1. Introduction
The use of attenuated bacteria as vaccine delivery vehicles for heterologous antigens has been studied extensively in both animals and humans. Attenuated Salmonella is an attractive choice due to its ability to stimulate both cell and humoral-mediated immunity against a heterologous antigen and thus provide protection against pathogen challenge [1–3] when given orally. An optimal live oral Salmonella vaccine would retain its ability to colonize and invade host lymphoid tissues while remaining completely avirulent [4]. Several Salmonella strains with various modifications have been created in an attempt to balance safety and immunogenicity (see reviews [1, 3, 5]), however, there is a need for improved strains.
A safe vaccine should carry multiple attenuating mutations. Some target genes for attenuation are those involved in the synthesis of lipopolysaccharide (LPS). LPS is a structural component of the gram-negative outer membrane. Full length (smooth) LPS is comprised of three components – lipid A, also a potent activator of the innate immune response, the core oligosaccharide, and O-antigen, a polymer of repeating sugar units.
Four LPS genes that have been shown previously to be useful deletion targets for Salmonella vaccines are galE, pmi, rfaH and rfc. Two of these genes, galE and pmi, are involved in synthesizing sugars required for core and/or O-antigen synthesis. GalE is a UDP-galactose epimerase that inter-converts UDP-glucose and UPD-galactose, an essential part of core sugar and O-antigen. This mutant synthesized core-defective LPS in the absence of galactose but made normal LPS when galactose was available in the growth media [6, 7]. S. Typhimurium galE strains are avirulent and immunogenic [6]. However, this same mutation, transferred to S. Typhi, was not attenuated and was poorly immunogenic in humans [8].
The pmi gene codes for phosphomannose isomerase, which converts fructose-6-P to mannose-6-P [9]. The deletion mutant is unable to synthesize the O-antigen due to unavailability of mannose, an O-antigen sugar. When the mutant is grown in the presence of mannose, the smooth LPS phenotype is exhibited. S. Typhimurium strains with a pmi deletion are attenuated and immunogenic [10]. We constructed and evaluated a Δpmi mutant in our lab, confirming previous results [10–12]. This mutant was attenuated as expected, but also induced antibodies that cross-reacted with outer membrane proteins from other Salmonella serovars and other gram-negative bacteria [12]. A pmi deletion in Typhi has not yet been evaluated in humans. Note that both galE and pmi mutant strains, if grown in the presence of galactose or mannose, respectively, transiently express LPS before colonizing the gut-associated lymphoid tissue (GALT) or organs [11, 13]. Because these mutants are essentially wild-type at the time of immunization and become attenuated in host tissues, we have termed this effect regulated delayed attenuation.
RfaH is a transcriptional anti-terminator required for the synthesis of many virulence determinants including O-antigen, core sugar, capsular polysaccharide, and Vi antigen [14, 15]. An rfaH deletion mutant, described as “gently rough”, exhibited some deep-rough characteristics, i.e. lack of O-antigen and outer core, sufficient attenuation, susceptibility to detergents and to some antibiotics, but still proved to be attenuated and immunogenic [14, 16].
Rfc (Wzy) is the O-antigen polymerase that, in conjunction with Wzx (transporter), Wzz (length determinant) and WbaP (O-antigen synthesis initiation), synthesizes, assembles, and transports the O-antigen to the periplasm, where WaaL (ligase) ligates O-antigen to lipid A to form complete LPS [17–19]. rfc mutants exhibit a characteristic “semi-rough” phenotype, making lipid A and core capped with a single O-antigen subunit. An S. Typhimurium rfc mutant administered orally to mice was avirulent and protective against oral challenge with wild-type S. Typhimurium [10].
Unlike pmi and galE mutants, strains with mutations in rfaH or rfc do not transiently express full length O-antigen and therefore may be impaired in their ability to invade and colonize host tissues effectively. A tightly regulated araC PBAD activator-promoter has been used extensively in our lab to regulate gene expression [12, 20]. In a previous study, we replaced the rfaH promoter with araC PBAD and the resulting strain exhibited arabinose-dependent O-antigen synthesis [21]. The promoter substitution mutation was then introduced into attenuated S. Typhimurium strain χ9241 (ΔpabA ΔpabB ΔasdA). The resulting strain was found to be superior to a χ9241 ΔrfaH strain for delivery of the pneumococcal protein PspA, eliciting significantly greater protection against challenge with virulent Streptoccocus pneumoniae [21].
In this study, we evaluated whether this regulated delayed attenuation approach would provide advantages when used with rfc. We replaced the rfc promoter with an araC PBAD promoter to create arabinose inducible production of Rfc and thus regulate rfc expression to mimic transient expression of smooth LPS. We evaluated a series of rfc promoter mutations in vitro and introduced one of them into vaccine strain χ9241. The resulting strain was evaluated for its ability to deliver the heterologous protein PspA to mice.
2. Materials and Methods
2.1. Bacterial strains, plasmids, media, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. S. Typhimurium cultures were grown at 37°C in LB broth [22], nutrient broth (NB) (Difco) or on LB agar with or without 0.1% arabinose. Selenite broth, with or without supplements, was used for enrichment of S. Typhimurium from mouse tissues. Diaminopimelic acid (DAP) was added (50 μg/ml) for the growth of Δasd strains [23]. LB agar containing 5% sucrose was used for sacB gene-based counterselection in allelic exchange experiments. S. pneumoniae WU2 was cultured on brain heart infusion agar containing 5% sheep blood or in Todd-Hewitt broth plus 0.5% yeast extract, and was kept at −80°C for future use. MOPS minimal medium [24] with/without 10 μg/ml p-aminobenzoic acid (pABA) was used to confirm the phenotype of ΔpabA ΔpabB mutants.
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Description | Source |
|---|---|---|
| χ3761 | S. Typhimurium UK-1, Wild-type | [46] |
| χ9241 | ΔpabA1516 ΔpabB232 ΔasdA16 ΔaraBAD23 ΔrelA198::araC PBAD lacI TT |
[31] |
| χ9659 | ΔPrfc173::TT araC PBAD rfc | χ3761 |
| χ9736 | ΔPrfc174::TT araC PBAD rfc | χ3761 |
| χ9737 | ΔPrfc175::TT araC PBAD rfc | χ3761 |
| χ9944 | Δrfc-48 | χ3761 |
| χ9430 | ΔpagL7 | χ3761 |
| χ9885 | ΔpabA1516 ΔpabB232 ΔasdA16 ΔaraBAD23 ΔrelA198::araC PBAD lacI TT Δrfc-48 |
χ9241 |
| χ9853 | ΔpabA1516 ΔpabB232 ΔasdA16 ΔaraBAD23 ΔrelA198::araC PBAD lacI TT ΔPrfc174::TT araC PBAD rfc |
χ9241 |
| χ9761 | Δ(galE-uvrB)-1005 ΔmsbB48 ΔfliC2426 ΔpefA1225 ΔfimA2119 ΔfimH1019 ΔagfBAC811 |
Lab stock |
| E. coli K-12 | ||
| χ7213 |
thi-1 thr-1 leuB6 glnV44 fhuA21 lacY1 recA1 RP4-2-Tc::Mu[λ pir] ΔasdA4 Δ(zhf-2::Tn10) |
[47] |
| χ7232 |
endA1 hsdR17 (rK− mK+) glnV44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 λ pir deoR (ϕ80dlac Δ(lacZ)M15) |
[47] |
| S. pneumoniae WU2 | Wild-type virulent, encapsulated type 3 | [48] |
| Suicide vectors | ||
| pRE112 | sacB mobRP4 R6K ori Cm+ | [49] |
| pYA4284 | pRE112, ΔpagL7 | Lab stock |
| pYA4717 | Constructed for rfc-48 deletion | pRE112 |
| pYA4297 | Constructed for Prfc173 promoter deletion and araC PBAD promoter insertion ctcgag AGGA gtcattATGa | pRE112 |
| pYA4298 | Constructed for Prfc174 promoter deletion and araC PBAD promoter insertion ctcgag AGGA gtcattGTG | pRE112 |
| pYA4299 | Constructed for Prfc175 promoter deletion and araC PBAD promoter insertion ctcgag GAGG gtcattGTG | pRE112 |
| Recombinant Plasmid | ||
| pYA3700 | TT araC PBAD cassette plasmid; Apr | [50] |
| pYA3493 | Plasmid Asd+; pBRori β-lactamase signal sequence-based periplasmic secretion plasmid | [30] |
| pYA4088 | 852 bp DNA encoding the α-helical region of PspA from aa 3–285 in pYA3493 | [31] |
The Shine-Dalgarno sequence and start codon for each rfc allele are indicated in uppercase, bold letters.
