Abstract
Lysophosphatidic acid (LPA) is a bioactive lipid with a plethora of biological functions including roles in cell survival, proliferation, and migration. Although high-performance liquid chromatography electrospray ionization tandem mass spectrometry (HPLC ESI-MS/MS) technology has been used to measure the levels of LPA in human blood, serum and plasma, current methods cannot readily detect the minute levels of LPA from cell culture. In this study, a modified HPLC ESI-MS/MS method with enhanced sensitivity was developed, which allows accurate measurements of LPA levels with a limit of quantitation at approximately 10 femtomoles. The method was validated by quantitation of LPA levels in the media of previously characterized cell lines ectopically expressing autotaxin. Specifically, autotaxin overexpression induced an increase in the 16:0, 18:2, 18:1, 18:0, and 20:4 subspecies of LPA, but not the 22:6 LPA subspecies. Lastly, this HPLC ESI-MS/MS method was cross-validated via biological assays previously utilized to assay LPA levels. Hence, this HPLC ESI-MS/MS method will allow researchers to measure in vitro LPA levels and also distinguish between specific LPA subspecies for the delineation of individual biological mechanisms.
Introduction
Lysophosphatidic acid (LPA) is a glycerophospholipid that acts as an extracellular signaling molecule and plays important roles in a variety of cellular functions.1 LPA is composed of a single, variable length acyl chain, a glycerol backbone and a phosphate head group. LPA interacts with specific G protein-coupled receptors to activate signaling cascades that play important roles in wound healing, cancer, atherosclerosis, and obesity. There are several routes of LPA synthesis, but the ecto-nucleotide pyrophosphatase/phosphodiesterase enzyme autotaxin (NPP2/ATX) is the major source of LPA production.2 Originally identified as an autocrine motility factor in melanoma cells, the discovery of the lysoPLD function of autotaxin led to a better understanding of LPA production and function.3–5 The generation of LPA by autotaxin occurs by the hydrolysis of the choline group at the sn-3 position of lysophosphatidylcholine (LPC).
High-performance liquid chromatography electrospray ionization tandem mass spectrometry (HPLC ESI-MS/MS) has gained momentum as an invaluable tool in the quantitative and qualitative analysis of lipid species because of its superior sensitivity, versatility, and the relatively small amount of labor required for accurate analysis. However, a major hurdle for the versatility of lipid analysis via HPLC ESI-MS/MS is the requirement of different solvent conditions and HPLC separation conditions for different lipid classes. The need for different solvent conditions for different lipid classes not only makes the analysis more cumbersome, but also results in a significant loss of valuable instrument time as every solvent exchange requires a significant period of time for system equilibration. Additionally, use of different solvents induces greater variability in the acquired data adding increased difficulty for appropriate comparisons between lipid classes from the same sample. A variety of HPLC ESI-MS/MS methods have been developed to undertake LPA measurements from tissue, blood, plasma, and serum samples, but most are specific for separation and detection of LPA with little to no application in the detection of other lipid classes.6–10 Additionally, most of these methods demonstrate poor quality peaks or inadequate separation.7–9,11,12 Other HPLC solvent systems reported for LPA, including the most recent HPLC ESI-MS/MS method, have the potential for ion suppression due to the inclusion of triethyl ammonium acetate as a modifier.6 This ion suppression will require regular and thorough cleaning of the instrument before further analysis of other lipid classes, thereby decreasing sample throughput. Some HPLC methods have even required pre-separation of LPA using thin-layer chromatography, a highly labour intensive method requiring the use of radiolabeled samples.8 There are also methodological issues for the extraction of LPA in these reported protocols as most have relied on the use of very low pH conditions, which may lead to the acid-catalysed hydrolysis of LPC to LPA and thereby provide artificially inflated LPA measurements.8 Even after acidic extractions, the limit of quantitation for LPA using these mass spectrometric methods have been in the low picomole or high femtomole range.9,10 While this sensitivity is adequate for quantitation for the high LPA levels of in vivo biological samples, greater sensitivity is required for in vitro LPA measurements as well as a reliable, non-volatile pre-separation and accurate extraction protocol.
In this study, we have developed a HPLC ESI-MS/MS method that not only avoids the induction of artificially enhanced LPA levels from acidic extraction protocols, but also a provides limit of quantitation significantly lower than the limits of detection for most published LPA methods. Additionally, the separation and detection utilized for LPA is compatible with the separation and detection of many other lipid classes. The method was validated for in vitro use by demonstrating increased levels of LPA in autotaxin overexpressing ovarian cancer cell lines. The biological role of these increased LPA levels was cross-validated using previously established proliferation and migration assays for measuring the levels of this bioactive lipid. Our modified HPLC ESI-MS/MS method will provide researchers with the ability to measure in vitro LPA levels as well as provide a more sensitive and accurate method for LPA quantitation in biological samples.
