Significance
Tumor suppressor Pbx-regulating protein-1 (Prep1) and myeloid ecotropic viral integration site-1 (Meis1) oncogene are transcriptional regulators, which bind to the same partner, pre–B-cell leukemia homeobox-1 (Pbx1). Meis1 overexpression induces tumorigenesis in Prep1i/i mouse embryonic fibroblasts, which is counteracted by Prep1 reexpression. The mechanism is unique: by binding to Pbx1, Prep1 regulates the stability of Meis1 and Pbx1. Influencing Meis1 stability, Prep1 controls the transcriptional landscape of Meis1 and hence, its tumorigenic activity. We also identify two novel Meis1 binding proteins, Ddx3x and Ddx5 RNA helicases, that are essential for cell proliferation and tumorigenesis, and their interaction with Meis1 is impaired at low Meis1 level. Thus, the level and function of three proteins (Prep1, Meis1, and Pbx1) of the same family are regulated by their stability, which depends on their interaction.
Abstract
Pbx-regulating protein-1 (Prep1) is a tumor suppressor, whereas myeloid ecotropic viral integration site-1 (Meis1) is an oncogene. We show that, to perform these activities in mouse embryonic fibroblasts, both proteins competitively heterodimerize with pre–B-cell leukemia homeobox-1 (Pbx1). Meis1 alone transforms Prep1-deficient fibroblasts, whereas Prep1 overexpression inhibits Meis1 tumorigenicity. Pbx1 can, therefore, alternatively act as an oncogene or tumor suppressor. Prep1 posttranslationally controls the level of Meis1, decreasing its stability by sequestering Pbx1. The different levels of Meis1 and the presence of Prep1 are followed at the transcriptional level by the induction of specific transcriptional signatures. The decrease of Meis1 prevents Meis1 interaction with Ddx3x and Ddx5, which are essential for Meis1 tumorigenesis, and modifies the growth-promoting DNA binding landscape of Meis1 to the growth-controlling landscape of Prep1. Hence, the key feature of Prep1 tumor-inhibiting activity is the control of Meis1 stability.
Two highly related members of the three amino acid loop extension (TALE) family of transcription factors play opposing roles in tumorigenesis. Myeloid ecotropic viral integration site-1 (Meis1) and Pbx-regulating protein-1 (Prep1, also known as pKnox1) belong to the MEINOX (a contraction of Meis and Knox) subfamily of the TALE family of homeoproteins that also includes the PBC (Pbx class) proteins. They are characterized by an atypical homeodomain harboring a TALE between the first and the second α-helices. Meis1 and Prep1 share highly conserved sequences in the domain required for the interaction with Pbx (HR1-HR2) and the homeodomain (HD) (1, 2). The Meis1- or Prep1-interacting domain in Pbx proteins is the PBC-A-PBC-B domain. These interactions can take place in both the absence and presence of DNA (3), and in fact, they are required for the transport of the complexes to the nucleus (4). In addition, they also regulate the stability of Pbx proteins (5). Furthermore, Meis1/Prep1-Pbx heterodimers form ternary complexes with anterior Hox proteins, modulating the specificity of Hox-dependent gene expression (6–8).
Meis1 cooperates with HoxA9 in leukemogenesis (9). Its overexpression accelerates the occurrence of HoxA9-induced acute myeloid leukemia (AML); moreover, MEIS1 is up-regulated in more than 80% of human AML and in acute lymphoid leukemia harboring mixed lineage leukemia (MLL) translocations (10, 11). Finally, Meis1 overexpression is a rate-limiting factor for the development of MLL-induced leukemias (12). In addition to leukemia, MEIS1 expression seems to be relevant in leiomyosarcomas (13), ovarian and uterine cancers (14), and neuroblastomas and medulloblastomas (15). All these evidences indicate that Meis1 acts as an oncogene to create and/or maintain a cellular context required for tumor formation.
In contrast to Meis1, Prep1 exerts a tumor-suppressive function. Indeed, the rare homozygous hypomorphic Prep1i/i mice that survive embryonic lethality (and express 2% Prep1 mRNA and 2–10% protein compared with WT) (16) as well as the heterozygous Prep1i/+ mice show a tumor-prone phenotype and develop pretumoral lesions or solid tumors late in life (17). In agreement with this phenotype, Prep1 haploinsufficiency accelerates Myc-driven lymphomagenesis in EμMyc mice. In fact, Prep1 overexpression not only does not accelerate the onset of HoxA9-induced leukemia but also slightly delays its occurrence (18). Moreover, tissue microarray immunohistochemistry revealed that PREP1 is absent or underexpressed in more than 70% of human tumors (17). In fact, in mouse and human cells in culture, Prep1 deficiency leads to genomic instability and accumulation of DNA damage, a condition that favors malignancy (19).
In the present work, we have confronted the opposing roles of Meis1 and Prep1 in tumorigenesis. (i) Meis1, on its own, can transform mouse embryonic fibroblasts (MEFs) but only in the absence of Prep1 (confirming the tumor-suppressive function of Prep1); however, growth of the Meis1-transformed MEFs is inhibited by Prep1 reexpression. (ii) Both Meis1 oncogenic and Prep1 tumor-suppressive activities require pre–B-cell leukemia homeobox-1 (Pbx1). (iii) Prep1 controls Meis1a-transforming activity by controlling its level through a Pbx1-dependent posttranslational mechanisms. (iv) Prep1 inhibits Pbx1-dependent Meis1a interaction with Ddx3x and Ddx5, two RNA helicases essential for the proliferation of Meis1a-induced tumors. (v) In addition to competing for Pbx1, Prep1 also reduces the range of Meis1 target genes partially by competitive binding to the same genes when overexpressed. This alteration results in a shift of target genes from Meis1-dependent, avian erythroblastosis oncogene B (ERBB)-dependent growth stimulation to Prep1-dependent, cell cycle-controlling gene categories.
Results
Meis1 Transforms Prep1i/i but Not Prep1+/+ MEFs.
The growth rate of independent cultures of primary Prep1i/i and WT MEFs was absolutely identical (Fig. S1A). However, immortalized Prep1i/i MEFs proliferated faster than WT (19) (Fig. S1B). Moreover, in primary WT MEFs, Meis1a overexpression had the same effect as RasV12 (i.e., blocked growth, possibly inducing oncogene-induced senescence) (Fig. S1C). However, Meis1a overexpression in the same WT MEFs induced cell growth when coinfected with another oncogene (i.e., RasV12 or Myc) (Fig. S1C), a feature expected for an oncogene (20). It must be noticed that Meis1 transforms primary hematopoietic progenitors only in collaboration with HoxA9 (18).
In p53−/− primary MEFs, in fact, Meis1a induced cell growth on its own (Fig. S1D). Surprisingly, in immortalized p53+/+ but Prep1i/i MEFs, overexpression of Meis1a robustly increased cell number (Fig. S1E). We conclude, therefore, that induction of cell growth is a property of overexpressed Meis1a but that this effect is evident only in the absence of p53 or Prep1.
We have used MEFs to measure Meis1a-dependent oncogenic transformation, looking for the loss of contact inhibition in monolayer, growth of cellular foci, and anchorage-independent growth in soft agar (21, 22).
Primary WT MEFs are not transformed by a single oncogene (20), and in fact, Meis1 or RasV12 did not transform primary MEFs (Fig. S1C). On Meis1a or Meis1a-HoxA9 retroviral transduction, immortalized passage-35 Prep1+/+ MEFs did not form as many transformed foci as passage-35 Prep1i/i cells (Fig. 1 A and B). Untransduced cells did not form any colony in agar. In agreement with this observation, individual infections with RasV12 or Myc retroviruses transformed both immortalized Prep1+/+ and Prep1i/i MEFs but at different rates (Fig. S2). Because the Meis1 ortholog Prep1 acts as a tumor suppressor (17, 19), we conclude that, in the absence of Prep1, a second oncogene (HoxA9, Rasv12, or Myc) is no longer required, allowing efficient transformation by Meis1 alone. In addition, the soft agar colonies formed by Prep1i/i cells were bigger than those formed by Prep1+/+ cells (Fig. 1B). The higher transforming activity of Meis1 in Prep1i/i cells agrees with the absence of the tumor-suppressive activity of Prep1. We also tested PREP1 tumor-suppressive function in the human neuroblastoma cell line IMR32. We screened a panel of eight different human brain tumor cell lines and selected the IMR32 cell line, which has the highest MEIS1 expression level (Fig. S3A), because in these cells, MEIS1 is amplified (23). We either overexpressed or down-regulated PREP1 in these cells using a specific vector or shRNA, respectively (Fig. S3B). As shown in Fig. S3C, PREP1 overexpression decreased, whereas its down-regulation increased the growth rate of IMR32 cells. Moreover, PREP1 level modulated the foci formation capacity and soft agar growth of the cells (Fig. S3 D and E). The number of foci in PREP1-overexpressing cells decreased, whereas it increased in PREP1 down-regulated cells (Fig. S3D). In agreement with these data, the number and the size of the soft agar colonies decreased on PREP1 overexpression and increased on PREP1 knockdown (Fig. S3E). These data provide additional support for the tumor-suppressive function of PREP1 in tumor cells overexpressing MEIS1.
