Significance
Toll-like receptors (TLRs) play key roles in innate immune recognition of pathogens leading to the activation of innate host defenses and sometimes to immunopathology. In humans, there are 10 identified TLR members, designated TLRs 1–10. Of those, TLR10 remains the only one without a defined ligand or function. We now provide previously unidentified evidence that TLR10 plays a role in innate immune responses following influenza viral infection. Influenza viruses are subtyped on the hemagglutinin (H) and neuraminidase (N) proteins. Compared with seasonal influenza virus H1N1, the highly pathogenic avian influenza virus H5N1 is a more potent inducer of TLR10 expression. Given the importance of innate immune sensing receptors in host defense and pathogenesis, evidence of a functional role for TLR10 in influenza infection suggests that this receptor probably plays a role in a range of other viral and perhaps other microbial diseases.
Abstract
Toll-like receptors (TLRs) play key roles in innate immune recognition of pathogen-associated molecular patterns of invading microbes. Among the 10 TLR family members identified in humans, TLR10 remains an orphan receptor without known agonist or function. TLR10 is a pseudogene in mice and mouse models are noninformative in this regard. Using influenza virus infection in primary human peripheral blood monocyte-derived macrophages and a human monocytic cell line, we now provide previously unidentified evidence that TLR10 plays a role in innate immune responses following viral infection. Influenza virus infection increased TLR10 expression and TLR10 contributed to innate immune sensing of viral infection leading to cytokine induction, including proinflammatory cytokines and interferons. TLR10 induction is more pronounced following infection with highly pathogenic avian influenza H5N1 virus compared with a low pathogenic H1N1 virus. Induction of TLR10 by virus infection requires active virus replication and de novo protein synthesis. Culture supernatants of virus-infected cells modestly up-regulate TLR10 expression in nonvirus-infected cells. Signaling via TLR10 was activated by the functional RNA–protein complex of influenza virus leading to robust induction of cytokine expression. Taken together, our findings identify TLR10 as an important innate immune sensor of viral infection and its role in innate immune defense and immunopathology following viral and bacterial pathogens deserves attention.
Pattern recognition receptors (PRRs) play a key role in recognizing pathogen-associated molecular patterns (PAMPs) on microbes leading to the initiation and orchestration of innate and adaptive immune responses. Toll-like receptors (TLRs) are an important group of PRRs; their activation leads to the induction of interferons (IFNs) and cytokines, which play a major role in host defense and pathogenesis (1). TLRs are expressed on a variety of cell types, including macrophages (2), and play a key role in the innate immune recognition of viruses, e.g., influenza viruses (3, 4). TLR signaling activates a cascade of intermediates, including adaptor proteins, protein kinases, and effector transcription factors, which result in the production of type I IFN (e.g., IFN-β) inducing an antiviral state in cells, providing an important first line of defense against virus infection. Other proinflammatory cytokines, e.g., tumor necrosis factor (TNF)-α and interleukin (IL)-6 are also triggered by TLR signaling and are important determinants of the balance between beneficial host innate immune responses and immunopathology.
Presently there are 10 known TLR members, TLRs 1–10, identified in humans. TLRs are a family of transmembrane receptors consisting of an N-terminal extracellular domain (ECD) with multiple leucine-rich repeats, linked by a transmembrane domain (TMD) to a cytosolic signaling domain called Toll/IL-1R (TIR) (5). Among the 10 different TLRs, TLR10 remains an orphan receptor without a known agonist or function. A major constraint to research on TLR10 has been the lack of a suitable mouse model because TLR10 is a pseudogene in mice due to sequence gaps and multiple retroviral insertions (6).
Recently, studies have reported genetic polymorphisms of TLR10 in humans in association with diverse diseases, including Crohn’s disease (7), asthma (8), urothelial bladder cancer (9), and nasopharyngeal cancer (10). Furthermore, TLR10 expression is also reported to be induced in response to reactive oxygen species in hypoxic cells (11). These data may suggest a role of TLR10 in the pathogenesis of different diseases, but the mechanisms remain obscure.