2.2. Plasmids and mutant strain construction
DNA manipulations were carried out as previously described [25]. Electroporation was used to transform E. coli and S. enterica. Transformants were selected on LB agar plates containing appropriate antibiotics. Strains were grown on LB agar plate without supplement to select for bacteria harboring the Asd+ plasmid. The primers used in this work are listed in Table 2. A 500-bp DNA fragment containing the region upstream of the rfc promoter (Prfc) was PCR amplified using the S. Typhimurium χ3761 genome as template with primers Rfc1-FXmaI-pstI and Rfc1-RPstI (Table 2). The amplicon was digested with PstI and ligated into the PstI site of vector pYA3700 (Table 1). This placed the promoter just upstream of araC. Primer T4TT-R, which binds to the T4 transcriptional terminator antisense sequence present downstream of the PstI site in pYA3700, and primer Rfc1-RPstI were used to screen plasmid isolates for inserts in the correct orientation. This intermediate plasmid was digested with XhoI and KpnI at restriction sites that lie downstream of the araC PBAD cassette. Three different 0.5 kb PCR fragments of the rfc gene were amplified from the S. Typhimurium χ3761 genome, using three different upstream primers, Rfc2-FXhoI, Rfc2-1, and Rfc2-2, and the same downstream primer Rfc2-RkpnI. Each PCR fragment was engineered to encode a different Shine-Dalgarno (SD) sequence and/or start codon (Table 1) due to the differences in the upstream primers. The PCR fragments were digested with XhoI and KpnI and inserted into the intermediate plasmid described above. The three resulting constructs were confirmed by DNA sequence analysis. Then, 2.5-kb DNA fragments encoding araC PBAD rfc and rfc 5′ and 3′ flanking regions were excised from each of the plasmids by digestion with KpnI and XmaI and inserted into pRE112, resulting in plasmids pYA4297, pYA4298, and pYA4299. To construct the Δrfc-48 deletion, two pairs of primers Rfc-1F/Rfc-1R and Rfc-2F/Rfc-2R were used to amplify approximately 300-bp fragments upstream and downstream of rfc, respectively, from the χ3761 genome. The two PCR fragments were purified from agarose gels and were used as template at a 1:1 molar ratio for joining by PCR using primers Rfc-1F and Rfc-2R. The resulting PCR product was digested with KpnI and XmaI and ligated into plasmid pRE112, digested with the same two enzymes, resulting in plasmid pYA4717, which carries a deletion of the entire rfc gene from ATG to TAA. The mutations were introduced into S. Typhimurium χ3761 by allelic exchange using the four suicide vectors pYA4717, pYA4297, pYA4298 and pYA4299 to generate χ9944, χ9659, χ9736, χ9737, respectively. The Δrfc-48 and ΔPrfc174 mutations were also introduced into S. Typhimurium strain χ9241 to yield strains χ9885 and χ9853, respectively. The presence of both ΔpabA1516 and ΔpabB232 mutations in S. Typhimurium strains χ9241, χ9885 and χ9853 were verified by the inability of the strains to grow in MOPS minimal medium without p-aminobenzoate. The presence of ΔasdA16 mutation was confirmed by PCR and by the strains’ inability to grow in media without DAP. The ΔaraBAD23 mutation was verified by PCR and by its white colony phenotype on MacConkey agar supplemented with 1% arabinose. LPS profiles of Salmonella strains were examined by the methods of Hitchcock and Brown [26] using cultures standardized based on OD600.
Table 2.
Primers used in this work
| Primer name | Sequence 5′-3′ |
|---|---|
| Rfc1-FXmaI-PstI | ACTGCCTGCAGCCCGGGTCTTTCTGTTCTACAGAACC |
| Rfc1-RPstI | ACTGCCTGCAGAGATTCATCATGAGGTTCCC |
| Rfc2-FXhoI | ATGCACTCGAG AGGACTCTATATGCTTATAATTTC |
| Rfc2-kpnI | ACGGAGGTACCCTCTCTGAACTCCATCAACAC |
| Rfc2-1 | ATGCACTCGAGAGGACTCTATGTGCTTATAATTTC |
| Rfc2-2 | ATGCACTCGAGAGGCTCTATGTGCTTATAATTTC |
| T4TT-R | ATCACAATTCTAGGATAGAAT |
| Rfc-1F | CTGCCTGCAGAAGTATGTCGCGGCACGATG |
| Rfc-1R | ATAACTTACCTGCAGGATAGAGCCTTTAGAAAAAATG |
| Rfc-2F | GGCTCTATCCTGCAGGTAAGTTATACGGCGGCAATGC |
| Rfc-2R | ATCTCCCGGG GTTCGTTTAAACCTGTTTCAC |
2.3. P22 transduction studies
P22HT int [27] was propagated on S. Typhimurium strain χ9430 carrying the integrated suicide plasmid pYA4284, which confers chloramphenicol resistance. Strains to be tested were grown overnight in NB at 37°C. Cultures were diluted 1:100 into fresh, prewarmed nutrient broth with or without 0.1% arabinose and grown at 37°C to an OD600 of 0.9. Then, 10 μl of phage was added to 1 ml of cells (5×108 CFU) and the mixture was incubated at room temperature for 30 min, centrifuged and resuspended in 200 μl of buffered saline with gelatin (BSG) [28]. A 100 μl aliquot was spread onto LB agar plates containing 15 μg/ml chloramphenicol and incubated overnight at 37°C. Colonies were counted the following day. We performed this experiment three times. Duplicate samples were plated for each repetition.
2.4. Minimum inhibitory concentration (MIC) test
The MICs of different antimicrobial substances were determined using 96-well microtitre plates [29]. Two-fold serial dilutions of the bile salt deoxycholate (0.3 to 40 mg/ml) and polymyxin B (0.075 to 4.7 μg/ml) were made down the plates. Bacteria were grown until they reached an OD600 of 0.8–0.9 in NB with or without 0.1% arabinose and washed in PBS. Cells were diluted to 1.0×105 to 1.0×106 CFU in NB with or without arabinose. Then, 0.1 ml of the diluted cell suspension was added to each well. The microtitre plates were incubated overnight at 37°C. The optical density of each culture was determined using a SpectraMax M2e (Molecular Devices, CA) plate reader. The threshold of inhibition was 0.1 at OD600. Actual titers were determined by spreading culture dilutions onto LB agar plates followed by overnight incubation at 37°C. Assays were repeated at least three times.