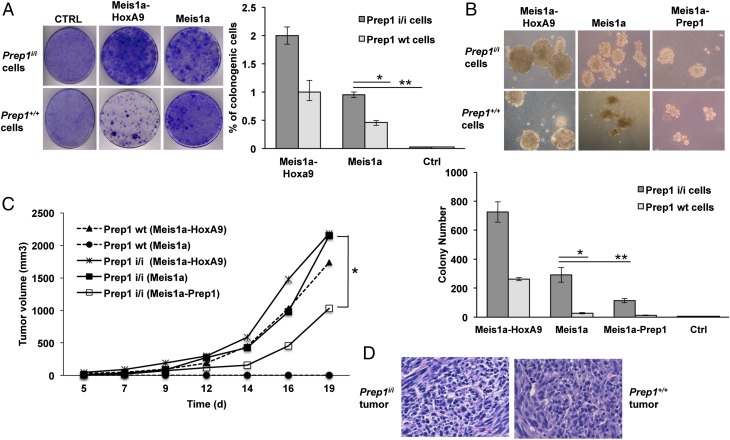
Fig. 1.
Effect of Meis1a on transformation and tumorigenicity of Prep1i/i and WT MEFs. (A) Meis1a alone induces colony formation in Prep1i/i MEFs. Colony formation assay of MEFs retrotransduced as indicated. Left shows as example plates seeded with 5 × 103 cells. The percentage of the colonogenic cells from two independent triplicate experiments is shown in Right (*P < 0.01; **P < 0.001). (B) Prep1 inhibits colony formation by Meis1a. The size (Upper) and the number (Lower) of colonies formed in soft agar by 1 × 105 Prep1i/i and WT MEFs per 6-cm plate and infected with the indicated retroviruses are shown (*P < 0.01; **P < 0.001). Data represent the mean of three independent triplicate experiments. (C) Prep1i/i and WT MEFs retrovirally transduced with the indicated vectors were s.c. injected into nude mice (1 × 106 cells per animal), and tumor volume was monitored. Lines represent the average of five animals per group. Differences between Prep1i/i-overexpressing Meis1a and Meis1a-Prep1 groups were statistically significant (*P < 0.05). (D) Examples of histological (H&E) analysis of Prep1i/i-overexpressing Meis1a and Prep1+/+-overexpressing Meis1a-HoxA9 tumors. Ctrl, control.
We then analyzed the effect of restoring Prep1 level in Prep1i/i MEFs. Colony formation in soft agar was significantly inhibited by Prep1 reexpression (Fig. 1B).
To definitively confirm the differential effect of Meis1 and Prep1 in tumorigenesis, we have tested the in vivo growth of the various cell lines by injecting them into immunodeficient nu/nu mice. First, s.c. transplantation of the infected cells revealed that only Prep1i/i (and not Prep1+/+) Meis1a-overexpressing cells formed tumors in mice within the time frame of the experiment (Fig. 1C). Second, Prep1+/+ cells required at least two oncogenes (Meis1 and HoxA9) for the tumorigenesis (Fig. 1C). Third, Prep1 reexpression remarkably decreased tumor growth rate of the Prep1i/i Meis1a-transformed cells (Fig. 1C). Histopathological examination of sections showed sarcoma-like tumors, with no major differences between the two genotypes (Fig. 1D). We conclude that Meis1a fully transforms MEFs but only in the absence of Prep1 and that Prep1 can inhibit the growth of Meis1a-dependent tumors.
Reexpression of Prep1 in Meis1a-Overexpressing Prep1i/i Cells Decreases Meis1a Stability.
Prep1i/i MEFs essentially express very low level of Meis1a (Fig. 2A) and no Prep1 (16). However, they robustly expressed the transduced FLAG-Meis1a. On Prep1 reexpression, the level of the transduced FLAG-Meis1a decreased (Fig. 2A). This data suggests that Prep1 affects Meis1a level by a posttranscriptional mechanism, because transduced Meis1a is expressed under a heterologous promoter. Indeed, the level of Meis1a is decreased by Prep1 overexpression when tested with both anti-FLAG and anti-Meis1 antibodies (Fig. 2A). Additionally, in fact, the decrease of Meis1a did not depend on a decrease of the level of its mRNA (Fig. 2B), confirming that Prep1 acts through a posttranscriptional mechanism. The observation was not limited to the MEFs because also PREP1 overexpression in IMR32 cells decreased the endogenous MEIS1A level (Fig. S4).
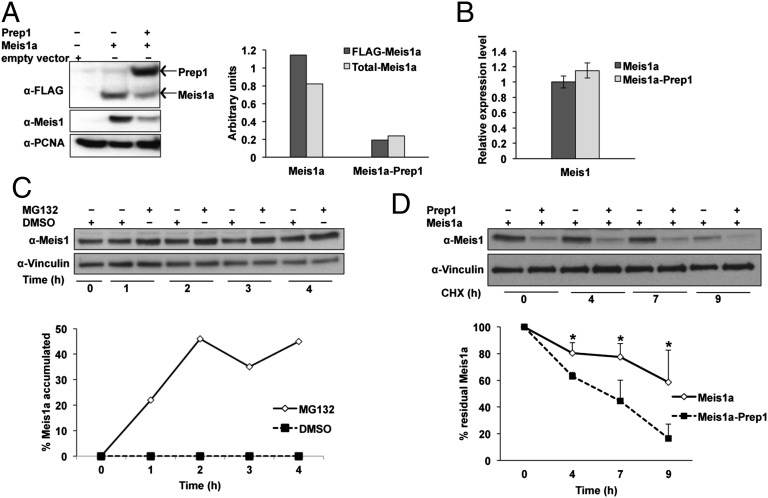
Fig. 2.
Ectopic expression of Prep1 destabilizes Meis1a in Prep1i/i MEFs. (A) Overexpression of Prep1 reduces the level of Meis1a in Meis1a-overexpressing Prep1i/i cells. Anti-FLAG antibody was used to check FLAG-Meis1a and FLAG-Prep1 expression in nuclear lysates of cells infected with Meis1a or Meis1a-Prep1. Anti-Meis1–specific antibody was used to check total Meis1a level in these cells. Proliferating cell nuclear antigen (PCNA) was used as a protein loading control. Bar graph shows densitometric analysis of Meis1a values normalized to the levels of PCNA. The data shown derive from a single experiment but are representative of at least three experiments showing the same result. (B) Effect of Prep1 overexpression on Meis1 mRNA level. Bar graph represents Meis1 transcript level measured by real-time PCR in Meis1a and Meis1a-Prep1–overexpressing Prep1i/i MEFs. The data shown derive from three independent triplicate experiments. (C) Meis1a degradation is proteasome-dependent in the presence of reexpressed Prep1. Exponentially growing passage-35 Prep1i/i MEFs transduced with both Meis1a and Prep1 were treated with 20 μM MG132 for the indicated time windows. Total extracts were processed for immunoblotting using anti-Meis1 antibody. Vinculin was used as a protein loading control. The graph represents the percentage of Meis1a accumulation on proteasome inhibition. (D) Prep1 overexpression decreases Meis1a stability. Passage-35 MEFs were treated with 10 μg/mL CHX and collected at the indicated time points. A specific Meis1 antibody was used for immunoblotting. The graph shows the percentage of residual Meis1a at various times after the addition of CHX. The levels at t = 0 are considered 100%. The data represent the average of three experiments. Error bar indicate SD (*P < 0.05).
We have tested the stability of Meis1a in Prep1-overexpressing cells using the proteasome inhibitor MG132 and the protein translation inhibitor cycloheximide (CHX). In the presence of MG132, the level of Meis1a in Prep1i/i cells overexpressing Meis1a-Prep1 increased with the time (Fig. 2C), indicating that Meis1a undergoes proteasomal degradation. Treatment of the Meis1a and Meis1a-Prep1 transduced Prep1i/i cells with CHX allowed us to compare the extent of degradation of Meis1a in the absence and the presence of Prep1. In the presence of Prep1, the steady state level of Meis1a decreased about fourfold (Fig. 2A) because of at least a threefold faster degradation rate, which was shown in the presence of CHX (Fig. 2D).
Pbx-Interacting Domain of Prep1 Is Required to Inhibit the Growth of Meis1a-Transformed MEFs.
Pbx is the most frequent interactor of Prep1 at the genomic level in mouse embryos (24). We, therefore, explored whether the Pbx-interacting region (HR1-HR2) of Prep1 or its other domains were required for the tumor growth. Different FLAG-Prep1 deletion mutants were constructed and are outlined in Fig. 3A.