One study has suggested that TLR10 cooperates with TLR2 in sensing triacylated lipopeptides and recruits myeloid differentiation factor 88 (MyD88) to the activated receptor complex (12). However, they showed that native TLR10 coexpressed with TLR2 as a heterodimer in a human colonic epithelial cell line did not respond to lipopeptide stimulation. Response only occurred in an artificial situation, when TLR2 was coexpressed with a chimeric receptor, in which the ECD and TMD of TLR1 was replaced with that of TLR10.
Macrophages are key sentinel cells of host defense and are abundantly found in all tissues of the body, including the alveolar spaces of the lung. Our previous studies have demonstrated that influenza A virus replicates and induces proinflammatory cytokine responses and paracrine cytokine cascades in human peripheral blood monocyte-derived macrophages and alveolar epithelial cells (13–15).
Defining the mechanisms of host defense after virus infection is important for the development of novel therapeutic options (16). In this study, we report the induction of TLR10 expression in human macrophages in response to influenza A virus infection. Influenza viruses are subtyped on the hemagglutinin (H) and neuraminidase (N) proteins. Compared with seasonal influenza virus H1N1, the highly pathogenic avian influenza virus H5N1 is a more potent inducer of TLR10 expression. TLR10 was found to be involved in a novel innate immune sensing and signaling pathway contributing to cytokine induction following influenza A virus infection. We used dual-luciferase and influenza ribonucleoprotein (RNP) assays to demonstrate the critical involvement of TLR10 in cytokine production induced by influenza A viruses. This study provides previously unidentified evidence suggesting a role of TLR10 in virus infection.
Results
TLR10 Is Expressed in Human Macrophages and Its Expression Is Enhanced by Influenza Virus Infection.
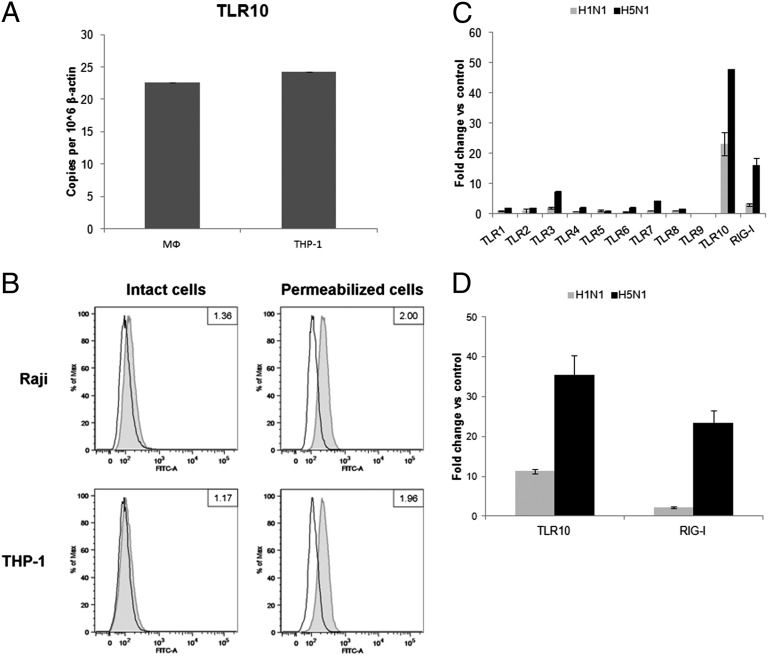
Among the 10 known human TLRs, TLR3, -7, and -8, together with retinoic acid-inducible gene 1 (RIG-I) are known to recognize influenza virus infection (17, 18). In this study, we have confirmed the basal expression levels of TLR10 mRNA in primary human monocyte-derived macrophages and a human monocytic cell line (THP-1) (Fig. 1A). TLR10 protein expression was also determined using flow cytometry (Fig. 1B). As previous data had demonstrated that TLR10 was expressed in cells of the B-cell lineage, Raji B cells were used as positive control for TLR10 staining (19). THP-1 cells expressed abundant TLR10 protein, at a level comparable to Raji B cells. TLR10 was predominantly expressed intracellularly, with very little or no detection on the surface of the intact cells (Fig. 1B).