Results and discussion
Retention time markers and linear signal response show increased HPLC ESI-MS/MS sensitivity for LPA quantitation
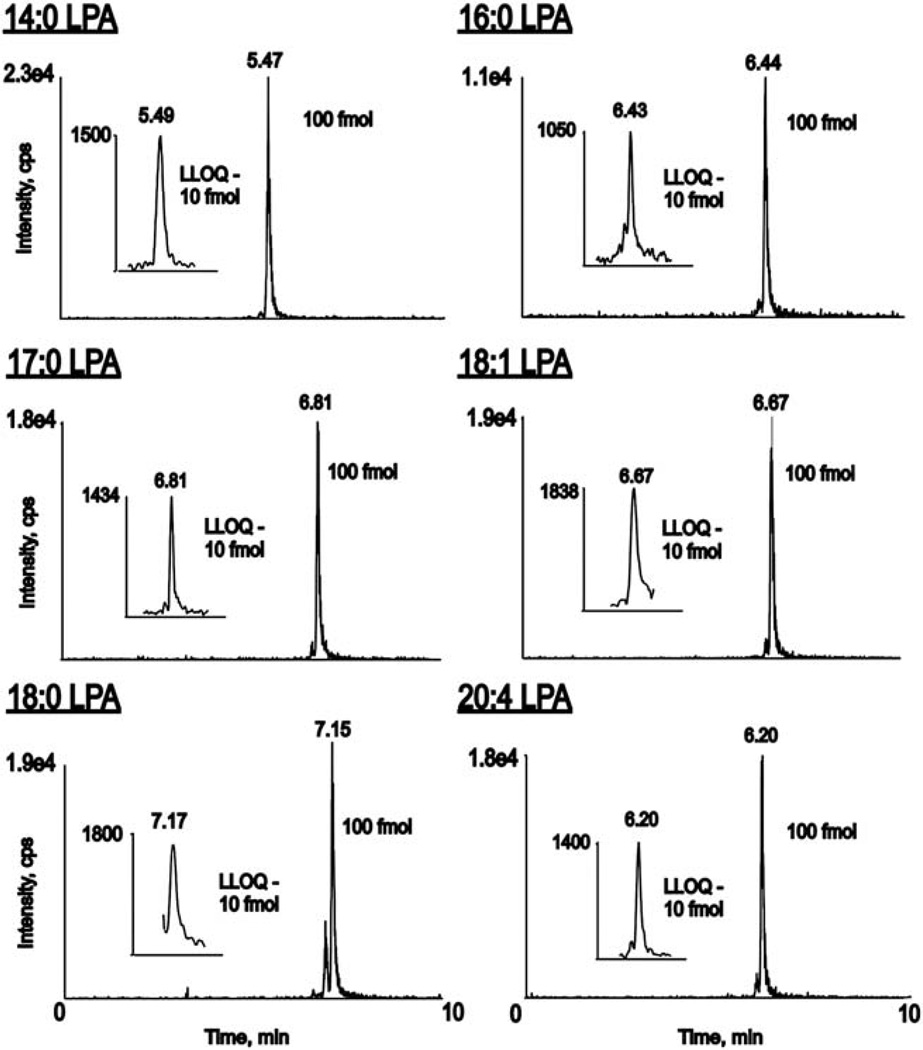
Previous HPLC ESI-MS/MS methods have been limited to HPLC solvents that have not been proven to be efficient in analysis of other lipid classes and limited in sensitivity allowing only for quantification of LPA from biological samples such as tissue, blood, plasma, or serum. The goal of this study was to produce an HPLC ESI-MS/MS method with a high degree of sensitivity to quantify LPA levels from cell culture and with solvent conditions suitable for analysis of other lipid classes. The method presented here, when used in the positive ionization mode is also applicable in the analysis of sphingosine, sphinganine, sphingosine-1-phosphate, sphinganine-1-phosphate, ceramides, monohexosyl ceramides, ceramide-1-phosphates, sphingomyelins, phosphatidylcholines, and also lysophosphatidylcholines. Using this new method, distinct chromatograms were produced (Fig. 1) to allow accurate identification of LPA species related to specific HPLC retention times (Table 1). Peak assignments were initially determined by the analysis of indicated LPA standards using the MRM transitions and mass spectrometry parameters detailed in Table 1. Since there are no commercially available standards for 18:2 LPA and 22:6 LPA, retention times for those species were estimated based upon our knowledge of the shifts in retention times due to increasing unsaturation and increasing chain length. These retention times were then validated as being accurate by carrying out enhanced product ion scans to identify the peaks eluting at the estimated retention times for 18:2 and 22:6 LPA from biological samples.
Figure 1.
Retention time standards are required for unambiguous peak assignment in the quantitation of LPA species during reverse phase HPLC ESI MS/MS analysis. An internal standard mixture of commercially available LPA standards produced distinct MRM chromatograms for 14:0 LPA, 16:0 LPA, 17:0 LPA, 18:1 LPA, 18:0 LPA and 20:4 LPA. Insets: LLOQ chromatograms for the respective LPA species.
Table 1.