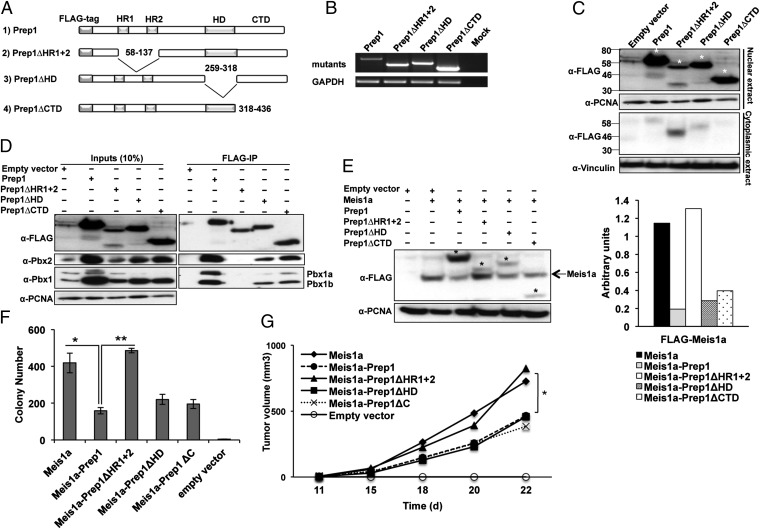
Fig. 3.
Identification of the Prep1 domain involved in inhibiting Meis1a-induced transformation. (A) Schematic representation of WT and mutant Prep1 proteins showing the positions of the Pbx interacting domain (HR1 and HR2), HD, and CTD. Blank spaces represent deletions. (B) RT-PCR analysis of different Prep1 mutants and GAPDH expression in Prep1i/i cells. (C) Intracellular location of the Prep1 mutants. Nuclear and cytoplasmic lysates were analyzed by immunoblotting with anti-FLAG antibody. PCNA and Vinculin were used as loading controls for nuclear and cytoplasmic lysates, respectively. Asterisks show the migration of the bands corresponding to FLAG-Prep1 and the mutants. (D) Pbx1 binding activity of the Prep1 mutants; 300 μg nuclear extracts of the Prep1i/i MEFs infected with FLAG-Prep1, Prep1 deletion mutants, or empty vector were immunoprecipitated using M2 anti-FLAG affinity resins and immunoblotted with the anti-Prep1–, anti-Pbx1–, and anti-Pbx2–specific antibodies. One-tenth of the lysates used for immunoprecipitation was loaded as input. (E) Effect of the Prep1 mutants on the level of Meis1a. Nuclear lysate was analyzed by immunoblotting using anti-FLAG antibody. PCNA was used as loading control. The migration of the FLAG-Meis1a band is shown by an arrow, and that band of FLAG-Prep1 and mutants is shown by asterisks. Bar graph shows the densitometric analysis. Meis1a values are normalized to the levels of PCNA. The data shown derive from a single experiment but are representative of at least three experiments showing the same result. (F) Effect of the Prep1 mutants on the colonogenic activity of Meis1; 1 × 105 Prep1i/i MEFs retrotransduced with Meis1a and Prep1 mutants were subjected to anchorage-independent soft agar growth assay. The number of colonies formed after 2 wk of culturing is shown (*P < 0.01, comparison with Prep1i/i MEFs reexpressing Prep1; **P < 0.0001, comparison with Prep1ΔHR1+2-overexpressing cells). Data represent the mean of three independent wells. Error bars indicate SD. (G) Effect of the Prep1 mutants on the tumorigenic activity of Meis1a. Prep1i/i MEFs overexpressing Meis1a-Prep1 or Prep1 mutants were s.c. transplanted into nude mice (1 × 106 cells per animal), and the tumor volume was monitored. Lines represent the average of five animals per group. Differences between Prep1i/i-overexpressing Meis1a and Meis1a plus Prep1, Prep1ΔHD, or Prep1ΔC groups are statistically significant (*P < 0.05).
The expression of each mutant was evaluated by semiquantitative RT-PCR (Fig. 3B) and immunoblotting (Fig. 3C) by infecting passage-35 Prep1i/i MEFs. Full-length and mutated proteins of the predicted molecular weight were observed by immunoblotting with an anti-FLAG antibody (Fig. 3C). Because the nuclear localization of Prep1 depends on the interaction with Pbx (4), the subcellular localization of the different mutant proteins was also tested by immunoblotting of nuclear and cytoplasmic extracts. Both Prep1ΔHD and Prep1Δ carboxy terminal domain (CTD) mutants were localized in the nucleus (Fig. 3C). Immunofluorescence assays confirmed these results (Fig. S5). The nuclear localization of the ΔHD and ΔCTD mutants indicates that the mutants are correctly folded, interact with Pbx, and are transported to the nucleus. Prep1ΔHR1+2 mutant was found in both the nucleus and the cytoplasm. In addition, we also tested the interaction of the mutant proteins with Pbx using anti-FLAG immunoprecipitation and Pbx1a, Pbx1b, and Pbx2 immunoblotting. In all cases, except with Prep1ΔHR1+2, the proteins coimmunoprecipitated Pbx. As expected, Prep1ΔHR1+2 did not interact (Fig. 3D). Thus, the partial presence of the ΔHR1+2 mutant in the nucleus, despite its inability to interact with Pbx, may suggest that additional mechanisms apart from the Pbx interaction can be involved in Prep1 nuclear localization.
It has been shown that Prep1i/i MEFs display an increased basal level of apoptosis and higher sensitivity to UV irradiation compared with WT (25). Moreover, Prep1 overexpression increases the sensitivity of the cells to genotoxic stress (26). Thus, we tested the effect of Meis1 and Prep1 mutants’ coexpression on the basal level of apoptosis in Prep1i/i MEFs. As shown in Fig. S6, cells overexpressing Meis1a-Prep1 or Meis1a-Prep1ΔC displayed a low-level increase of basal apoptosis compared with empty vector-transduced cells. Overexpression of Meis1a alone or together with other Prep1 derivatives had a lower effect. Thus, the marginal effect of Prep1 overexpression on apoptosis may have minimally affected the growth of the various cells.
To test the ability of the different Prep1 mutants to substitute for Prep1 in inhibiting the growth of Meis1a-transformed MEFs, passage-35 Prep1i/i MEFs overexpressing FLAG-Meis1a were retrovirally infected with different FLAG-Prep1 mutants. The expression of exogenous Meis1a and Prep1 mutants was checked by Western blotting with an anti-FLAG antibody (Fig. 3E). The decrease of FLAG-Meis1a was observed not only for the full-length protein but also for its mutants, with the exception of Prep1ΔHR1+2. The ability of ΔHD and ΔCTD mutants to interact with Pbx (see above) suggests that the Prep1-dependent Meis1a degradation relies on the formation of a Prep1–Pbx complex.
Meis1a-transformed Prep1i/i MEFs infected with full-length or the above Prep1 mutants’ retroviruses were subjected to anchorage-independent growth assays in soft agar. Full-length Prep1, Prep1ΔHD, and Prep1ΔCTD significantly reduced the number of soft agar colonies. The effect of full-length Prep1 was a threefold decrease, whereas the mutants showed a twofold effect. However, Prep1ΔHR1+2 showed no inhibitory effect (Fig. 3F). When the cells were injected in nu/nu mice, Prep1ΔHD and ΔCTD mutants still inhibited in vivo growth of Meis1a-transformed cells, unlike the Prep1ΔHR1+2 mutant that had no effect (Fig. 3G). These results indicate that the HR1+2 domain significantly contributes to Prep1 tumor-suppressive function, suggesting that the tumor-suppressive function is exerted by a Prep1–Pbx1 complex.
Both Growth of Meis1a-Transformed Prep1i/i MEFs and Its Inhibition by Prep1 Require Pbx1.
Both Meis1 and Prep1 interact with Pbx1 and Pbx2 through the conserved HR1+HR2 surface (4) and increase its stability (Fig. S7A). Prep1i/i MEFs overexpressing Meis1a or Meis1a-Prep1 were depleted of Pbx1 by one of two different specific shRNAs, resulting in 80–90% reduction of Pbx1 protein level. A control shRNA had no effect (Fig. 4A). Cells down-regulated for Pbx1 are, henceforth, indicated as Pbx1kd.
