore, we observed only a minor global effect of methylations on transcript stability in WT compared to ime4Δ/Δ strains, based on monitoring of expression levels across a time-course following transcriptional shut-off via thiolutin (data not shown). Our high-resolution yeast methylome and the yeast’s tractability will help test further hypotheses on methylation’s role in mRNA processing, export, localization, or translation.
Conclusion
The induction of the MIS complex during meiosis, the regulated temporal dynamics of the methylations, and the impaired progression through meiosis of the IME4 catalytic mutant strain all strongly suggest that m6A methylation plays a critical role in yeast meiosis. The striking similarities in methylation profiles and components between yeast and mammals suggests that yeast is a compelling model system for studying the role of methylations. Our high resolution dynamic maps will pave the way to a better understanding of the roles of RNA methylation and to advance this rapidly emerging field coined RNA epitranscriptomics (Saletore et al., 2012).
Experimental Procedures
m6A-seq and analysis
Meiosis was induced, and mRNA was extracted as detailed in (Agarwala et al., 2012). The m6A pull-down procedure and library preparation is described in Extended Experimental Procedures. We used a modified version of the approach described in (Dominissini et al., 2012) for aligning reads and detecting putative methylated sites based on (1) enrichment at a certain region within a gene, compared to background region and (2) absence of this enrichment in the input sample. This pipeline was extended to allow comparison of called peaks across multiple samples/conditions, based on overlapping genomic coordinates. De novo motif analysis and secondary structure analysis was performed as described in (Dominissini et al., 2012).
In vitro transcription of IME4-independent peaks
150-nt long DNA templates tiling across 17 genes containing IME4-independent peaks were synthesized on a CustomArray 12K Microarray using a B3 Synthesizer (CustomArray, Bothell, WA), and subjected to in-vitro transcription.
Methylation classifier
A logistic regression classifier was trained to discriminate between a set of RGAC sites and non-methylated RGAC counterparts. Features used were relative position within the gene, nucleotide composition of positions −4 to +5 with respect to the methylated position, and predicted secondary structure strength. A 10-fold cross validation scheme was designed, and implemented using the RWeka package (Hornik et al., 2009) in R.
RNA affinity chromatography and mass spectrometry
Three RNA baits, each comprising a ~120 nt long sequence containing only a single adenine were in vitro transcribed from dsDNA templates, either in the presence of ATP or N6-methyl-ATP. These were exposed to proteins isolated from meiotic cultures in prophase arrest. Isolated proteins were labeled via iTRAQ and subjected to Mass-Spectrometry as described in (Mertins et al., 2013).
Additional Methods
Spread meiotic nuclei were prepared using the method described in (Falk et al., 2010). TLC analysis was carried out as in (Zhong et al., 2008); mRNA was purified with the Dynabeads mRNA purification system (Invitrogen) and analyzed on cellulose plates (20cm × 20cm) from EMD.
Accession Numbers: Sequencing data have been deposited into the Gene Expression Omnibus (GEO, accession number GSE51583).
See Extended Experimental Procedures for full details.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwala S, Blitzblau H, Hochwagen A, Fink G. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS genetics. 2012;8 doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z, Button J, Grierson D, Fray R. Yeast targets for mRNA methylation. Nucleic acids research. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nature reviews Molecular cell biology. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Bokar JA. Grosjean Henri., editor. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. Fine-Tuning of RNA Functions by Modification and Editing. 2005;12:141–177. [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA (New York, NY. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science (New York, NY. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Bringmann P, Luhrmann R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS letters. 1987;213:309–315. doi: 10.1016/0014-5793(87)81512-0. [DOI] [PubMed] [Google Scholar]
- Clancy M, Shambaugh M, Timpte C, Bokar J. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic acids research. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers RC, Friderici KH, Rottman FM. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5' terminus. Biochemistry. 1975;14:4367–4374. doi: 10.1021/bi00691a004. [DOI] [PubMed] [Google Scholar]
- Dimock K, Stoltzfus CM. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. 1977;16:471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nature protocols. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Falk JE, Chan AC, Hoffmann E, Hochwagen A. A Mec1- and PP4-dependent checkpoint couples centromere pairing to meiotic recombination. Developmental cell. 2010;19:599–611. doi: 10.1016/j.devcel.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci U S A. 2011;108:14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornik K, Buchta C, Zeileis A. Open-Source Machine Learning: R Meets Weka. Computational Statistics. 2009;24:225–232. [Google Scholar]
- Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, Frye M. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. The Journal of cell biology. 2009;186:27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al. NSun2-Mediated Cytosine-5 Methylation of Vault Noncoding RNA Determines Its Processing into Regulatory Small RNAs. Cell reports. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang Y-G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature chemical biology. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science (New York, NY. 2001;292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nature biotechnology. 2013;31:458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Saletore Y, Zumbo P, Elemento O, Mason C, Jaffrey S. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3' UTRs and near Stop Codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Molecular systems biology. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P, Ludwiczak RL, Goodwin EC, Rottman FM. Context effects on N6-adenosine methylation sites in prolactin mRNA. Nucleic acids research. 1994;22:419–426. doi: 10.1093/nar/22.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P, Rottman FM. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science (New York, NY. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- Potashkin JA, Derby RJ, Spector DL. Differential distribution of factors involved in pre-mRNA processing in the yeast cell nucleus. Molecular and cellular biology. 1990;10:3524–3534. doi: 10.1128/mcb.10.7.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome biology. 2012;13:175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. Journal of molecular biology. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Molecular and cellular biology. 1992;12:1078–1086. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires J, Patel H, Nousch M, Sibbritt T, Humphreys D, Parker B, Suter C, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic acids research. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Billi AC, Hayes GD, Newman MA, Zuk O, Gabel H, Kamath R, Yacoby K, Chapman B, Garcia SM, et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013a;493:694–698. doi: 10.1038/nature11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Golan T, Hernandez-Hernandez A, Messer AR, Fukuda T, Kouznetsova A, Liu JG, Lilienthal I, Levy C, Ruvkun G. Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Molecular systems biology. 2013b;9:692. doi: 10.1038/msb.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B. 5'-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. 1976;15:397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
- Williams RM, Primig M, Washburn BK, Winzeler EA, Bellis M, Sarrauste de Menthiere C, Davis RW, Esposito RE. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc Natl Acad Sci U S A. 2002;99:13431–13436. doi: 10.1073/pnas.202495299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FH, et al. The YTH domain is a novel RNA binding domain. J Biol Chem. 2010;285:14701–14710. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. The Plant cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.