ABSciex 4000 QTRAP mass spectrometer settings and retention times for reverse phase chromatographic separation of LPA species DP: declustering potential, CE: collision energy, EP: entrance potential
| LPA Subspecies | Precursor Ion (m/z) (-ve ionization) |
Product Ion (m/z) (-ve ionization) |
DP | CE | EP | Retention Time (min) |
|---|---|---|---|---|---|---|
| 4:0 | 381.3 | 153.0 | −70.0 | −26.0 | −11.0 | 5.47 |
| 16:0 | 409.3 | 153.0 | −60.0 | −30.0 | −12.0 | 6.44 |
| 17:0 | 423.3 | 153.0 | −80.0 | −30.0 | −11.0 | 6.81 |
| 18:2 | 433.3 | 153.0 | −80.0 | −30.0 | −10.0 | 6.14 |
| 18:1 | 435.3 | 153.0 | −80.0 | −30.0 | −10.0 | 6.67 |
| 18:0 | 437.3 | 153.0 | −60.0 | −30.0 | −12.0 | 7.15 |
| 20:4 | 457.3 | 153.0 | −70.0 | −30.0 | −12.0 | 6.20 |
| 22:6 | 481.3 | 153.0 | −70.0 | −30.0 | −12.0 | 6.14 |
A previously published HPLC ESI-MS/MS method demonstrated what was termed “unidentified peaks” during chromatographic separation.10 The HPLC conditions reported here allow for a more reliable identification of LPA species by the elimination of these unidentifiable peaks. It also does not utilize undesirable solvent conditions that have plagued previous HPLC methods. For example, the highly acidic solvents used by some methods may result in on-column degradation of LPA and artificial conversion of LPC to LPA.11 Also, the highly organic solvents reported by other groups can affect the lifetime of the HPLC seals and are not functional for analysis of other lipid classes in the same sample run.9,12 Use of such lipid class specific HPLC methods significantly reduces the amount of data acquired from a single sample. The solvent system used in this study has previously been established for the study of sphingolipids, particularly ceramide, ceramide-1-phosphate, sphingosine, and sphingosine-1-phosphate.13 Therefore, this modified HPLC ESI-MS/MS method for LPA not only overcomes the undesirable HPLC conditions but also uses chromatographic separation that allows for other lipids to be efficiently analyzed during the same sample run.
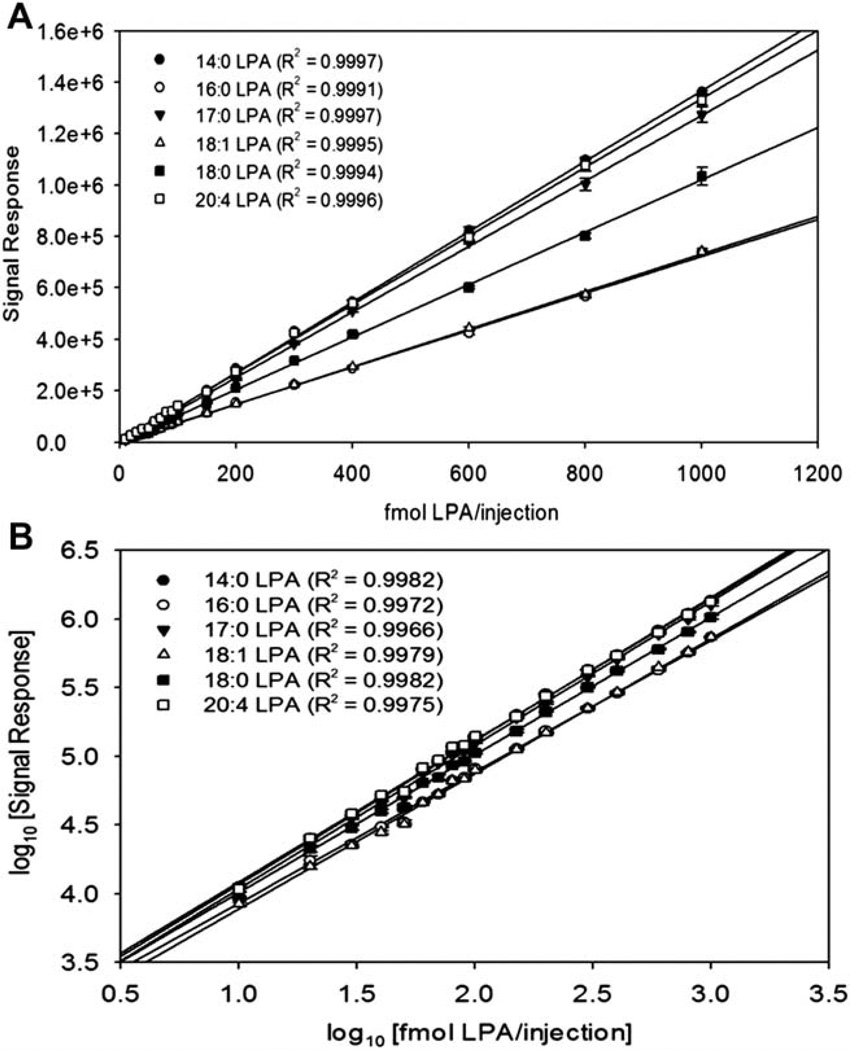
To investigate the sensitivity limits of this novel HPLC ESI-MS/MS method, a standard mixture of 14:0 LPA, 16:0 LPA, 17:0 LPA, 18:0 LPA, 18:1 LPA and 20:4 LPA was analyzed in concentrations from 10 fmols to 1 pmol per injection, and the signal response was measured as the area under the peak. When the results were plotted as signal response versus LPA concentration, a linear relationship was generated with correlation coefficients above 0.999 (Fig. 2A). When the results were plotted as a log-log plot of signal response versus LPA concentration, a linear relationship was generated with correlation coefficients above 0.997 (Fig. 2B). Intra-day and inter-day precision and accuracy of QC samples were within acceptable range (Table 2). The method produced a lower limit of quantitation of at least five times signal to noise ratio (in accordance to U.S. F.D.A. Guidelines for Bioanalytical Method Validation) of 10 femtomoles per injection for each chain length.14 This lower limit of quantitation conservatively represents a minimum of a fivefold increase in sensitivity from the most recent HPLC ESI-MS/MS LPA method and fifteen-fold increase in the remaining mass spectrometric reports in the literature for LPA analysis.6,10
Figure 2.