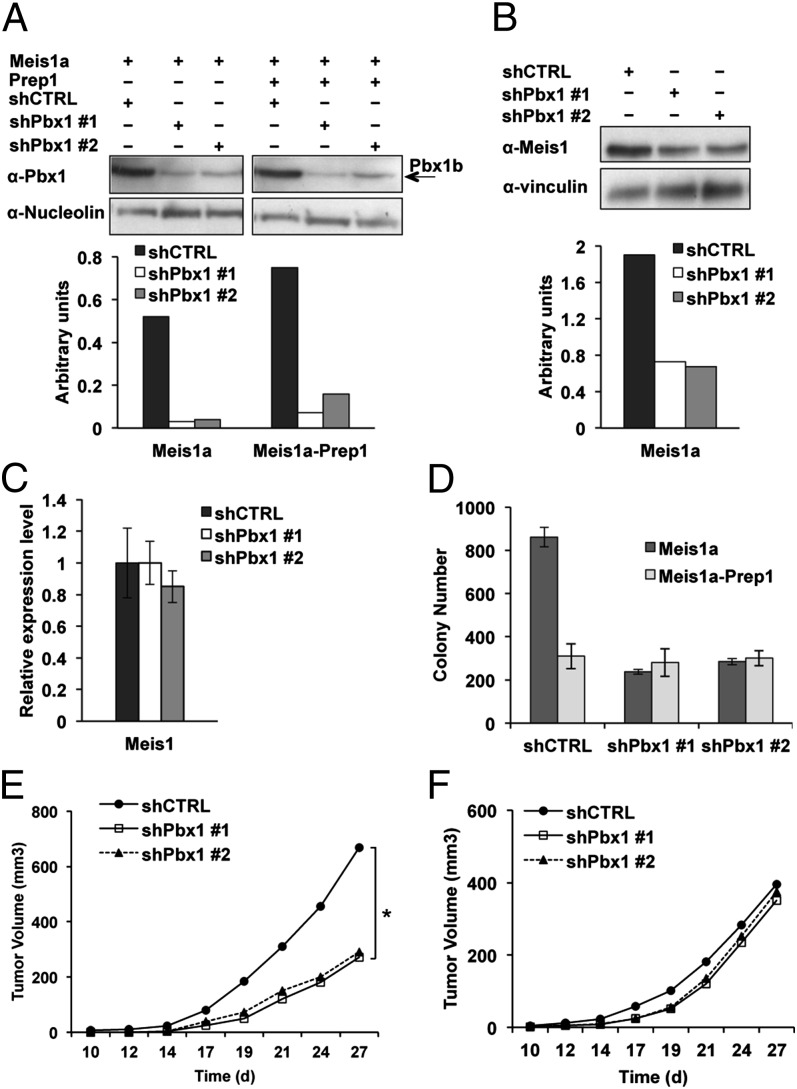
Fig. 4.
Pbx1 is required for Meis1a-mediated transformation and Prep1 inhibition of Meis1a tumorigenicity. (A) Down-regulation of Pbx1. The immunoblot shows the Pbx1 down-regulation efficiency in the nuclear lysate of Meis1a or Meis1a-Prep1–overexpressing Prep1i/i MEFs. Two different shRNAs were used against Pbx1, namely shPbx1 #1 and Pbx1 #2. Nucleolin was used for protein loading control. Quantification of the bands was done by densitometric analysis. (B) Effect of Pbx1 down-regulation on Meis1a level. Representative immunoblotting (Upper) and densitometric analysis (Lower) of total lysates treated with control or Pbx1-specific shRNAs. The graph shows the result of a single experiment, which is representative of at least three experiments giving the same results. Specific anti-Meis1 and Vinculin antibodies were used. (C) Effect of Pbx1 down-regulation on the level of Meis1 mRNA. The bar graph shows Meis1 transcript levels measured by real-time PCR in control or shPbx1-treated Meis1a-overexpressing cells. The data shown derive from three independent triplicate experiments. (D) Effect of Pbx1 down-regulation on the growth of cells in soft agar. Soft agar colony formation of Pbx1kd cells overexpressing Meis1a or Meis1a-Prep1 is shown relative to cells transduced with scrambled shRNA lentiviral vector; 1 × 105 cells per plate were used for each experimental point, and the experiment was performed in triplicate. Error bars indicate SDs of two independent experiments. (E and F) Effect of Pbx1 down-regulation on (E) in vivo tumor growth and (F) the Prep1 inhibition of tumor growth; 1 × 106 Pbx1kd cells overexpressing Meis1a or Meis1a-Prep1 were s.c. transplanted into nu/nu mice, and the tumor volume was monitored over time. Lines represent the average of five animals per group (*P < 0.05).
Pbx1kd cells displayed a decreased Meis1a protein level (Fig. 4B) but no decrease of Meis1 mRNA (Fig. 4C). Moreover, overexpression of tandem affinity purification (TAP) -Pbx1b in these cells sharply increased the Meis1a level (Fig. S7B). This data suggests that Meis1 is more stable in MEFs when they overexpress Pbx, possibly because Meis1 is in complex with Pbx1. To exclude the possibility that Meis1a level reduction was caused by its relocation, we checked the Meis1a level in nuclear and cytoplasmic extracts of Pbx1kd cells overexpressing Meis1a. As shown in Fig. S7C, in Pbx1kd cells Meis1a reduction was not caused by relocation, because there was no Meis1a in the cytoplasmic extract of these cells. Pbx1 down-regulation had no effect on Prep1 level in Prep1-overexpressed cells (Fig. S7D).
Pbx1kd-Meis1a–transformed cells proliferated almost as efficiently as the scrambled shRNA control cells (Fig. S7E). We also tested the effect of Pbx1 depletion on the basal level of apoptosis of Meis1a-transformed cells. As shown in Fig. S7F, down-regulation of Pbx1 did not induce apoptosis in the cells. However, it marginally decreased the apoptotic rate of these cells. We conclude that Pbx1 down-regulation does not affect either cell growth or apoptosis in this system. However, soft agar assays showed that the colony formation potential of Pbx1kd-Meis1a–transformed cells was substantially decreased (>69%) compared with the scrambled shRNA vector (Fig. 4D). In addition, nu/nu mice transplanted with Pbx1kd-Meis1a–transformed cells yielded smaller and slower-growing tumors compared with the control group (Fig. 4E). These effects were not observed in cells also overexpressing Prep1 (their tumorigenic activity was already lower) (Fig. 4 D and F), and in fact, in the absence of Pbx1, Prep1 showed no tumor-suppressive effect. Data in Fig. 4 E and F were part of the same experiment and are presented separately for the sake of clarity.
The fact that Meis1a does not transform Pbx1kd cells as efficiently as control indicates that Meis1a-mediated transformation is dependent on the presence of Pbx1 and further supports the idea that, not only in MLL-induced leukemia (12) but also in MEFs transformation, Meis1a functions in complex with Pbx. Likewise, the lack of the inhibitory effect of Prep1 on Pbx1kd-Meis1a–transformed cells shows that Prep1 inhibition of the Meis1a-induced transformation requires Pbx1. All these data suggest that Meis1a and Prep1 compete for Pbx1 and hence, that Pbx1 cooperates with an oncogene (Meis1) as well as a tumor suppressor (Prep1), confirming the previous data with Prep1ΔHR1+2 mutant (Fig. 3).
We conclude that Pbx1 plays an important role in both Meis1 tumorigenic and Prep1 tumor-suppressive activities, becoming a previously unidentified example of a single protein that can affect tumorigenesis in both directions. The underlying mechanism is, however, different from that mechanism of the Myc/Mad/Max system (Discussion).
Prep1 Competition for Pbx1b Binding Impairs Meis1a Interaction with Ddx RNA Helicases.
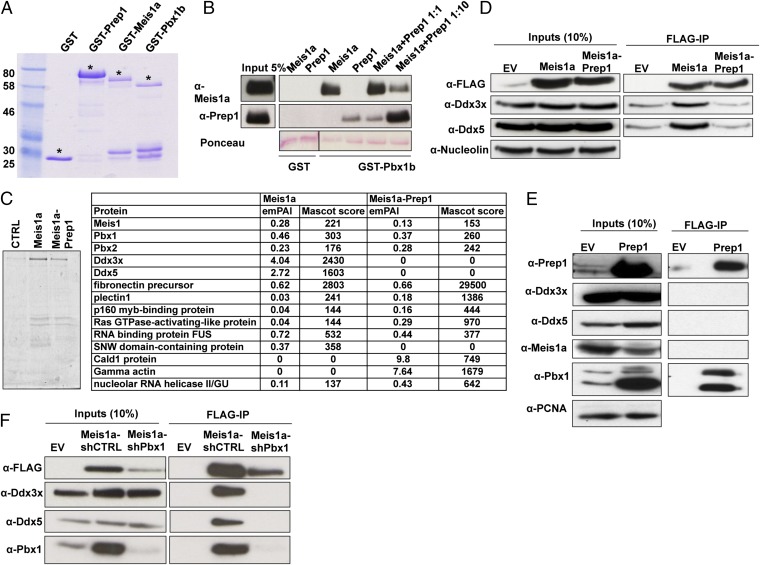
We directly tested competition between Prep1 and Meis1a for Pbx1b binding. We expressed GST-Prep1, Meis1, and Pbx1b in Escherichia coli and purified these proteins from bacterial lysate (Fig. 5A). Prep1 and Meis1a were cleaved from the GST-tag with PreScission protease, whereas GST-Pbx1b was used as bait in a pull-down assay with the purified, soluble Prep1 and Meis1a. GST was used as a negative control. As shown in Fig. 5B, a major Meis1a binding reduction was observed in the presence of a 10-fold excess of Prep1. Although not really quantitative, these results show that Prep1 competes with Meis1a to bind Pbx1b.
Fig. 5.