2.5. Determination of virulence in mice
Seven week old, female BALB/c mice were obtained from the Charles River Laboratories. The Arizona State University Animal Care and Use Committees approved all animal procedures. Mice were acclimated for 7 days after arrival before starting the experiments. For determination of the 50% lethal dose (LD50), bacteria were grown statically overnight at 37°C in LB broth, diluted 1:50 into fresh media containing 0.1% arabinose, and grown with aeration (180 rpm) at 37°C. When the cultures reached an OD600 = 0.8–0.9, they were harvested by room temperature centrifugation at 4,000 rpm, and normalized to the required inoculum densityin BSG by adjusting the suspension to the appropriate OD600 value. Groups of five mice each were infected orally with 20 μl containing various doses of S. Typhimurium χ3761 or its derivatives, ranging from 1.0×103 CFU to 1.0×109 CFU. Animals were observed for 4 weeks post infection, and deaths were recorded daily.
To evaluate colonization, mice were orally inoculated with 20 μl of BSG containing 1.0×109 CFU of each strain. At days 4 and 8 after inoculation, three animals per group were euthanized and their spleens and livers were collected. Each sample was aseptically weighed and then homogenized in a total volume of 1 ml BSG. Dilutions of 10−1 to 10−6 (depending on the tissue) were plated onto MacConkey agar and LB agar, each containing 0.1% arabinose, to determine the number of viable bacteria. If 0.1ml of homogenized organ yielded no colony on the LB plate, the remaining BSG suspension of each tissue sample was inoculated into selenite cysteine broth the next day. Samples that were positive by enrichment in selenite cysteine broth for 14 h at 37°C were recorded as < 10 CFU/g.
2.6. Immunogenicity of vaccine strains in mice
Recombinant attenuated Salmonella vaccine (RASV) strains were grown statically overnight in LB broth with 0.1% arabinose at 37°C. The following day, 2 ml of the overnight culture was inoculated into 100 ml of LB with 0.1% arabinose and grown with aeration at 37°C to an OD600 of 0.8 to 0.9. Cells were harvested by room temperature centrifugation at 4,000 rpm for 15 min and the pellet resuspended in 1 ml of BSG. Mice were orally inoculated with 20 μl of BSG containing 1×109 CFU of each strain on day 0 and boosted on day 28 with the same dose of the same strain. Blood was obtained by mandibular vein puncture at biweekly intervals. Blood was allowed to coagulate at 37°C for two hours. Following centrifugation, the serum was removed from the whole blood and stored at −20°C.
2.7. Antigen preparation
The rPspA protein was purified as described [30]. The rPspA clone, which encodes the α-helical region of PspA (aa 1–302) in pET20b, was a kind gift from Dr. Susan Hollingshead at the University of Alabama at Birmingham. S. Typhimurium LPS was purchased from Sigma. Salmonella outer membrane proteins (SOMPs) were prepared as described [30].
2.8. SDS-PAGE and western blot analyses
Protein samples were boiled for 5 min and then separated by SDS-PAGE. For western blotting, proteins separated by SDS-PAGE were transferred electrophoretically to nitrocellulose membranes. The membranes were blocked with 3% skim milk in 10 mM Tris-0.9% NaCl (pH 7.4) and incubated with rabbit polyclonal antibodies specific for PspA [31] or GroEL (Sigma, St. Louis, MO). The secondary antibody was an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Sigma). Immunoreactive bands were detected by the addition of BCIP/NBT solution (Sigma). The reaction was stopped after 2 min by washing with large volumes of deionized water.
2.9. Enzyme-linked immunosorbent assay (ELISA)
ELISA was used to assay serum antibodies against S. Typhimurium LPS, rPspA and SOMPs as previously described [32]. Color development (absorbance) was recorded at 405 nm using a SpectraMax M2e automated ELISA plate reader (Molecular Devices, CA). Absorbance readings 0.1 nm higher than PBS control values was considered positive.
2.10. Pneumococcal challenge
We assessed the protective efficacy of immunization with the attenuated Salmonella expressing pspA at week 8 by intraperitoneal (i.p.) challenge with 4×104 CFU of S. pneumoniae WU2 in 200 μl of BSG [33] The LD50 of S. pneumoniae WU2 in BALB/c mice was 2×102 CFU by i.p. administration (data not shown). Challenged mice were monitored daily for 30 days.
2.11. Statistical analysis
Antibody titers were expressed as means ± SEM and the relative immunoreactivity was expressed as an arithmetic mean. The means were evaluated with one or two-way ANOVA and Bonferroni’s Multiple Comparison Test for multiple comparisons among groups of antibody titers. Chi square test and the log-rank test were used to analyze the survival rate after challenge by WU2. P < 0.05 was considered statistically significant.
3. Results
3.1. Mutant construction and LPS phenotypes
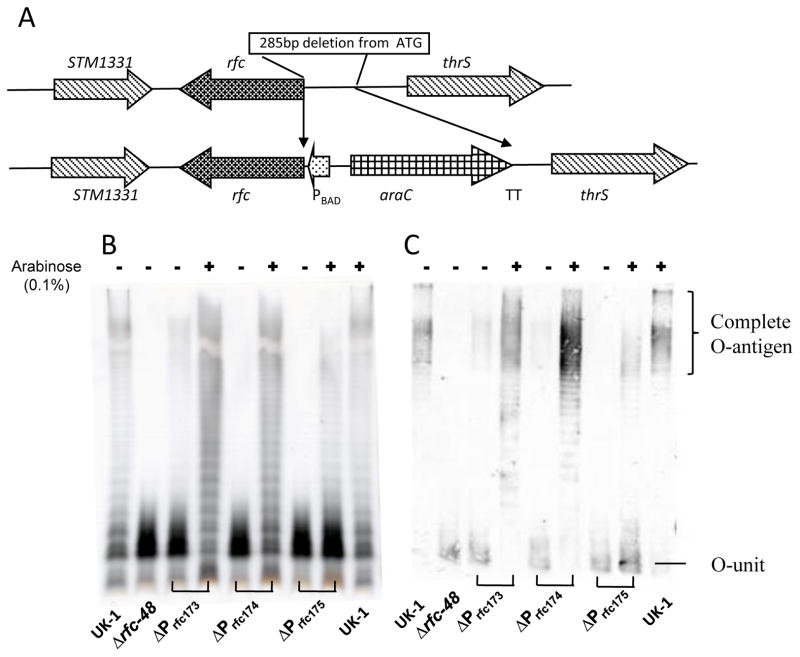
Mutants designed to produce various levels of Rfc were constructed as described in Materials and Methods. Figure 1A illustrates the chromosomal structure of the araC PBAD rfc mutant strains. Nucleotides (285) upstream from the rfc start codon were replaced with araC PBAD and either a different Shine-Dalgarno (SD) sequence, a changed ATG/GTG start codon, or both. Table 1 shows the SD and start codon sequence for each mutant strain.
Fig. 1.