The method of detection for LPA species shows a linear response in the range from 10 fmol to 1 pmol for LPA standards. An internal standard mixture of 14:0 LPA, 16:0 LPA, 17:0 LPA, 18:0 LPA and 20:4 LPA was made in concentrations varying from 10 fmol/inject to 1 pmol/injection. (A) The results were plotted as signal response (area under the peak) versus amount standard in fmol/injection. Data are the average of four separate sample injections ± SEM. (B) The results were plotted as log10 of signal response (area under the peak) versus log10 fmol of standard per injection in a log-log plot. Data are the average of 35 four separate sample injections ± SEM.
Table 2.
Intra-day and inter-day precision and accuracy
| LPA Subspecies | Nominal Concentration (nM) |
Intra-day precision and accuracy Mean (n = 4) (nM) |
Mean accuracy (%) | CV (%) | Inter-day precision Mean (n = 4 days) (nM) |
CV (%) |
|---|---|---|---|---|---|---|
| 14:0 | 20 | 20.7 | 103.7 | 2.3 | 21.4 | 7.0 |
| 40 | 41.8 | 104.5 | 6.8 | 42.1 | 10.0 | |
| 80 | 85.4 | 106.7 | 2.7 | 81.9 | 8.1 | |
| 100 | 101.8 | 101.8 | 10.3 | 98.4 | 11.7 | |
| 16:0 | 20 | 21.4 | 107.2 | 3.3 | 20.5 | 8.8 |
| 40 | 40.6 | 101.5 | 5.8 | 41.4 | 7.5 | |
| 80 | 75.1 | 93.8 | 4.9 | 76.0 | 7.2 | |
| 100 | 96.9 | 96.9 | 7.1 | 97.2 | 8.3 | |
| 17:0 | 20 | 21.7 | 108.7 | 6.6 | 21.7 | 4.8 |
| 40 | 42.3 | 105.9 | 4.1 | 42.4 | 6.1 | |
| 80 | 80.1 | 100.1 | 6.3 | 81.8 | 5.5 | |
| 100 | 103.4 | 103.4 | 3.2 | 101.0 | 6.6 | |
| 18:1 | 20 | 20.7 | 103.3 | 1.9 | 20.6 | 9.3 |
| 40 | 39.8 | 99.4 | 5.5 | 41.2 | 5.6 | |
| 80 | 76.3 | 95.4 | 3.8 | 82.0 | 6.9 | |
| 100 | 95.8 | 95.8 | 3.6 | 103.9 | 8.3 | |
| 18:0 | 20 | 22.3 | 111.7 | 4.1 | 21.6 | 6.4 |
| 40 | 43.5 | 108.7 | 3.5 | 42.1 | 5.6 | |
| 80 | 79.9 | 99.9 | 3.7 | 82.5 | 4.3 | |
| 100 | 100.3 | 100.3 | 3.9 | 100.2 | 4.7 | |
| 20:4 | 20 | 21.4 | 107.2 | 10.3 | 20.0 | 11.7 |
| 40 | 40.4 | 101.1 | 5.6 | 41.3 | 10.7 | |
| 80 | 75.2 | 94.1 | 6.0 | 83.2 | 10.6 | |
| 100 | 91.2 | 91.2 | 3.0 | 99.3 | 9.8 |
Non-acidified extraction methods produce quantifiable LPA levels
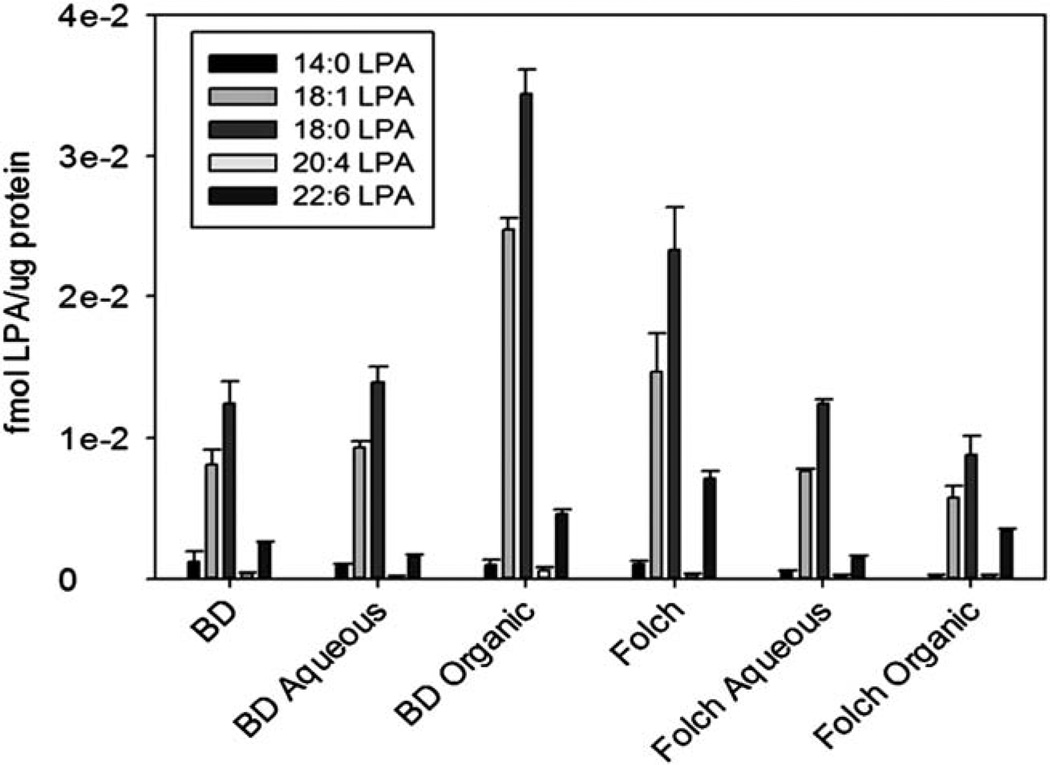
The reported HPLC ESI-MS/MS methods for quantitation of LPA have relied on extraction methods that required acidic treatments.9 Highly acidic conditions may contribute to the conversion of LPC to LPA, and thereby lead to LPA measurements that are not characteristic of actual sample levels. To address this issue, a number of extraction techniques without acidic treatment were tested on media samples. First, a direct media sample