Prep1 reexpression affects Meis1a interacting proteins. (A) Purification of full-length Meis1a and Prep1 by SDS-PAGE. GST constructs were coupled to glutathion-sepharose beads and run on 8% (vol/vol) SDS-PAGE. Asterisks show the migration of GST and each GST construct in Coomassie blue-stained gel. (B) Pull-down analysis of purified Meis1a and Prep1 by GST and GST-Pbx1b by immunoblotting and Ponceau staining (indicated). (C) TAP–MS analysis of the Meis1a interactome. To define the Meis1a interactome in the absence and presence of Prep1, TAP-Meis1a was expressed in Prep1i/i and Prep1i/i MEFs reexpressing Prep1 and purified using glutathione-sepharose beads. Then, the purified complexes were eluted by tobacco etch virus (TEV) protease cleavage. The remaining complex after TEV cleavage was further purified using calmodulin beads and eluted by boiling in SDS sample buffer. The final TAP eluate was separated on a 10% (vol/vol) SDS-PAGE and stained by colloidal Coomassie blue. The entire lane was cut out from the gel and divided into different zones. Proteins identified by liquid chromatography–MS-MS complex compositions from each zone are listed. Exponentially modified protein abundance index (emPAI) is indicated as a measure of relative quantitation of proteins in each sample. A purification from cells infected with TAP empty vector is presented as control. (D) Ddx proteins coimmunoprecipitate with Meis1a; 300 μg nuclear extracts of the Prep1i/i MEFs infected with either FLAG-Meis1a alone or FLAG-Meis1a and Prep1 was immunoprecipitated with M2 anti-FLAG affinity resins and immunoblotted with the anti-FLAG, anti-Ddx3x, and anti-Ddx5 specific antibodies. One-tenth of the lysates used for immunoprecipitation was loaded as input. (E) Ddx proteins do not coimmunoprecipitate with Prep1. FLAG-Prep1 was immunoprecipitated from 300 μg nuclear extracts of the Prep1i/i cells infected with FLAG-Prep1 using M2 anti-FLAG affinity resins. The Prep1, Ddx3x, Ddx5, Meis1a, and Pbx1 in the inputs and precipitated samples were identified by immunoblotting using appropriate antibodies. IP, immunoprecipitation. (F) Pbx down-regulation decreases the Meis1–Ddx interaction. FLAG-Meis1a was immunoprecipitated from 300 μg nuclear lysate of Prep1i/i cells overexpressing Meis1a-shCTRL or Meis1a-shPbx1 and immunoblotted with the anti-FLAG–, anti-Ddx3x–, anti-Ddx5–, and anti-Pbx1–specific antibodies; one-tenth of the lysates was loaded as input.
To gain insight into the molecular mechanisms of Meis1a oncogenic activity and its inhibition by Prep1, we analyzed the Meis1a interactome using TAP (22) in the absence or the presence of overexpressed Prep1. We introduced a TAP-Meis1a (Materials and Methods) expression vector in Prep1i/i cells (Prep1i/i TAP-Meis1a) or Prep1i/i cells reexpressing Prep1 (Prep1i/i TAP-Meis1a-Prep1). The TAP-purified proteins were run on SDS-PAGE and identified by MS. Fig. 5C shows all of the identified proteins with their exponentially modified protein abundance index and Mascot score. This analysis showed that Meis1a copurified with Pbx1 and Pbx2, which was expected. Moreover, Meis1a also copurified with Mybbp1a, a protein that has also been shown to interact with Prep1 through the HR1 domain (27, 28). No Prep1 peptides were identified in the analysis, confirming that Meis1a and Prep1 do not form a complex. Importantly, we also identified additional proteins that copurify only in the absence of Prep1 (Fig. 5C), particularly the two ATP-dependent RNA helicases Ddx3x and Ddx5. Using the anti-FLAG antibody, we confirmed that Ddx3x and Ddx5 coprecipitated with Meis1 and not Prep1-specific antibodies (Fig. 5 D and E). Moreover, Meis1a did not coprecipitated with Prep1 antibody (Fig. 5E).
To explain the impairment of the Ddx3x and Ddx5 interactions with Meis1a in the presence of Prep1, we checked whether Pbx1, the common interactor of Prep1 and Meis1a, is required for Ddx interaction with Meis1a, because Prep1 and Meis1a compete for Pbx1 binding (detailed above). Indeed, the Meis1a-Ddx coimmunoprecipitation disappeared in cells in which Pbx1 has been down-regulated with a specific shRNA (Fig. 5F). Thus, Pbx1 is required for Meis1a interaction with Ddx proteins.
Ddx3x and Ddx5 Depletion Impairs Cell Growth and Colony Formation in Soft Agar.
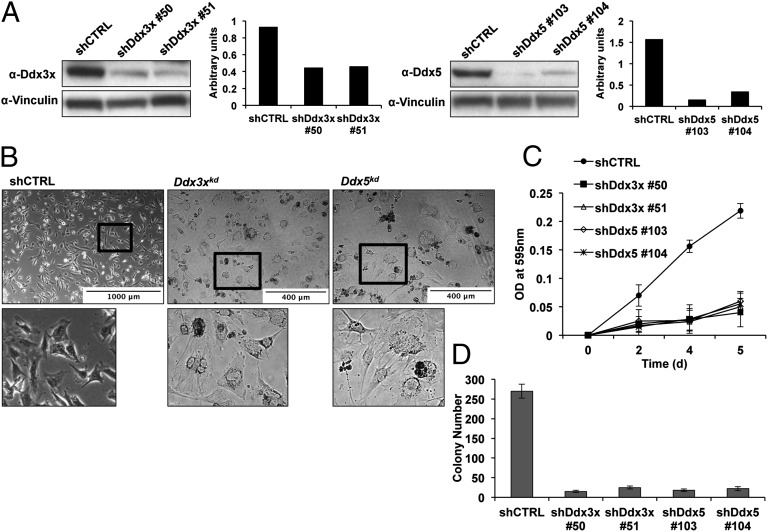
Ddx3 and Ddx5 are involved and required for tumorigenicity (29, 30). Indeed, depletion of Ddx3x and Ddx5 protein levels by ∼50% and ∼70%, respectively, using two independent shRNAs specifically targeting each gene (Fig. 6A) did not affect the expression of Meis1a or Pbx1 (Fig. S8) but changed the morphology of Meis1a-overexpressing Prep1i/i cells (but not control cells). The cells became abnormally flat, looking like serum-starved cells, and several round bodies appeared, possibly corresponding to cell carcasses (Fig. 6B). As expected (23, 31), Ddx down-regulation severely compromised the growth of Meis1a-transduced Prep1i/i cells (Fig. 6C). Ddx depletion also strongly reduced the Meis1a-dependent anchorage-independent growth in semisolid medium (Fig. 6D). Overall, these results indicate that both Ddx3x and Ddx5 have important roles in cell growth, and their down-regulation inhibits Meis1a-induced transformation in vitro.
Fig. 6.
Impaired growth and transformation activity of Ddx3x or Ddx5 depleted cells. (A) Down-regulation of Ddx3x and Ddx5. Prep1i/i cells overexpressing Meis1a were treated with Ddx3x, Ddx5, or scrambled shRNA retroviruses, resulting in efficient down-regulation as shown by immunoblotting. Vinculin was used as a loading control. Quantification of the bands was done by densitometric analysis. (B) Morphology of Ddx down-regulated cells. Scrambled, Ddx3xkd, and Ddx5kd shRNA-treated cells were photographed 1 wk after infection. Lower shows the zoom of selected areas from Upper. (C) Ddx down-regulation blocks the growth of Meis1a-transfected Prep1i/i MEFs; 5 × 104 Ddxkd cells were plated in six-well plates, fixed at the indicated time points, and stained with 0.1% crystal violet solution, and the OD at 595 nm was determined (SI Materials and Methods). The experiment was performed in triplicate. Error bars indicate SD. (D) Ddx down-regulation inhibits the colonogenic activity of Meis1a-transfected Prep1i/i MEFs. Soft agar colony formation of Ddx3xkd and Ddx5kd cells overexpressing Meis1a is shown relative to cells transduced with scrambled lentiviral vector; 1 × 105 cells per plate were used for each experimental point, and the experiment was performed in triplicate. Error bars indicate SD of two independent experiments.
Overexpression of Prep1 Specifically Affects the Fractions of Meis1 Binding Sites That Are Occupied only on Meis1a Overexpression.