Arabinose regulation of rfc. (A) Map of deletion-insertion mutations resulting in arabinose-regulated rfc expression. (B) LPS phenotypes of wild-type S. Typhimurium χ3761 and the indicated isogenic derivatives. LPSs from different mutant strains grown in nutrient broth with (+) or without (−) 0.1% arabinose were silver stained after separation by 12% SDS-PAGE. (C) Western blots of LPS preparations from panel B. The blots were probed with anti-Salmonella group B antibodies.
The effects of the rfc and arabinose-regulated rfc mutations on the LPS phenotypes were revealed by SDS-PAGE with silver staining (Fig. 1B) and by western blotting against Salmonella Group B O-antigen (Fig. 1C). Deletion of rfc resulted in expected LPS semi-rough phenotype, but two of the three promoter-replacement mutants produced low levels of complete O-antigen when grown in NB without arabinose. Growth in the presence of arabinose resulted in synthesis of full-length O-antigen in the araC PBAD rfc strains, but not in the χ9944 (Δrfc-48) strain. Strain χ9736 (ΔPrfc174::TT araC PBAD rfc, or ΔPrfc174) and χ9659 (ΔPrfc173::TT araC PBAD rfc, or ΔPrfc173), when grown with arabinose, produced an LPS pattern similar to the wild-type Salmonella UK-1. However, strain χ9659 also produced complete O-antigen when grown in the absence of arabinose, detectable in both silver-stained gels and by western blot (Fig. 1B) indicating leaky expression of rfc. Strain χ9737 (ΔPrfc175::TT araC PBAD rfc, or ΔPrfc175), when grown in the absence of arabinose, did not produce any detectable full-length O-antigen. However, this strain did not produce wild-type amounts of O-antigen when grown in the presence of arabinose. Strain χ9736 (ΔPrfc174::TT araC PBAD rfc) produced low levels of full-length O-antigen detectable only by western blot.
To determine if the O-side chains are present, albeit at a level non-detectable by silver staining or western blotting, we tested the transduction efficiency of each strain using the S. Typhimurium O-antigen specific phage P22, which cannot infect rfc mutants [10]. Strains were grown in NB with or without arabinose and used as recipients for transduction assays. Strains χ9659 (ΔPrfc173), χ9736 (ΔPrfc174) and χ9737 (ΔPrfc175) were sensitive to transduction when grown with or without arabinose (Table 3), confirming existence of the complete O-antigen under both conditions. But fewer transductants were obtained for all three promoter mutants when they were grown in NB without arabinose, indicating a decrease in O-antigen synthesis during arabinose-free growth. The number of transductants obtained for each strain roughly mirrored the amount of O-antigen detected for each strain (Fig. 1B, 1C). As expected, no transductants of Δrfc-48 mutant were found, confirming that P22 transduction requires multiple O-unit O-antigens to infect.
Table 3.
Minimum inhibitory concentrations (MIC) of antibiotic substances, transduction efficiency and virulence of S. Typhimurium strain χ3761 and its rfc mutant derivatives
| Strain | 0.1% arabinose | P22 transductantsa (M±SEM) | MIC DOCb Bile (mg/ml) | MIC Polymyxin B (μg/ml) | LD50 (CFU) |
|---|---|---|---|---|---|
| χ9659 (ΔPrfc173) | − | 2292±229 | 2.5 | 0.3 | NDc |
| + | 3250±357 | 10 | 0.59 | 5.0 × 105 | |
| χ9736 (ΔPrfc174) | − | 1892±191 | 2.5 | 0.15 | >1.0 × 109 |
| + | 3425±135 | 10 | 0.59 | >1.0 × 109 | |
| χ9737 (ΔPrfc175) | − | 600±113 | 2.5 | 0.15 | NDc |
| + | 1908±196 | 5 | 0.59 | ||
| χ9944 (Δrfc-48) | − | 0 | 2.5 | 0.15 | >1.0 × 109 |
| + | 0 | 2.5 | 0.15 | >1.0 × 109 | |
| χ3761 (rfc+) | − | 4430±352 | 10 | 0.59 | 1.0×104 |
| + | 4300±306 | 10 | 0.59 |
The phage lysate used for transduction was grown on a chloramphenicol-resistant strain. Transduction was performed as described in the Materials and Methods section. The results reflect the number of chloramphenicol resistant colonies obtained after transduction.
deoxycholate.
Not determined.
3.2. Phenotypes of mutant strains
Rough mutants display a pleiotropic phenotype. Three common features are increased susceptibility to environmental factors, loss of cell surface organelles, and decreased ability to colonize the mouse intestine [34]. We began our analysis of these mutants with in vitro assays examining their susceptibility to different environmental factors such as the bile salt deoxycholate and the antimicrobial peptide polymyxin B. The results are summarized in Table 3. The deoxycholate MIC for the wild-type strain, χ3761, was about four-fold higher than that for the Δrfc-48 mutant, χ9944 and the arabinose-regulated mutants, χ9659 (ΔPrfc173), χ9736 (ΔPrfc174), χ9737 (ΔPrfc175) when grown in absence of arabinose (Table 3).
The results of the MIC to polymyxin B were consistent with the levels of LPS observed in Figure 1. In the presence of arabinose, the arabinose-regulated rfc mutants behaved identically to the wild-type strain (MIC = 0.59 μg/ml); while in the absence of arabinose, the MIC was reduced four-fold compared to the wild-type strain (MIC = 0.15 μg/ml), except for χ9659 (ΔPrfc173). This strain, which appeared to have leaky control over rfc expression as shown in Figure 1, had an MIC closer to the wild-type control. The MIC for strain χ9944 (Δrfc-48), in which the rfc gene was deleted, is similar to the levels of the promoter mutation strains under non-inducing conditions (MIC = 0.15 μg/ml) regardless of whether arabinose was present.
3.3. Virulence in BALB/c mice
Groups of female BALB/c mice received graded doses of various strains orally and were monitored for 30 days after inoculation. The LD50s for the strains were shown in Table 3. The LD50 of χ3761 (1.0 × 104) in mice was similar to that previously observed [21]. The mutant strain χ9944 (Δrfc-48) was avirulent by the oral route, as previously reported [10]. The LD50 of χ9659 (ΔPrfc173), which exhibited leaky O-antigen synthesis (Fig. 1), was 50-fold higher than that of χ3761 when it was grown in arabinose-containing LB prior to feeding. However, strain χ9736 (ΔPrfc174) was totally avirulent (LD50 > 109 CFU), even when grown with arabinose prior to oral administration to mice.
Mice that survived infection with the different mutants were challenged orally with 105 LD50 (1.0 × 109 CFU) of wild-type S. Typhimurium 30 days after administration of the attenuated strains. All five mice immunized with 1.0 × 109 χ9736 (ΔPrfc174) were resistant to challenge. However, only two of five mice immunized with 1.0 × 109 χ9944 (Δrfc-48) survived after challenge by wild-type S. Typhimurium. These data indicate that χ9736 (ΔPrfc174) was sufficiently attenuated but still retained its immunogenic characteristics.