injection following polarity adjustment to starting HPLC conditions was tested, but this method proved to be insufficient due to a lack of sample concentration step during extraction (data not shown). Next, a simple lysophospholipid extraction for HPLC ESI-MS/MS analysis that consists of a single methanol solvent dilution and single step of centrifugation was utilized as previously reported.15 While this extraction was suitable for LPA extraction of in vivo samples, it was unsuccessful at providing sufficient recovery of the minute levels of LPA from in vitro samples because this protocol also lacked a sample concentration step (data not shown). A solid-phase extraction protocol utilizing a reversed phase silica based column was also tested, but did not provide sufficient results due to an inability to efficiently remove LPA from the column during the final elution step (data not shown). Finally, two non-acidified phase break extractions termed modified Bligh-Dyer and modified Folch extractions were tested and provided reliable recovery of LPA at levels above the limits of quantitation (Fig. 3). The Bligh-Dyer extraction resulted in recoveries of 82% for the internal standard 17:0 LPA, 83% for 14:0 LPA, 84% for 16:0 LPA, 82% for 18:0 LPA, 83% for 18:1 for LPA and 80% for 20:4 LPA. The Folch extraction process on the other hand gave 72% recovery for the internal standard 17:0 LPA. 75% for 14:0 LPA, 73% for 16:0 LPA, 72% for 18:0 LPA, 73% for 18:1 LPA, and 73% for 20:4 LPA. However, the Folch extraction technique demonstrated a more reproducible recovery of naturally occurring LPA species. Additionally, the combined organic and aqueous fractions approximately equalled the total Folch recovery (Fig. 3). Therefore, the total Folch extraction can be used with this modified HPLC ESI-MS/MS method due to its consistent recovery, simple protocol, and lack of producing artificial LPA contamination from acidic conditions.
Figure 3.
Non-acidic extraction methods for in vitro LPA quantitation showed that organic-aqueous phase break protocols provided sufficient recovery for HPLC ESI-MS/MS analysis for 1% FBS overnight treated DOV13-Zeo cells. BD - combined layers of Bligh-Dyer, BD Aqueous - aqueous layer of Bligh-Dyer, BD Organic- organic layer of Bligh-Dyer, Folch - combined layers of Folch, Folch Aqueous - aqueous layer of Folch, Folch Organic - organic layer of Folch. Extraction protocols are detailed in Experimental. Data represents n = 3 ± SEM. Results are normalized to protein levels from sample.