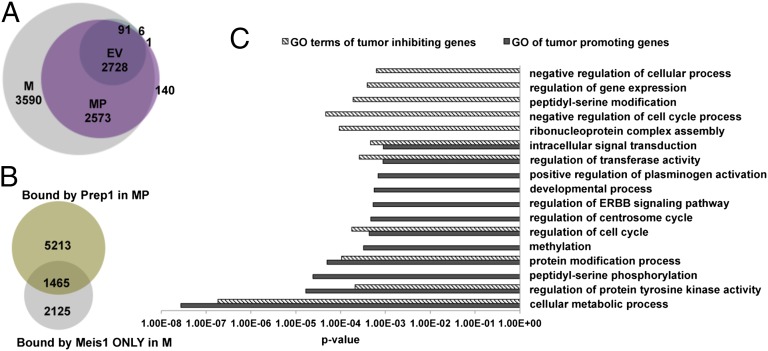
Because the tumorigenic effect of Meis1a is dependent on its overexpression, the inhibition by Prep1 should be acting on functions that are unique of overexpressed Meis1a. We, therefore, performed Meis1 ChIP-seq analysis in Prep1i/i cells bearing an empty vector (EV sample), Prep1i/i-overexpressing Meis1a (M sample), or Prep1i/i-overexpressing both Meis1a and Prep1 (MP sample). As shown in Fig. 7A, Meis1 bound to 2,825 genes in Prep1i/i MEFs (EV sample); on overexpression of Meis1a, the number increased to 8,981 (M sample). Overexpression of both Meis1a and Prep1 (MP sample) reduced the number to 5,441 (reduction of 39.4%). Overlap of these data shows that the reduction of the Meis1 binding gene caused by Prep1 was totally at the expense of those extra peaks bound under conditions of Meis1a overexpression (Fig. 7A) but not the sites bound by Meis1 in the parental cells (EV sample).
Fig. 7.
ChIP-seq analyses show that Prep1 competes with Meis1 at the genomic level and decreases the number of Meis1 binding genes. (A) Venn diagram showing the number of Meis1 binding sites in the EV, M cells, and MP cells. (B) Venn diagram showing the overlap between the sites bound by Prep1 in MP cells and the Meis1 sites bound by Meis1 only in M cells. (C) GO analysis of tumor-promoting and -inhibiting genes.
In addition, in MP cells overexpression of Prep1 prevented the binding of Meis1 to 3,590 genes, which were otherwise bound in the M cells; of these genes, 1,465 (40.8%) were directly occupied by Prep1, whereas the remaining 2,125 genes (59.2%) were not bound by either Prep1 or Meis1 in the MP cells (Fig. 7B). The 1,465 genes must include those genes that affect Meis1 oncogenic activity, and we have called them the tumor-inhibiting signature. However, 3,590 genes bound by Meis1 in the M cells must include those genes that mediate Meis1 tumorigenicity and hence, are called the tumor-promoting signature.
Gene Ontology (GO) analysis (Fig. 7C), showed that the tumor-promoting signature was uniquely enriched in tumor-linked categories, like the regulation of the ERBB pathway, the regulation of centrosome cycle, and peptidyl-serine phosphorylation, whereas the tumor-inhibiting signature was uniquely enriched for categories like the negative regulation of the cell cycle, which are easily recognizable as tumor-suppressing. In addition, Prep1 also competed with Meis1 for regulating different cellular processes, such as protein tyrosine kinase activity and regulation of cell cycle.
Discussion
Here, we show that Prep1 and Meis1a compete for Pbx1 to prevent and induce tumorigenesis, respectively, highlighting the unique feature of Pbx1 that assumes a tumor suppressor or oncogenic role, depending on the transcriptional partner. We also highlight that the oncogenic–nononcogenic choice depends on the relative levels of Meis1a and Prep1, which in turn, depend on the extent and nature of the interactions with the common partner Pbx1. In the presence of excess Prep1, Pbx1 is squelched, and Meis1a is destabilized. This change tips the balance against tumorigenesis. Other levels of competition have been unraveled, in which Prep1 prevents binding of Meis1 to genes only bound by the ectopic conditions without affecting those genes bound at the endogenous level.
This feature is not MEF- or mouse-specific; also, the human IMR32 neuroblastoma cell line is transformed by MEIS1 gene amplification (32), and in these cells, the decrease of MEIS1A induced by PREP1 modulates the tumorigenic potential (Fig. S3).
Two sets of data may seem to decrease the novelty of these findings. First, the Prep/Pbx/Meis system is reminiscent of the Myc/Max/Mad network, in which Max can act as an oncogene when bound to Myc or inhibit tumorigenesis when bound to Mad. However, in this case, a major role is played by the transactivation domain of Myc and Mad and not by the control of proteins stability (33). Second, the Meis1 CTD is required for HoxA9 cooperation in leukemogenesis (18, 34, 35). However, swapping the Prep1 with the Meis1 CTD has failed to convert Prep1 into an HoxA9-cooperating oncogene (36), which succeeds, however, when a full-length Prep1 is lengthened with the Meis1-CTD that induces Meis1-specific gene sets (36). However, these data do not prove that the central mechanism is transcriptional, because no studies of protein levels were carried out and there is peculiarity of the Prep1-Meis1 chimeric protein carrying both CTDs. Other than the difference of the cellular system studied (hematopoietic cells vs. fibroblasts), in the present study, Meis1 transforms Prep1i/i fibroblasts without a cooperating HoxA9 oncogene.
We show that Meis1 can act as a bona fide oncogene in fibroblasts and that its oncogenic capacity is strongly attenuated by its paralog Prep1. Therefore, Meis1 does not only function as an HoxA9-collaborating oncogene in leukemia (18) but also, may have independent activities in different tumor types.
Transformation of primary rodent fibroblasts requires at least two oncogenes or one oncogene in the absence of a tumor suppressor gene (20, 37). Accordingly, Meis1 alone transforms primary p53−/− MEFs but requires RasV12 or Myc to transform WT cells (Fig. S1 C and D). However, immortalized MEFs lacking Prep1 (Prep1i/i) are transformed by Meis1a alone as efficiently as when coexpressed with HoxA9. This feature seems to be somewhat specific, because HoxA9 neoplastic transformation of Prep1i/i cells requires a second cooperating oncogene. However, Prep1i/i MEFs are transformed by other single oncogenes, like RasV12 and Myc.
Meis1 alone transforms primary p53−/− but not Prep1i/i MEFs (Fig. S1 C and D). Hence, the lack of either p53 or Prep1 tumor suppressors favors Meis1-dependent tumorigenesis but in different ways.
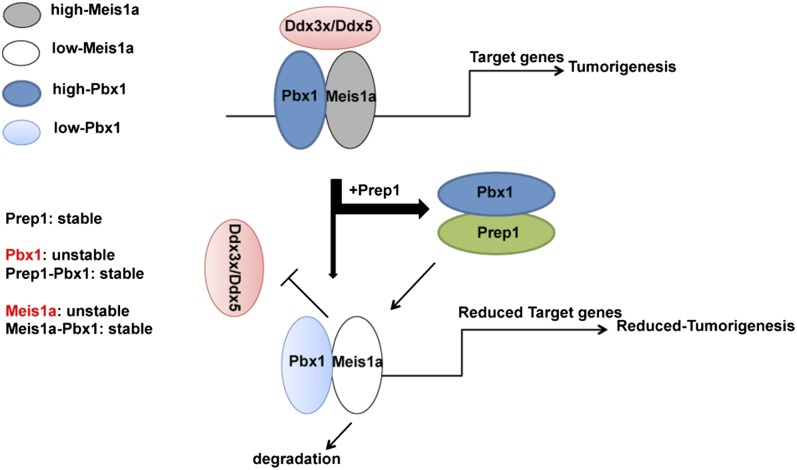
The inhibiting effect of Prep1 reexpression (Fig. 1C) allows investigating the mechanisms involved in the tumor-suppressive function of Prep1 as well as the oncogenic activity of Meis1. Previous data (4) show that Prep1 controls the stability of Pbx1. In addition, we now show that the absence of Prep1 induces a concurrent Pbx1 decrease by a posttranscriptional mechanism (Fig. S7A). However, Prep1 overexpression reduces Meis1a, again by a posttranscriptional protein degradation mechanism (Fig. 2). Because both Prep1 and Meis1a form stoichiometric complexes with Pbx1, overall, the data indicate that Prep1 stabilizes Pbx1, whereas Pbx1 stabilizes Meis1a. Possibly, only the dimeric forms of Meis1a are stable. Therefore, under conditions of low Pbx1 or when Pbx1 is squelched by excess Prep1, Meis1a is reduced. Fig. 8 details these concepts in a schematic form. (i) Prep1 and Meis1a compete with each other for binding Pbx1 (Fig. 5B) and form a dimeric complex. (ii) By squelching Pbx1, excess Prep1 reduces the level of Meis1a protein (not mRNA), which acquires a shorter proteasome-dependent half-life. Conversely, Meis1a level increases in the absence of Prep1. These effects act not only at the level of the endogenously encoded protein but also, on the ectopically expressed protein. (iii) Pbx1 knockdown also reduces Meis1a level. Hence, overexpression or absence of any of the three proteins determines the presence, nature, and amount of the formed dimer. (iv) Thus, Meis1a stability, which is Prep1- and Pbx1-dependent, determines Meis1a level and the extent of tumor formation. (v) The level of Meis1a–Pbx1 complexes determines the extent of interaction with specific tumorigenic partners (i.e., the newly identified Ddx3x and Ddx5) as well as the regulation of an Meis1-specific set of tumor-promoting genes.
Fig. 8.