3.4. Expression of the pneumococcal gene pspA in RASV strain χ9241 derivatives carrying different rfc mutations
Based on the above results, we decided to focus on the ΔPrfc174::TT araC PBAD rfc mutant. We wanted to evaluate the ability of strains carrying the Δrfc-48 and ΔPrfc174::TT araC PBAD rfc mutations, in a genetic background that included additional attenuating mutations, to elicit an effective immune response against a vectored antigen in mice. Mutations were introduced into attenuated S. Typhimurium strain χ9241 (ΔpabA1516 ΔpabB232 ΔasdA16 ΔaraBAD23 ΔrelA198::araC PBAD lacI TT) [31, 32]. In addition to the pabA and pabB mutations that confer attenuation, strain χ9241 also encodes an arabinose-regulated lacI gene. Synthesis of LacI represses transcription from promoters such as Ptrc that carry a lacO sequence. This system was designed to prevent the stress of high-level expression of heterologous antigens transcribed from Ptrc during in vitro growth and the early stages of host infection. Once inside the host, where free arabinose is not available, the antigen gene is expressed [31].
The χ9241 derivatives χ9885 (Δrfc-48) and χ9853 (ΔPrfc174) were constructed by using suicide plasmids pYA4717 and pYA4298, respectively, as described in Materials and Methods. The pYA4088 plasmid (Asd+ plasmid that encodes pspA under transcriptional control of the LacI-repressible Ptrc promoter) or the empty vector pYA3493, was electroporated into both mutants and the parent strain. We then confirmed arabinose-regulated PspA and LacI synthesis. We found that all strains carrying plasmid pYA4088 synthesized LacI, but no PspA when arabinose was present and synthesized PspA, but no LacI, when grown in the absence of arabinose (supplementary Figure 1), as we have previously observed [21]. All strains synthesized similar amounts of PspA and LacI (supplementary Fig. 1). No PspA was detected in χ9241(pYA3493), which does not carry the pspA gene.
3.5. Colonization of mouse tissues and immune responses in mice after oral immunization with RASV expressing PspA
The observation that the χ9736 (ΔPrfc174) was avirulent when given orally yet was capable of immunizing mice against a χ3761 challenge suggests that the strain retained a limited ability to colonize either or both the intestinal or systemic immune tissues of the mice. To confirm that these mutations in the χ9241 background still retained the ability to colonize systemic immune tissues of the mice, groups of mice were orally inoculated with 1.0 × 109 cells of various strains harboring pYA4088, and bacterial colonization of spleen and liver was enumerated at 4 and 8 days post inoculation (Fig. 2).
Fig. 2.

Colonization of mouse spleens and livers by attenuated S. Typhimurium harboring plasmid pYA4088 growing in LB broth containing 0.1% arabinose. Shown is spleen (A) and liver (B) colonization by the indicated strains in BALB/c mice at 4 and 8 days post-inoculation. The horizontal lines represent the means, and the error bars represent standard errors of the means.
Strains χ9241(pYA4088) and χ9853(pYA4088) (ΔPrfc174) colonized the spleen and liver, and reached high numbers in both tissues (approximately 106 CFU per gram of tissue), with a slight reduction at eight days. The titers in spleen and liver for strain χ9885(pYA4088) (Δrfc-48) were significantly lower than the other two strains at both time-points tested. There was a slight reduction in tissue colonization by χ9853(pYA4088) (ΔPrfc174) compared to its parent strain χ9241(pYA4088), but the difference was not statistically significant.
3.6. Effect of Δrfc and ΔPrfc174 mutations on immunogenicity of RASV strains
Mice were inoculated orally (1.0×109 CFU) with one of four strains harboring either pYA4088 or the empty vector pYA3493. Mice were boosted with a similar dose of the same strain 4 weeks later. This experiment was performed twice, 5 mice per group were involved in the first experiment, and 6–8 mice per group were used in the second experiment. The results from both experiments were similar and have been pooled for analysis.
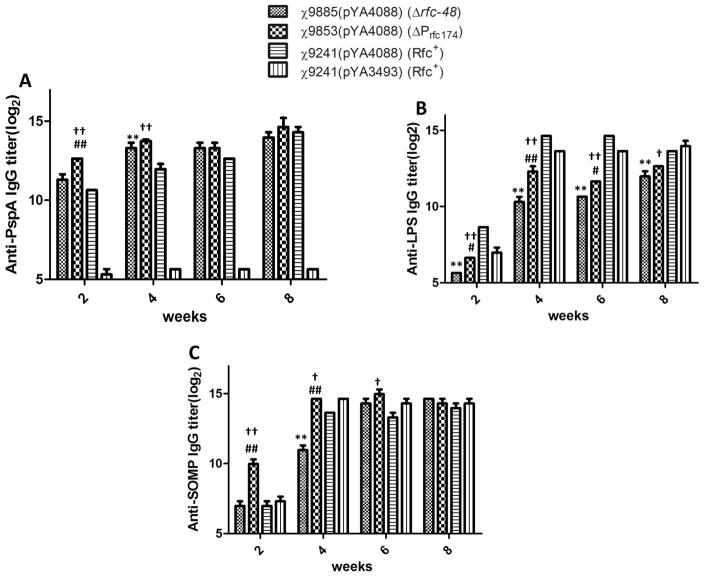
High serum IgG titers against rPspA (Fig. 3A) were observed 2 weeks after the primary immunization in mice inoculated with χ9241(pYA4088), χ9885(pYA4088) (Δrfc-48) and 9853(pYA4088) (ΔPrfc174). There was a significant difference between the serum IgG titers from the mice immunized with χ9241(pYA4088) and χ9853(pYA4088) (ΔPrfc174). Anti-LPS titers were low, but detectable for all strains at 2 weeks, and reached a maximum by 8 weeks (Fig. 3B). Anti-LPS titers from mice immunized by χ9885(pYA4088) (Δrfc-48) and χ9853(pYA4088) (ΔPrfc174) were significantly lower than those of mice immunized by χ9241(pYA4088 or pYA3493) during all time points measured. The anti-SOMP responses developed faster for the ΔPrfc174 strain than that for the other groups, with significantly higher titers by week 2 (Fig. 3C).
Fig. 3.
Serum IgG responses in immunized and control mice. Total serum IgGs specific for rPspA (A), S. Typhimurium LPS (B), and SOMP (C) were measured by ELISA. The data represent reciprocal anti-IgG antibody levels in pooled sera from mice orally immunized with attenuated Salmonella carrying either pYA4088 (pspA) or pYA3493 (control) the indicated number of weeks after immunization. The error bars represent variations between triplicate wells. The mice were boosted at week 4. **: χ9885(pYA4088) vs. χ9241(pYA4088), P < 0.001; #: χ9885(pYA4088) vs. χ9853(pYA4088), P < 0.01; ##: χ9885(pYA4088) vs. χ9853(pYA4088), P < 0.001; ††: χ9853(pYA4088) vs. χ9241(pYA4088), P < 0.001; †: χ9853(pYA4088) vs.χ9241(pYA4088), P < 0.01.
By week 4, the serum anti-rPspA IgG antibody levels of mice immunized with χ9885(pYA4088) (Δrfc-48) was significantly higher than mice immunized with χ9241(pYA4088). The titers were slightly lower than those in mice immunized with χ9853(pYA4088) (ΔPrfc174), but the difference was not significant. The anti-LPS IgG titers in χ9853(pYA4088) (ΔPrfc174) immunized mice were significantly lower than that in mice immunized with χ9241(pYA4088). All groups except those immunized with the Δrfc strain developed similar, high titers against SOMPs. No anti-PspA IgG was detected in mice immunized with strain χ9241(pYA3493).
After the second immunization at week 4, no significant boosting of serum antibody responses to either rPspA or LPS or SOMP was observed for any group of immunized mice, except in the case of the anti-SOMP titers for the Δrfc-48 strain χ9885(pYA4088) (Δrfc-48). This strain achieved anti-SOMP titers similar to all other strains by week 6 (Fig. 3C). The serum immune responses peaked at 6 weeks with no significant changes observed at week 8.