Autotaxin overexpression induces an increase in specific LPA species
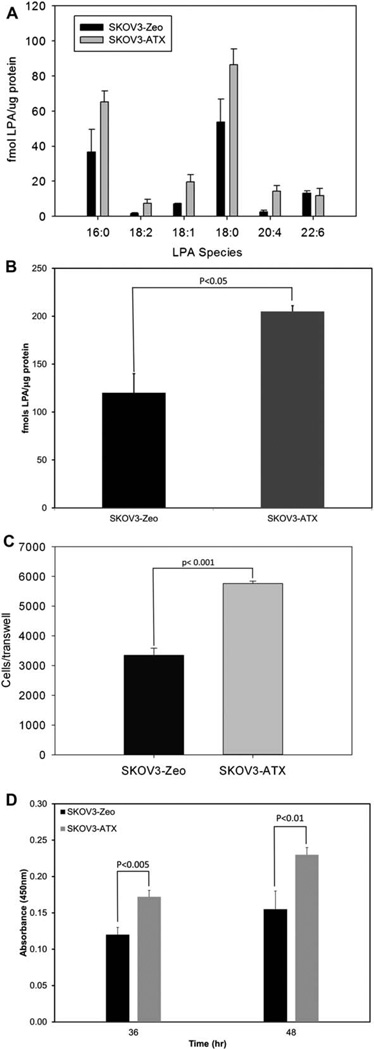
With the optimal extraction procedure for LPA determined, the protocol was fully operational for validation using biological applications. In this regard, we chose to examine the production of LPA in autotoxin overexpressing cells as compared to vector control cells. Fig. 4A shows that the 16:0, 18:1, 18:2, 18:0, and 20:4 LPA subspecies were significantly increased in the media from autotoxin overexpressing cells, but importantly, the 22:6 LPA subspecies remained relatively unchanged. Hence, autotaxin increases the levels of specific chain lengths of LPA. Furthermore, the total levels of LPA increased two-fold in autotaxin-overexpressing cells, as indicated in Fig. 4B.
Figure 4.
Comparison of control vector versus autotaxin-overexpressing clones show increased levels of LPA in autotaxin-overexpressing cell lines. (A) HPLC ESI-MS/MS analysis of LPA subspecies shows increases in 16:0, 18:2, 18:1, 18:0, and 20:4 LPA levels, but not 22:6 LPA, between control vector and autotaxin-overexpressing clones. A non-acidified Folch extraction was analyzed on overnight 1% FBS experimental protocol detailed in Experimental. Data represents n = 3 ± SEM. Results are normalized to protein levels from sample. (B) HPLC ESI-MS/MS analysis detailed in (A) shows an approximate two-fold increase in total LPA levels between control vector and autotaxin-overexpressing clones. Data represents n = 3 ± SEM. Results are normalized to protein levels from sample. (C) Conditioned media from autotaxin-overexpressing clones corresponded to increased cellular migration of SKOV3-Zeo when compared to conditioned media from control vector cell lines. Media from overnight 1% FBS experimental protocol was used as a chemo-attractant for cellular migration assay as detailed in Experimental. Data represents n = 4 ± SEM. (D) Conditioned media from autotaxin- overexpressing clones is associated with increased cellular proliferation when compared to media from control vector cell line. Cellular proliferation was calculated by subtracting Abs450nm of the blank media and baseline reading from the Abs450nm of 36 h and 48 h time points. Data represents n = 5 ± SEM. p-values were calculated as student’s t-test using SigmaPlot v. 12.0 (SyStat).
Prior to the development of this HPLC ESI-MS/MS method for quantitation of LPA from cell culture, quantitation of LPA production relied on the effect of exogenously introduced LPA on biological assays such as cellular migration and proliferation.16,17 Therefore, the observed increases in LPA levels produced by autotaxin overexpression as assayed by HPLC ESI-MS/MS were compared to the biological effect of these increased levels of LPA cellular migration and proliferation. Specifically, media from either SKOV3-Zeo or SKOV3-ATX was used as a chemoattractant in cellular migration assays. Media from SKOV3-ATX produced a two-fold increase in cellular migration when compared to media from SKOV3-Zeo, which correlated to the increase in LPA levels measured by HPLC ESI-MS/MS (Fig. 4C). To further validate the increase in LPA levels in SKOV3-ATX media, the effect of SKOV3-ATX and SKOV3-Zeo media on cellular proliferation was measured. Media from autotaxin overexpressing cells induced an increased in cellular proliferation as compared to SKOV3-Zeo media, which also correlated to the total increase in LPA levels measured by HPLC ESI-MS/MS (Fig. 4D). Overall, the increased levels of biological activity correlated to the increased levels of LPA measured using HPLC ESI-MS/MS, which demonstrates that the method developed in this study accurately reflects the levels of LPA.