Proposed model of how Prep1 attenuates Meis1a-induced tumorigenesis. We propose that Meis1a stability, which is Prep1- and Pbx1-dependent, determines Meis1a level and the extent of tumor formation. Prep1 attenuates Meis1a tumorigenic activities by squelching Pbx1 from Pbx1–Meis1a complexes. This alteration leads to proteasome-dependent degradation of Meis1a, impairment of its interaction with specific tumorigenic partners, like Ddx3x and Ddx5, and deregulation of an Meis1a-specific set of tumor-promoting genes.
Pbx1, therefore, is central in both Meis1 tumorigenic and Prep1 tumor-suppressive functions. Although the Pbx1–Meis1a complex induces tumorigenesis, Pbx1–Prep1 complexes prevent it. Thus, Pbx1 becomes an oncogene or a tumor suppressor in Meis1-dependent tumorigenesis, depending on the choice of its transcriptional partner. Prep1 posttranscriptionally determines the level of expression of Meis1a, hence determining the fate of the cells. Prep1-dependent posttranscriptional regulation of Meis1a is, therefore, central in the choice of the tumor cell fate.
In the scheme in Fig. 8, the tumorigenic decision depends only on the level of Meis1a. However, several data indicate a high degree of complexity. For example, Prep1 overexpression changes dramatically the genomic landscape of potential Meis1-regulated genes (Fig. 7). For instance, Prep1 overexpression counteracts the targets of overexpressed Meis1 but does not affect the basic Meis1 targets. Prep1 reprogramming of Meis1-overexpressing cells cancels specific target gene sets that favor tumorigenesis, like the ERBB signaling pathway, while identifying others that control tumor cell proliferation (like regulation of the cell cycle) (Fig. 7C).
In this work, we also identify two novel Meis1a interactors, the Ddx3x and Ddx5 RNA helicases (Fig. 5 C–F). Ddx3x and Ddx5 specifically interact with Meis1a and not Prep1. Therefore, the inhibition of these interactions by Prep1 is not through competition but possibly caused by the decrease of Meis1a induced by Prep1 overexpression. The interaction requires Pbx1 and in fact, disappears when Pbx1 is down-regulated (Fig. 5F). The disappearance of the Meis1a–Ddx interaction correlates well with the loss of tumorigenic activity in cells in which Prep1 is also overexpressed. Prep1 may dissociate Ddx3x and Ddx5 from the Pbx1–Meis1a complex by squelching Pbx1.
The interaction with the RNA helicases seems to be important for Meis1a tumorigenesis. Indeed, down-regulation of Ddx3x or Ddx5 inhibits the growth of the Meis1a-overexpressing cells and blocks soft agar colonies formation, which is in agreement with literature data causally connecting the helicases to neoplastic transformation (27, 28), transcription, and cell cycle regulation (23, 38). In fact, Ddx helicases interact with different transcription factors and act as transcriptional coactivators/corepressors (38–42). Thus, Meis1a might recruit Ddx3x and Ddx5 to the regulatory regions of its target genes or sequester them, preventing the interaction with other genes.
The ability of Meis1 to interact with Pbx proteins is essential for the induction and maintenance of MLL-mediated myeloid transformation (12). In this system, both the deletion of the HR1+2 domain of Meis1 (35) and the depletion of Pbx1 (12) abolish Meis1 oncogenic activity. Defining the function of different domains of a protein provides mechanistic insights. However, such information is missing for the tumor-suppressive functions of Prep1. In fact, the HR1+2 domain of Prep1 is indispensable for its tumor-suppressive function (Fig. 3 F and G). Deletion of the HR1+2 domain prevented Prep1 inhibition of Meis1-induced anchorage-independent cell growth and tumor formation. However, Prep1ΔHD and Prep1ΔC mutants still sustain Pbx1 interaction and inhibit Meis1-induced tumorigenesis. The requirement for the HR1+2 domain indicates that the inhibitory form of Prep1 is the Prep1–Pbx1 complex.
Meis1a stability may be a pharmacological target in tumors. Because the Prep1 HR1+2 domain is not only required to both squelch Pbx1 and induce Meis1a instability (Fig. 3D), it might be useful to reduce Meis1a level and tumorigenesis. In fact, protein domain or drugs able to interfere with the formation of a Pbx1–Meis1a complex, like recombinant HR1+2 domain from both Prep1 and Meis1a, may become possible drugs. Whether Meis1a HD and C-terminal domains might be useful in blocking Meis1-depending tumorigenesis by interfering with the Ddx interactions remains to be established. Attempts to synthesize PBX-DNA–inhibiting drugs have been published (43).
Because MEIS1 is implicated in neuroblastoma (15), our data in IMR32 cells can suggest a potential therapeutic role for MEIS1 and PREP1 in this malignancy. Indeed, MEIS1 tumorigenicity is partially rescued by PREP1 reexpression, which, therefore, can partially revert the tumorigenicity. The effect of PREP1 reexpression might be important for future therapeutic developments, because many human tumors express very low levels of PREP1 (17).
Pbx1 is not the unique partner of Prep1 and Meis1. However, Prep1 is the most frequent interactor of Pbx1 in DNA binding in mouse embryos, and the Prep1–Pbx complexes account for a very large percentage of Prep1 binding sites in vivo. However, Meis1 is a less frequent partner of Pbx1 in the mouse embryo (about one-third as frequent) (24). However, our in vivo data show that, in MEFs, Meis1 requires Pbx1 for oncogenic transformation. Indeed, Pbx1 depletion in Prep1i/i cells overexpressing Meis1 attenuates their tumorigenic potential as assayed by soft agar colonies and size of tumors in transplanted mice (Fig. 4E). Moreover, Prep1 is unable to rescue the residual Meis1-induced tumorigenicity in the Pbx1 down-regulated cells. In fact, cells coexpressing Meis1a and Prep1 show the same tumorigenic potential in Pbx1-depleted and control cells (Fig. 4F), further supporting the idea that Prep1 tumor-suppressive function requires Pbx1. Thus, Pbx1 becomes a previously unidentified example of a molecule that modulates tumorigenesis in both directions based on whether it interacts with the Meis1 oncogene or the Prep1 tumor suppressor.
ChIP-seq data have highlighted an interesting relationship between Prep1 and Meis1 binding sites. On overexpression, Meis1 binds to a larger number of binding sites than in nonoverexpressing cells. However, this feature is counteracted by the concurrent overexpression of Prep1 (Fig. 7A). In fact, Prep1 overexpression alters the genomic binding pattern of Meis1 by strongly decreasing the set of sites bound when it is overexpressed without affecting those sites bound in parental, nonoverexpressing cells. A fraction of these genes is bound by Prep1, whereas the rest remains unbound. This alteration is probably caused by the decrease of Meis1 in Prep1-overexpressing cells. These data would argue that Meis1 and Prep1 have common binding sites, which can be competitively bound. This concept will require deeper analysis.
In conclusion, we have presented evidence for a complex mechanism of tumorigenesis by members of the TALE family of transcription factors. Our data show that both Meis1 oncogenic and Prep1 tumor-suppressing activities depend on Pbx1 and hence, that Pbx1 is in the unique position to be both an oncogene and a tumor suppressor. Moreover, we confirm the tumor-suppressive function of Prep1 and show that the Meis1-dependent tumorigenicity is exerted and regulated primarily at the level of the Prep1-mediated stability of the oncogene. However, Prep1 can also use a broad range of mechanisms to suppress Meis1 oncogenicity from competition for Pbx1, which regulates Meis1a protein level and its interaction with Ddx, to limiting the range of Meis1 target genes and possibly, their deregulation.
Materials and Methods
Mice.
Prep1i/i mice have been described (16). Seven-week-old athymic nu/nu nude mice (Harlan Inc.) were used for tumorigenicity assays (Institutional Animal Care and Use Committee Project 110/11).
Cells.
Experiments were performed on the IMR32 cell line (ATCC), primary p53−/− and WT MEFs, and primary or passage-35 Prep1i/i and WT MEFs taken from E14.5 embryos. IMR32 cells were cultured in MEM supplemented with 10% (vol/vol) heat-inactivated FBS, 1% (vol/vol) nonessential amino acids, and 1 mM sodium pyruvate. MEFs were cultured in DMEM supplemented with 10% (vol/vol) heat-inactivated FBS.
Soft Agar Assay and Allograft Study.
In total, 1 × 105 cells per plate were seeded in 35-mm soft agar dishes in triplicate. Colonies were scored and counted after 12 d. For tumorigenicity assays, athymic nude mice were s.c. inoculated with 1 × 106 cells. Tumor growth and animal survival were monitored.
Colony Formation Assay.
For the colony formation assay, 3 × 103 MEFs or 2 × 104 IMR32 cells were plated into 100-mm plates, and medium was changed every 3 d. After 2 wk, colonies were fixed, stained with crystal violet, and counted.
MG132 and CHX Treatments.
Exponentially growing passage-35 Prep1i/i MEFs induced with Meis1a-Prep1 were treated with MG132 or CHX. A time course was performed by incubating cells for 1, 2, 3, and 4 h with 20 μM MG132 or 4, 7, and 9 h with 10 μg/mL CHX at 37 °C. Cells were lysed at the end of indicated time points, and total extracts were processed for immunoblotting.