3.7. IgG isotype analyses
The serum immune responses to rPspA were further examined by measuring the levels of IgG isotype subclasses IgG1 and IgG2a. Th1-biased immune responses are typically observed after immunization with attenuated Salmonella strains. Although the IgG2a levels were always higher than IgG1 levels during each time period checked, the levels of anti-rPspA IgG1 and IgG2a isotypes antibodies gradually increased (Fig. 4). The ratio of IgG2a to IgG1 was 16:1 for χ9885(pYA4088) (Δrfc-48)-immunized mice and 2–4:1 for χ9241(pYA4088) or χ9853(pYA4088) (ΔPrfc174)-immunized mice.
Fig. 4.
Serum IgG1 and IgG2a responses to rPspA. The data represent ELISA results determining the level of IgG1 and IgG2a subclass antibody to rPspA in the sera of BALB/c mice orally immunized with χ9241(pYA4088), χ9885(pYA4088) (Δrfc-48), or χ9853(pYA4088) (ΔPrfc174) the indicated number of weeks after immunization. The error bars represent the standard deviations.
3.8. Evaluation of protective immunity
To examine the ability of RASV-rPspA vaccines to protect against pneumococcal infection, mice were challenged i.p. with 4.0 × 104 CFU (200 LD50) of S. pneumoniae WU2 4 weeks after they were boosted. The pspA gene carried by plasmid pYA4088 was derived from S. pneumoniae strain RX1 and is cross-reactive with PspA produced by S. pneumoniae WU2. Immunization with any of the pspA-expressing strains provided significant protection against challenge compared with the control group receiving χ9241(pYA3493) (empty vector) (Table 4). However, the protection afforded by χ9885(pYA4088) (Δrfc-48) and χ9853(pYA4088) (ΔPrfc174) was significantly higher than χ9241(pYA4088). There was no significant difference in protection afforded by strains χ9885(pYA4088) (Δrfc-48) and χ9853(pYA4088) (ΔPrfc174). All of the mice that died in these experiments succumbed within 4 days of the challenge.
Table 4.
Oral immunization with rPspA-expressing S. enterica serovar Typhimurium χ9241(pYA4088) vaccine and its derivatives (pYA4088) protect BALB/c mice against challenge with virulent S. pneumoniae strain WU2
| Vaccine | Genotype | pspA expressiona | Days to death | Survival/totalb | Percent protectionb |
|---|---|---|---|---|---|
| χ9853 | ΔPrfc174 | + | 2, 3, 4, 4, 4 | 6/11 | 55% |
| χ9885 | Δrfc-48 | + | 2, 3, 4 | 10/13 | 77% |
| χ9241 | Rfc+ | + | 2, 2, 3, 3, 4, 4, 4 | 4/11 | 36% |
| χ9241 | Rfc+ | − | 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 3 | 0/11 | 0 |
+, rPspA expressed; -, rPspA not expressed.
Mice were challenged with 4 × 104 CFU of S. pneumoniae WU2 (200 times LD50) 4 weeks after the second oral immunization. Mortality was monitored for 2 weeks after pneumococcal challenge. Differences between groups were determined using Chi square test and the log-rank test. All vaccine groups were significantly different from the χ9241(pYA3493) control (P < 0.002) by two methods. Chi square test: χ9885(pYA4088) vs. χ9853(pYA4088), P = 0.1219; χ9885(pYA4088) vs. χ9241(pYA4088), P < 0.0001; χ9853(pYA4088) vs. χ9241(pYA4088), P = 0.005. The log-rank test: no significant difference between vaccine strains (pYA4088).
4. Discussion
The greatest challenge in the development of live Salmonella vaccines is to maintain a balance between safety and immunogenicity [3, 35]. After oral immunization, a RASV must be able to withstand acidic, osmotic, and enzymatic stresses as well as surviving the host defenses of bile, antimicrobial peptides, and innate immunity. The ideal RASV strain will be capable of tolerating the aforementioned stressors while remaining avirulent and capable of eliciting a robust immune response. Smooth LPS provides Salmonella with a covalently attached, protective envelope conferring resistance to a variety of environmental stresses encountered both within and outside a mammalian host thereby enhancing virulence. Strains with mutations that eliminate LPS O-antigen are typically less immunogenic, a phenotype associated with their failure to colonize the intestinal tract and invade intestinal mucosal cells [36, 37]. It is therefore advantageous for vaccine strains to regulate the O-antigen synthesis, producing the full-length LPS during immunization, when it will encounter stresses imposed by the host, and then shutting off O-antigen polymerization in vivo after the bacteria have colonized the host lymphoid tissues [3].
Structural rough mutants have been considered to be inappropriate live vaccine carriers due to their hyperattenuation and poor colonization [38, 39]. In this work, we applied the regulated delayed attenuation approach to produce strains with arabinose-regulated synthesis of Rfc, the polymerase required for production of full-length O-antigen, and compared arabinose-regulated rfc expression strains to a Δrfc strain and wild-type strain in in vitro assays. As previously observed in other arabinose regulated expression systems [21], the SD sequence and start codon greatly affected the tight control of expression of rfc under the araC PBAD promoter. In the case of arabinose-regulated rfc expression, based on the LPS profile silver staining and western blot analysis using Salmonella anti-LPS sera, only the strain containing the ΔPrfc174::TT araC PBAD rfc construct was unable to produce complete O-antigen in the absence of arabinose, and able to produce the full-length O-antigen in the presence of arabinose (Fig. 1). The strain containing ΔPrfc173 was leaky for expression of rfc, leading to some polymerization of O-antigen without arabinose, and ΔPrfc175 exhibited tight control of rfc expression (Fig. 1). When grown without arabinose, production of full length O-antigen in the strain carrying the ΔPrfc175 mutation, undetectable by gel or western blot (Fig. 1), was revealed by the P22 transduction assay (Table 3). However the number of P22 transductants obtained for all three promoter mutants was reduced when they were grown in the absence of arabinose compared to the number obtained when the strains were grown with arabinose.
As mentioned above, an ideal Salmonella vaccine strain should exhibit wild-type abilities to withstand stresses and host defenses encountered after mucosal immunization while remaining avirulent. When the ΔPrfc174 mutant was grown in the presence of arabinose, its phenotype was similar to that of its wild-type parent χ3761 (Fig. 1 and 2 and Table 3), though it was totally attenuated (LD50 > 109) for virulence in mice (Table 3). In the absence of arabinose, this mutant was more susceptible to deoxycholate and polymyxin B than the wild-type parent χ3761.
It is also important that live RASV strains carry multiple attenuating mutations. Therefore, we evaluated the effect of the ΔPrfc174 mutation in the context of an attenuated vaccine strain, χ9241, and introduced plasmid pYA4088 to allow expression of the heterologous antigen gene, pspA. The ΔPrfc174 strain χ9853(pYA4088) colonized host lymphoid tissues as well as the parent strain χ9241(pYA4088) (Fig. 2), while the Δrfc strain, χ9885(pYA4088), colonized poorly. It is interesting to note that the numbers we obtained in the spleen for χ9885(pYA4088) (Δrfc-48) are similar to numbers previously reported for an rfc mutant strain with no additional attenuating mutations [10].