Experimental
Materials
SKOV3-Zeo and SKOV3-ATX cell lines were established by transfection of SKOV-3 cells with pcDNA3.1/Zeo or pcDNA3.1/Zeo-ATX (kindly provided by J Aoki, Kawasaki Medical School, Okayama, Japan) The transfected cells were selected with zeocin (500 ng ml−1). Individual zeocin-resistant colonies were isolated by ring cloning and expanded sequentially in 24-well, 6-well and 60-mm plates. Expression of ATX protein was confirmed by immunoblotting. The ATX-positive clones were maintained as table lines in RPMI (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 2% penicillin/streptomycin (Bio Whittaker). SKOV3-Zeo and SKOV3-ATX clone cell lines were cultured under 5% CO2 at 37 °C with routine passage every 2–3 days. LPA standards (14:0, 16:0, 17:0, 18:0, 18:1, 20:4) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Ammonium formate (Fluka) were purchased from Sigma- Aldrich (St. Louis, MO, USA). HPLC grade methanol, HPLC grade chloroform and ACS grade formic acid (EMD Chemicals) were purchased from VWR (Bridgeport, NJ, USA). Cellular Proliferation Reagent WST-1 was purchased from Roche (Indianapolis, IN, USA).
HPLC ESI-MS/MS conditions
LPA species were separated using a Kinetex 2.6µ C18 100 Å50 × 2.1 mm reverse phase column on a Shimadzu 20-AD Series HPLC and subjected to mass spectrometric analysis using a 4000 QTRAP (ABSCIEX). Mass spectrometry parameters were as follows: Polarity-Negative, Ion Source: Electrospray, Q1 Resolution: Low, Q3 Resolution: Unit, Collision Activated Dissociation: High, MCA: No, Curtain Gas: 15.0 psi, Ion Source Temperature: 400.0 °C, Nebulizer Gas: 30.0 psi, Turbo Gas: 70.0 psi, Interface Heater: On, Ion Spray Voltage −4500.0 V, Collision Cell Exit Potential: −9.0. MRM transitions with corresponding declustering potentials, collision energies and collision exit potentials are listed in Table 1.
HPLC conditions were as follows: Total Flow: 300 µl min−1, Injection Volume: 10 µl, Column Oven: 50.0 °C. Solvents for reverse phase HPLC separation were: Solvent A, 58:41:1 methanol/water/formic acid and solvent B, 99:1 methanol/formic acid. Both solvents contained 5 mM ammonium formate. Solvent conditions for HPLC separations were: 100% Solvent A from 0–1 min, linear increase in minutes 1–7 from 100% Solvent A to 100% Solvent B, 100% Solvent B from minutes 7–8, immediate switch from 100% Solvent B to 100% Solvent A at minute 8, 100% A for minutes 8–10.
1% FBS serum experiments
SKOV3-Zeo and SKOV3-ATX cells were plated in 10 cm dishes and grown to 80% confluency in 10% FBS supplemented media. Cells were transferred to 1% FBS supplemented media overnight. Media was aliquoted into glass screw-top vials for appropriate extraction protocols and biological assays. Cells were harvested in 200 µl cold PBS. Results for HPLC ESI-MS/MS analysis were normalized to µg protein from harvested cells using a Bradford assay.
LPA extraction procedures
A modified Bligh-Dyer method was used to extract LPA from sample media. The extraction protocol was as follows: 100 µl of sample media, 1 ml chloroform, 500 µl methanol, 250 µl dH2O and 100 fmol 17:0 LPA standard were combined, vortexed, sonicated, and then centrifuged at 4000 rpm for 10 min. The aqueous (top) layer was transferred to a clean glass tube, dried and resuspended in 100 µl methanol for mass spectrometry analysis and is represented as “Bligh-Dyer Aqueous”. Organic (bottom) layer from the same extraction was transferred to a separate clean glass vial, dried and resuspended in 100 µl methanol for mass spectrometry analysis and is represented as “Bligh-Dyer Organic”. After a separate Bligh-Dyer extraction was performed, the aqueous and organic layers were combined, dried and resuspended in 100 µl methanol for mass spectrometry analysis and are represented as “Bligh-Dyer”.
A modified Folch method was also attempted for LPA extraction. The protocol was as follows: 200 µl sample + 2.5 ml chloroform + 1.25 ml methanol + 250 µl dH2O and 100 fmol 17:0 LPA standard were combined, vortexed, sonicated, and then centrifuged at 4000 rpm for 10 min. The aqueous (top) layer was transferred to a clean glass tube, dried and resuspended in 100 µl methanol for mass spectrometry analysis and is represented as “Folch Aqueous”. Organic (bottom) layer from the same extraction was transferred to a separate clean glass vial, dried and resuspended in 100 µl methanol for mass spectrometry analysis and is represented as “Folch Organic”. After a separate Folch extraction was performed, the aqueous and organic layers were combined, dried and resuspended in 100 µl methanol for mass spectrometry analysis and are represented as “Folch”.