Coimmunoprecipitation.
Preparation of nuclear and cytoplasmic protein extract was performed as described (28); 300 μg nuclear lysate was immunoprecipitated using M2 anti-FLAG magnetic beads (Sigma-Aldrich) following the manufacture’s protocol. Nuclear lysate from cells infected with EV was used as a negative control.
ChIP-seq and Data Analysis.
ChIP-seq on the various MEF lines was performed and analyzed using standard protocols (24). We used anti-Prep1 antibody and anti-Meis1/2 antibodies from Santa Cruz. ChIP DNA was sequenced on Illumina HiSeq 2000G. Single-end 50-bp reads were mapped with BWA software against the mm9 version of the mouse genome. The alignments were then used for peak calling. The data shown were obtained by selecting peaks with a P value < 10−6 and a false discovery rate < 0.5%. Venn diagrams were generated using BioVenn software (44). The original data have been deposited in the National Center for Biotechnology Information’s GEO database and are accessible under accession no. GSE54221.
GO Analysis.
GO term analysis was done with GOrilla, comparing the lists of genes with Prep1 or Meis1 binding sites against the list of all nuclear genes in Ensembl v63, with a P value cutoff of 10−3.
Statistical Analysis.
All values are expressed as mean ± SD. Statistical analyses were done by two-tailed Student t test, unless differently specified.
Pull-Down Assay.
Details of plasmid construction and pull-down assay are described in SI Materials and Methods.
TAP and MS Analysis.
Details of plasmid construction, TAP purification, and MS analysis are provided in SI Materials and Methods.
SI Materials and Methods describes in detail antibodies, shRNAs, plasmids, infections, cell growth, and apoptosis assays. Gene expression analysis is also described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Giorgio Iotti, Stefano Casola, and Eric C. W. So for stimulating scientific discussions. We also thank all members of the Istituto Fondazione Italiana per la Ricerca sul Cancro di Oncologia Molecolare (IFOM) Proteomic, Genomic and Imaging facilities for their constant and expert help. L.D. was supported by the Fondazione Umberto Veronesi (FUV) Fellowship. This work was supported by grants from the Italian Association for Cancer Research (AIRC), Cariplo, and the Italian Ministry of Health. We also thank the Italian National Research Council (CNR) Medical Research in Italy grant for the contribution.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54221).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321200111/-/DCSupplemental.
References
- 1.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291(2):193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 2.Longobardi E, et al. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev Dyn. 2014;243(1):59–75. doi: 10.1002/dvdy.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci USA. 1997;94(26):14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 1999;13(8):946–953. doi: 10.1101/gad.13.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longobardi E, Blasi F. Overexpression of PREP-1 in F9 teratocarcinoma cells leads to a functionally relevant increase of PBX-2 by preventing its degradation. J Biol Chem. 2003;278(40):39235–39241. doi: 10.1074/jbc.M304704200. [DOI] [PubMed] [Google Scholar]
- 6.Ferretti E, et al. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127(1):155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 7.Ferretti E, Schulz H, Talarico D, Blasi F, Berthelsen J. The PBX-regulating protein PREP1 is present in different PBX-complexed forms in mouse. Mech Dev. 1999;83(1-2):53–64. doi: 10.1016/s0925-4773(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19(7):5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroon E, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence HJ, et al. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13(12):1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 11.Imamura T, et al. Frequent co-expression of HoxA9 and Meis1 genes in infant acute lymphoblastic leukaemia with MLL rearrangement. Br J Haematol. 2002;119(1):119–121. doi: 10.1046/j.1365-2141.2002.03803.x. [DOI] [PubMed] [Google Scholar]
- 12.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21(21):2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baird K, et al. Gene expression profiling of human sarcomas: Insights into sarcoma biology. Cancer Res. 2005;65(20):9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 14.Crijns AP, et al. MEIS and PBX homeobox proteins in ovarian cancer. Eur J Cancer. 2007;43(17):2495–2505. doi: 10.1016/j.ejca.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Spieker N, et al. The MEIS1 oncogene is highly expressed in neuroblastoma and amplified in cell line IMR32. Genomics. 2001;71(2):214–221. doi: 10.1006/geno.2000.6408. [DOI] [PubMed] [Google Scholar]
- 16.Ferretti E, et al. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol Cell Biol. 2006;26(15):5650–5662. doi: 10.1128/MCB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longobardi E, et al. Prep1 (pKnox1)-deficiency leads to spontaneous tumor development in mice and accelerates EmuMyc lymphomagenesis: A tumor suppressor role for Prep1. Mol Oncol. 2010;4(2):126–134. doi: 10.1016/j.molonc.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21(1):224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iotti G, et al. Homeodomain transcription factor and tumor suppressor Prep1 is required to maintain genomic stability. Proc Natl Acad Sci USA. 2011;108(29):E314–E322. doi: 10.1073/pnas.1105216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 21.MacPherson I, Montagnier L. Agar suspension culture for the selective assay of cells transformed by polyoma virus. Virology. 1964;23:291–294. doi: 10.1016/0042-6822(64)90301-0. [DOI] [PubMed] [Google Scholar]
- 22.Freedman VH, Shin SI. Cellular tumorigenicity in nude mice: Correlation with cell growth in semi-solid medium. Cell. 1974;3(4):355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 23.Jalal C, Uhlmann-Schiffler H, Stahl H. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 2007;35(11):3590–3601. doi: 10.1093/nar/gkm058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penkov D, et al. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep. 2013;3(4):1321–1333. doi: 10.1016/j.celrep.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 25.Micali N, Ferrai C, Fernandez-Diaz LC, Blasi F, Crippa MP. Prep1 directly regulates the intrinsic apoptotic pathway by controlling Bcl-XL levels. Mol Cell Biol. 2009;29(5):1143–1151. doi: 10.1128/MCB.01273-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Micali N, et al. Down syndrome fibroblasts and mouse Prep1-overexpressing cells display increased sensitivity to genotoxic stress. Nucleic Acids Res. 2010;38(11):3595–3604. doi: 10.1093/nar/gkq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Díaz VM, et al. p160 Myb-binding protein interacts with Prep1 and inhibits its transcriptional activity. Mol Cell Biol. 2007;27(22):7981–7990. doi: 10.1128/MCB.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Díaz VM, Bachi A, Blasi F. Purification of the Prep1 interactome identifies novel pathways regulated by Prep1. Proteomics. 2007;7(15):2617–2623. doi: 10.1002/pmic.200700197. [DOI] [PubMed] [Google Scholar]
- 29.Botlagunta M, et al. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27(28):3912–3922. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Causevic M, et al. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene. 2001;20(53):7734–7743. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- 31.Lai MC, Chang WC, Shieh SY, Tarn WY. DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol. 2010;30(22):5444–5453. doi: 10.1128/MCB.00560-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones TA, Flomen RH, Senger G, Nizetić D, Sheer D. The homeobox gene MEIS1 is amplified in IMR-32 and highly expressed in other neuroblastoma cell lines. Eur J Cancer. 2000;36(18):2368–2374. doi: 10.1016/s0959-8049(00)00332-4. [DOI] [PubMed] [Google Scholar]
- 33.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 34.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106(1):254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mamo A, et al. Molecular dissection of Meis1 reveals 2 domains required for leukemia induction and a key role for Hoxa gene activation. Blood. 2006;108(2):622–629. doi: 10.1182/blood-2005-06-2244. [DOI] [PubMed] [Google Scholar]
- 36.Bisaillon R, Wilhelm BT, Krosl J, Sauvageau G. C-terminal domain of MEIS1 converts PKNOX1 (PREP1) into a HOXA9-collaborating oncoprotein. Blood. 2011;118(17):4682–4689. doi: 10.1182/blood-2011-05-354076. [DOI] [PubMed] [Google Scholar]
- 37.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 38.Schröder M. Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochem Pharmacol. 2010;79(3):297–306. doi: 10.1016/j.bcp.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 39.Fuller-Pace FV. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34(15):4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin S, Rossow KL, Grande JP, Janknecht R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 2007;67(16):7572–7578. doi: 10.1158/0008-5472.CAN-06-4652. [DOI] [PubMed] [Google Scholar]
- 41.Rossow KL, Janknecht R. Synergism between p68 RNA helicase and the transcriptional coactivators CBP and p300. Oncogene. 2003;22(1):151–156. doi: 10.1038/sj.onc.1206067. [DOI] [PubMed] [Google Scholar]
- 42.Wortham NC, et al. The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERalpha-positive breast cancer. Oncogene. 2009;28(46):4053–4064. doi: 10.1038/onc.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji T, Lee M, Pruitt SC, Hangauer DG. Privileged scaffolds for blocking protein-protein interactions: 1,4-disubstituted naphthalene antagonists of transcription factor complex HOX-PBX/DNA. Bioorg Med Chem Lett. 2004;14(15):3875–3879. doi: 10.1016/j.bmcl.2004.05.068. [DOI] [PubMed] [Google Scholar]
- 44.Hulsen T, de Vlieg J, Alkema W. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.