Fig. 1.
Expression of TLR10 in primary human macrophages and THP-1 cells. (A) Basal mRNA expression of TLR10 in primary human macrophages (MФ) and THP-1 cells determined by RT-PCR. (B) Determination of TLR10 protein by flow cytometry in intact or permeabilized human Raji and THP-1 cell lines. Cells were stained with either FITC-conjugated isotype control antibody (open histogram) or FITC-conjugated anti-human TLR10 antibody (gray tinted histogram). Data shown are the ratio value of a signal-to-isotype control based on the median of FITC fluorescence intensity. Changes in expression level of TLR10 in response to infection by H1N1 and H5N1 influenza A viruses with MOI of 2 at 6 h after infection compared with mock infection in (C) primary human macrophages and (D) THP-1 cells. Results shown are representative of biological replicates performed in three independent experiments and error bars indicate SD of technical triplicates.
The expression level of TLRs 1–10 and RIG-I after influenza A infection of primary human macrophages was investigated (Fig. 1C). Compared with mock-infected cells, the relative fold expression of TLR10 mRNA increased markedly following influenza A H1N1 (∼20-fold increase) or H5N1 (>40-fold increase) infection. These findings were confirmed by the marked increase in number of TLR10 cDNA copies following H1N1 or H5N1 infection (Fig. S1A). Similarly, influenza A infection markedly increased TLR10 mRNA expression in THP-1 cells (Fig. 1D). Thus, both cell types were used for subsequent experiments in this study.
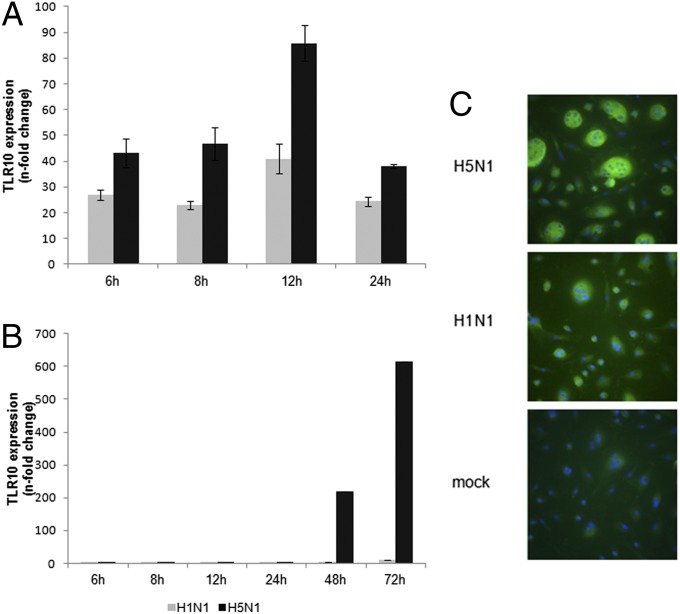
Next, the kinetics of TLR10 expression induced by influenza A virus was measured over a time course of influenza infection in macrophages. Expression of TLR10 was markedly up-regulated by H5N1 virus infection at a multiplicity of infection (MOI) of 2 at 6 h postinfection (Fig. 2A). Expression of TLR10 was further increased at 12 h after H5N1 virus infection. Infection by H5N1 virus at lower MOI (MOI = 0.001) led to strong induction of TLR10 at 48 h postinfection with a further increase at 72 h postinfection (Fig. 2B). Human H1N1 virus induced a similar TLR10 expression pattern but to a lesser intensity compared with H5N1 virus (Fig. 2 A and B). Induction of TLR10 expression was also detected in human alveolar epithelial cells (A549 cells) in response to influenza A virus infection (Fig. S1B). TLR10 protein expression following influenza A virus infection was demonstrated using immunofluorescence staining. Mock-infected human macrophages only showed very little or no immunostaining for TLR10. Increased TLR10 protein expression was observed in H5N1- and H1N1-infected cells, with H5N1 virus inducing more protein expression compared with H1N1 virus (Fig. 2C). Specificity of TLR10 antibody was confirmed using Western blotting on wild-type and TLR10-overexpressed THP-1 cells (Fig. S2).