Cellular migration assay
Cellular migration was measured in transwell chambers (pore size 8.0 µm; Corning Incorporated, Corning, NY) according to protocol in previous literature.16 In short, transwells were coated with 0.1 mg ml−1 collagen 1 and placed in lower chamber containing media from 1% FBS experiment detailed above. SKOV3-Zeo cells suspended in serum-free RPMI containing 0.1% fatty acid-free BSA were added to the upper chamber at 2.5 × 104 cells/well. Cells were allowed to migrate for 6 h under 5% CO2 at 37 °C,. Top of insert filter surface was washed with PBS and then stained with 0.1% crystal violet for 10 min. Filter surface was washed three times with PBS, and nonmigrated cells were removed from the top filter surface with a cotton swab. Migrated cells were stained by the crystal violet and counted under a microscope.
Conclusions
Measurement of LPA in vitro has proven to be unreliable until the development of the HPLC ESI-MS/MS method described herein. Previous HPLC ESI-MS/MS methods have focused on biological samples, which contain high levels of LPA, and which have also suffered from unfavorable HPLC conditions, hazardous and inaccurate extraction protocols, and inadequate sensitivity levels. This new HPLCESI-MS/MS will offer researchers a valuable tool for determination of the activities of LPA subspecies for the purpose of delineating biological mechanisms and pre-clinical drug development for diseases such as diabetes, atherosclerosis, and cancer. The increased sensitivity levels will also provide clinicians with better ability to quantitate LPA with the goal of early detection of disease states. Overall, the HPLC ESI-MS/MS method presented in this study supplies a new tool for further exploration of the biological functions of LPA.
Acknowledgements
This work was supported by research grants from the Veteran’s Administration (VA Merit Review I to C.E.C. and a Research Career Scientist Award to C.E.C.); the National Institutes of Health HL072925 (C.E.C.), CA117950 (C.E.C.), CA154314 (C. E.C.), CA154314 (C.E.C.), NH1C06-RR17393 (to Virginia Commonwealth University for renovation), 2R01CA102196 (X.F.); a National Research Service Award-T32 Postdoctoral Fellowship in Wound Healing (D.S.W.); a Career Development Award from the Department of Veterans Affairs (D.S.W.); a National Training Grant-T32 in Functional Lipidomics in Cardiovascular and Respiratory Diseases 5T32HL094290-02 (J.A.M.); and a research grant from ONO Pharmaceutical Company, Ltd (to C.E.C.).
Notes and references
- 1.Mills G, Moolenaar W. Nat. Rev. Cancer. 2003;3:582. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 2.Moolenaar W. J. Cell Biol. 2002;158:197. doi: 10.1083/jcb.200206094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohn EC, Hollister GH, DiPersio JD, Wahl S, Liotta LA, Schiffmann E. Int. J. Cancer. 1993;53:968. doi: 10.1002/ijc.2910530618. [DOI] [PubMed] [Google Scholar]
- 4.Tokumura A, Majima E, Kariya Y, Tominaga K, Kogure K, Yasuda K, Fukuzawa K. J. Biol. Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- 5.Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, Yamori T, Mills G, Inoue K, Aoki J, Arai H. J. Cell Biol. 2002;158:227. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aaltonen N, Laitinen J, Lehtonen M. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2010;878:1145–1152. doi: 10.1016/j.jchromb.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 7.Yoon H, Kim H, Cho S. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2003;788:85. doi: 10.1016/s1570-0232(02)01031-0. [DOI] [PubMed] [Google Scholar]
- 8.Xiao YJ, Schwartz B, Washington M, Kennedy A, Webster K, Belinson J, Xu Y. Anal. Biochem. 2001;290:302. doi: 10.1006/abio.2001.5000. [DOI] [PubMed] [Google Scholar]
- 9.Baker DL, Desiderio DM, Miller DD, Tolley B, Tigyi GJ. Anal. Biochem. 2001;292:287–295. doi: 10.1006/abio.2001.5063. [DOI] [PubMed] [Google Scholar]
- 10.Shan L, Jaffe K, Li S, Davis L. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2008;864:22–28. doi: 10.1016/j.jchromb.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 11.Meleh M, Pozlep B, Mlakar A, Meden-Vrtovec H, Zupancic-Kralj L. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2007;858:287–291. doi: 10.1016/j.jchromb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Murph M, Tanaka T, Pang J, Felix E, Liu S, Trost R, Godwin A, Newman R, Mills G. Methods Enzymol. 2007;433:1. doi: 10.1016/S0076-6879(07)33001-2. [DOI] [PubMed] [Google Scholar]
- 13.Wijesinghe D, Allegood J, Gentile L, Fox T, Kester M, Chalfant C. J. Lipid Res. 2010;51:641. doi: 10.1194/jlr.D000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brodie R, Hill H. Validation issues arising from the new FDA guidance for industry on bioanalytical method validation. Pergamon, Braunschweig, Germany [etc.]: 2002. [Google Scholar]
- 15.Zhao Z, Xu Y. J. Lipid Res. 2010;51:652–659. doi: 10.1194/jlr.D001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Z, Cheng C, Zhang H, Subler M, Wu J, Mukherjee A, Windle J, Chen C, Fang X. Mol. Biol. Cell. 2008;19:5435–5445. doi: 10.1091/mbc.E08-03-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo C, Luttrell LM, Price DT. J. Urol. 2000;163:1027. [PubMed] [Google Scholar]