The most surprising result we obtained in this study was the observation that the Δrfc mutant χ9885(pYA4088) performed as well as the (ΔPrfc174) strain χ9853(pYA4088) and the Rfc+ strain χ9241(pYA4088) Despite the fact that χ9885(pYA4088) (Δrfc-48) was a poor colonizer of spleen and liver tissues compared to the other two strains (Fig. 2), immunized mice developed high titers against PspA and achieved levels of protection against S. pneumoniae challenge similar to the other two strains (Table 4). This is in contrast to our previous study in which we introduced ΔrfaH and araC PBAD rfaH mutations into the χ9241(pYA4088) background [21]. In that study, the ΔrfaH mutant colonized poorly, induced significantly lower anti-PspA titers and significantly less protection, although some protection was observed.
The basis for the ability of χ9885(pYA4088) (Δrfc-48) to elicit a protective response while being impaired for colonization is not clear. However, this result is not without precedent. For example, rpoS and relA spoT mutants of S. Typhimurium are immunogenic and protective against challenge with virulent S. Typhimurium and yet these mutants colonize no better than χ9885(pYA4088) (Δrfc-48) [40, 41]. What is not clear, either from this study or the studies just cited, is what impact poor colonization will have on the duration of immunity where it is likely that a strong cellular response is required. We did not include measurement of cellular responses in our experimental design because antibody responses to PspA are adequate for protection against S. pneumoniae [42]. With regard to the observed protection, it is possible that while colonization was not robust, presentation of PspA may have been enhanced in the Δrfc mutant due to its lack of full-length O-antigen. In addition, the robust immune response to PspA may have been due, at least in part, to the fact that PspA is a highly immunogenic protein [43]. It is not clear whether or not our results would have been different if we had used a less immunogenic test protein. Alternatively, it was recently shown that mesenteric lymph nodes are an important barrier to systemic disease caused by Salmonella [44] and, along with the spleen, can serve as a site for induction of systemic immunity [45]. In light of our current findings that the Δrfc mutant strain χ9885(pYA4088) was a poor colonizer of spleen and liver (Fig. 2), yet was able to induce systemic and protective immunity (Fig. 3, Table 4), it is of interest for us to examine the ability of our vaccine strains to colonize the mesenteric lymph nodes in future studies. If χ9885(pYA4088) can colonize the mesenteric lymph nodes as well or better than χ9241(pYA4088) and χ9853(pYA4088), this may provide a clear understanding of our results with respect to the relationship between colonization and induction of the immune response which would aid greatly in the design of future vaccine strains.
Supplementary Material
Supplementary Fig. 1. PspA and LacI synthesis is regulated by 0.1% arabinose. The Western blots show the synthesis of PspA in S. Typhimurium strains χ9885(pYA4088) (Δrfc48), χ9853(pYA4088) (ΔPrfc174), χ9241(pYA4088) (Rfc+), and χ9241(pYA3493). The bacteria were grown in LB broth with (+) or without (−) 0.1% arabinose overnight at 37°C. Equal number of cells from each culture were pelleted, suspended in loading buffer, and boiled. After centrifugation, equal volumes were subjected to SDS-PAGE in triplicate gels. Each gel was transferred to nitrocellulose and probed with a different polyclonal antibody specific for either PspA, LacI or GroEL. GroEL was used as a standardization marker. Relevant portions of each blot are shown.
Acknowledgments
We thank Shifeng Wang for his suggestions on plasmid construction. We are grateful to Kenneth L. Roland for critically editing the manuscript. This work was supported by grant 37863 from the Bill and Melinda Gates Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheminay C, Hensel M. Rational design of Salmonella recombinant vaccines. Int J Med Microbiol. 2008 Jan;298(1–2):87–98. doi: 10.1016/j.ijmm.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Cardenas L, Clements JD. Oral immunization using live attenuated Salmonella spp as carriers of foreign antigens. Clin Microbiol Rev. 1992 Jul;5(3):328–42. doi: 10.1128/cmr.5.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtiss R., III Bacterial infectious disease control by vaccine development. J Clin Invest. 2002 Oct;110(8):1061–6. doi: 10.1172/JCI16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medina E, Guzman CA. Use of live bacterial vaccine vectors for antigen delivery: potential and limitations. Vaccine. 2001 Feb 8;19(13–14):1573–80. doi: 10.1016/s0264-410x(00)00354-6. [DOI] [PubMed] [Google Scholar]
- 5.Bueno SM, Gonzalez PA, Kalergis AM. Use of genetically modified bacteria to modulate adaptive immunity. Curr Gene Ther. 2009 Jun;9(3):171–84. doi: 10.2174/156652309788488587. [DOI] [PubMed] [Google Scholar]
- 6.Germanie R, Furer E. Immunity in experimental salmonellosis .2. basis for avirulence and protective capacity of galE mutants of Salmonella typhimurium. Infect Immun. 1971;4(6):663. doi: 10.1128/iai.4.6.663-673.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson G, Manning PA. Galactose epimeraseless (GalE) mutant G30 of Salmonella typhimurium is a good potential live oral vaccine carrier for fimbrial antigens. FEMS Microbiol Lett. 1985;28(3):317–21. [Google Scholar]
- 8.Hone DM, Attridge SR, Forrest B, Morona R, Daniels D, Labrooy JT, et al. A galE-via (Vi-antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect Immun. 1988 May;56(5):1326–33. doi: 10.1128/iai.56.5.1326-1333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins LV, Hackett J. Sequence of the phosphomannose isomerase encoding gene of Salmonella typhimurium. Gene. 1991 Jul 15;103(1):135–6. doi: 10.1016/0378-1119(91)90406-2. [DOI] [PubMed] [Google Scholar]
- 10.Collins LV, Attridge S, Hackett J. Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect Immun. 1991 Mar;59(3):1079–85. doi: 10.1128/iai.59.3.1079-1085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Wang S, Scarpellini G, Gunn B, Xin W, Wanda SY, et al. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc Natl Acad Sci U S A. 2009 Jan 13;106(2):593–8. doi: 10.1073/pnas.0811697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtiss R, III, Zhang X, Wanda SY, Kang Ho Young, Konjufca V, Li Y, Gunn B, Wang S, Scarpellini G Lee In Soo. Virulence mechanisms of bacterial pathogens. 4. ASM Press; Washington, DC: 2007. Induction of host immune responses using Salmonella-vectored vaccines; pp. 297–313. [Google Scholar]
- 13.Hone D, Morona R, Attridge S, Hackett J. Contruction of defined galE mutants of Salmonella for use as vaccines. J Infect Dis. 1987 Jul;156(1):167–74. doi: 10.1093/infdis/156.1.167. [DOI] [PubMed] [Google Scholar]
- 14.Nagy G, Danino V, Dobrindt U, Pallen M, Chaudhuri R, Emody L, et al. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect Immun. 2006 Oct;74(10):5914–25. doi: 10.1128/IAI.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109(2):193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 16.Nagy G, Palkovics T, Otto A, Kusch H, Kocsis B, Dobrindt U, et al. “Gently rough”: The vaccine potential of a Salmonella enterica regulatory lipopolysaccharide mutant. J Infect Dis. 2008 Dec 1;198(11):1699–706. doi: 10.1086/593069. [DOI] [PubMed] [Google Scholar]
- 17.Whitfield C. Biosynthesis of lipopolysaccharide o-antigens. Trends Microbiol. 1995 May;3(5):178–85. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 18.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran AX, Whitfield C. Encyclopedia of Microbiology. Oxford: Academic Press; 2009. Lipopolysaccharides (Endotoxins) pp. 513–28. [Google Scholar]
- 20.Curtiss R, III, Wanda S-Y, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, et al. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect Immun 2009. 2009 Mar 1;77(3):1071–82. doi: 10.1128/IAI.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong Q, Liu Q, Roland KL, Curtiss R., III Regulated delayed expression of rfaH in an attenuated Salmonella enterica serovar Typhimurium vaccine enhances immunogenicity of outer membrane proteins and a heterologous antigen. Infect Immun 2009. 2009 Dec 1;77(12):5572–82. doi: 10.1128/IAI.00831-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertani G. Studies on lysogenesis I.: The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 1951. 1951 Sep 1;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama K, Kelly SM, Curtiss R., III Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat Bio-technol. 1988 Jun;6(6):693–7. [Google Scholar]
- 24.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol 1974. 1974 Sep 1;119(3):736–47. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 26.Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver stained polyacrylamide gels. J Bacteriol. 1983;154(1):269–77. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 28.Curtiss RI. Chromosomal aberrations associated with mutations to bacteriophage resistance in Escherichia Coli. J Bacteriol. 1965;89(1):28. doi: 10.1128/jb.89.1.28-40.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protocols. 2008;3(2):163–75. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 30.Kang HY, Srinivasan J, Curtiss RI. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect Immun. 2002 Apr;70(4):1739–49. doi: 10.1128/IAI.70.4.1739-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin W, Wanda SY, Li Y, Wang S, Mo H, Curtiss R., III Analysis of type II secretion of recombinant pneumococcal PspA and PspC in a Salmonella enterica serovar Typhimurium vaccine with regulated delayed antigen synthesis. Infect Immun. 2008 Jul;76(7):3241–54. doi: 10.1128/IAI.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Wang S, Xin W, Scarpellini G, Shi Z, Gunn B, et al. A sopB deletion mutation enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Infect Immun. 2008 Nov;76(11):5238–46. doi: 10.1128/IAI.00720-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nayak AR, Tinge SA, Tart RC, McDaniel LS, Briles DE, Curtiss R., III A live recombinant avirulent oral Salmonella vaccine expressing pneumococcal surface protein A induces protective responses against Streptococcus pneumoniae. Infect Immun. 1998 Aug;66(8):3744–51. doi: 10.1128/iai.66.8.3744-3751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nevola JJ, Stocker BAD, Laux DC, Cohen PS. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect Immun. 1985;50(1):152–9. doi: 10.1128/iai.50.1.152-159.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon YM, Cox MM, Calhoun LN. Salmonella-based vaccines for infectious diseases. Expert Rev Vaccines. 2007 Apr;6(2):147–52. doi: 10.1586/14760584.6.2.147. [DOI] [PubMed] [Google Scholar]
- 36.Stocker BAD, Makela PH. Genetic determination of bacterial virulence, with special reference to Salmonella. Curr Top Microbiol Immunol. 1986;124:149–72. doi: 10.1007/978-3-642-70986-9_9. [DOI] [PubMed] [Google Scholar]
- 37.Stocker BAD, Makela PH. Genetics of (gram-negative) bacterial surface. P Roy Soc Lond B Bio. 1978;202(1146):5–30. doi: 10.1098/rspb.1978.0055. [DOI] [PubMed] [Google Scholar]
- 38.Nagy G, Pal T. Lipopolysaccharide: a tool and target in enterobacterial vaccine development. Biol Chem. 2008 May;389(5):513–20. doi: 10.1515/bc.2008.056. [DOI] [PubMed] [Google Scholar]
- 39.Nagy G, Emody L, Pal T. Strategies for the development of vaccines conferring broad- spectrum protection. Int J Med Microbiol. 2008 Jul;298(5–6):379–95. doi: 10.1016/j.ijmm.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Coynault C, Robbe-Saule V, Norel F. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (sigma (S)) regulon. Mol Microbiol. 1996 Oct;22(1):149–60. doi: 10.1111/j.1365-2958.1996.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 41.Na HS, Kim HJ, Lee HC, Hong Y, Rhee JH, Choy HE. Immune response induced by Salmonella typhimurium defective in ppGpp synthesis. Vaccine. 2006 Mar 15;24(12):2027–34. doi: 10.1016/j.vaccine.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 42.Briles DE, Hollingshead SK, King J, Swift A, Braun PA, Park MK, et al. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J Infect Dis. 2000 Dec;182(6):1694–701. doi: 10.1086/317602. [DOI] [PubMed] [Google Scholar]
- 43.McDaniel LS, McDaniel DO, Hollingshead SK, Briles DE. Comparison of the PspA sequence from Streptococcus pneumoniae EF5668 to the previously identified PspA sequence from strain Rx1 and ability of PspA from EF5668 to elicit protection against pneumococci of different capsular types. Infect Immun. 1998 Oct;66(10):4748–54. doi: 10.1128/iai.66.10.4748-4754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voedisch S, Koenecke C, David S, Herbrand H, Forster R, Rhen M, et al. Mesenteric lymph nodes confine dendritic cell-mediated dissemination of Salmonella enterica serovar Typhimurium and limit systemic disease in mice. Infect Immun. 2009 Aug;77(8):3170–80. doi: 10.1128/IAI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinoli C, Chiavelli A, Rescigno M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity. 2007 Dec;27(6):975–84. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Hassan JO, Curtiss R., III Control of colonization by virulent Salmonella typhimurium by oral immunization of chickens with avirulent Dcya Dcrp Salmonella typhimurium. Res Microbiol. 1990 Sep-Oct;141(7–8):839–50. doi: 10.1016/0923-2508(90)90119-b. [DOI] [PubMed] [Google Scholar]
- 47.Roland K, Curtiss R, III, Sizemore D. Construction and evaluation of a Δcya Δcrp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 1999 Jul-Sep;43(3):429–41. [PubMed] [Google Scholar]
- 48.Briles DE, King JD, Gray MA, McDaniel LS, Swiatlo E, Benton KA. PspA, a protection-eliciting pneumococcal protein: Immunogenicity of isolated native PspA in mice. Vaccine. 1996 Jun;14(9):858–67. doi: 10.1016/0264-410x(96)82948-3. [DOI] [PubMed] [Google Scholar]
- 49.Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998 Jan 30;207(2):149–57. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 50.Sun W, Wang S, Curtiss R., III Highly efficient method for introducing successive multiple scarless gene deletions and markerless gene insertions into the Yersinia pestis chromosome. Appl Environ Microbiol. 2008 Jul;74(13):4241–5. doi: 10.1128/AEM.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. PspA and LacI synthesis is regulated by 0.1% arabinose. The Western blots show the synthesis of PspA in S. Typhimurium strains χ9885(pYA4088) (Δrfc48), χ9853(pYA4088) (ΔPrfc174), χ9241(pYA4088) (Rfc+), and χ9241(pYA3493). The bacteria were grown in LB broth with (+) or without (−) 0.1% arabinose overnight at 37°C. Equal number of cells from each culture were pelleted, suspended in loading buffer, and boiled. After centrifugation, equal volumes were subjected to SDS-PAGE in triplicate gels. Each gel was transferred to nitrocellulose and probed with a different polyclonal antibody specific for either PspA, LacI or GroEL. GroEL was used as a standardization marker. Relevant portions of each blot are shown.