Fig. 2.
Kinetics of influenza A virus-induced TLR10 expression in human macrophages. Expression of TLR10 in human macrophages infected by H1N1 or H5N1 influenza A viruses at (A) MOI of 2 or (B) 0.001 compared with mock infection at different postinfection time were assessed by RT-PCR. The result of one representative experiment from three independent experiments with three donors is shown. Error bars indicate SD of three technical triplicates. (C) TLR10 protein expression (FITC-green) was detected using immunofluorescent staining. Cells were counterstained with DAPI (blue) and viewed in a fluorescent microscope (magnification 400×). Multinucleate giant cells are seen, especially in H5N1 virus infected cells.
The difference between two viruses on TLR10 induction is much more pronounced following low MOI infection (Fig. 2B) and its expression was shown to have a correlation with virus replication and production, especially at the low MOI (Fig. S3B). In single cycle infection experiments (MOI = 2), although the two viruses had comparable viral replication (Fig. S3A), H5N1 virus remained a more potent inducer of TLR10.
Mechanism of TLR10 Induction by Influenza A Virus Infection.
Induction of TLR10 is dependent on virus replication.
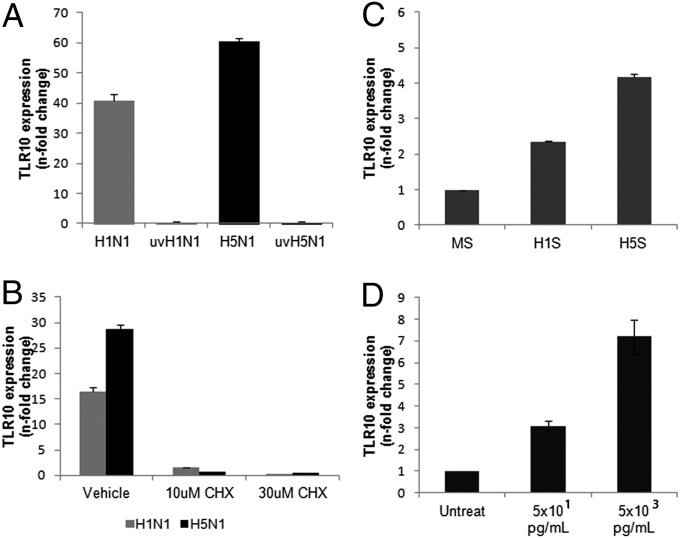
To determine if the induction of TLR10 in infected cells is dependent on virus replication, identical doses of infectious or UV-inactivated influenza virus were added to human macrophages. UV-inactivated influenza virus did not induce TLR10 expression compared with live virus infection at 6 h after infection, suggesting that the expression of TLR10 is dependent on virus replication (Fig. 3A).
Fig. 3.
Mechanism of TLR10 induction by influenza A virus infection. (A) TLR10 expression in cells infected by UV-irradiated H1N1 (uvH1N1) and H5N1 (uvH5N1) influenza A viruses at MOI of 2 were compared with nonirradiated virus infection at 6 h postinfection. (B) Suppression of TLR10 expression in H1N1 and H5N1 virus infected cells (MOI of 2) by treatment with protein synthesis inhibitor, cycloheximide (CHX). (C) Induction of TLR10 expression in uninfected human macrophages after challenged with H1N1 supernatant (H1S) and H5N1 supernatant (H5S) compared with mock supernatant (MS) at 3 h after stimulation. (D) Induction of TLR10 by TNF-α in a dose-dependent manner. Data shown are representative of biological replicates performed in three independent experiments and error bars indicate SD of technical triplicates.
Induction of TLR10 mRNA is dependent on de novo protein synthesis.
To determine if TLR10 expression is induced directly by virus transcription or via the autocrine/paracrine feedback effects from other mediators or by viral protein synthesis, the protein synthesis inhibitor, cycloheximide (CHX) was used to treat the cells before and after virus infection (Fig. 3B). Induction of TLR10 by both influenza A viruses at 6 h after infection was markedly suppressed in the presence of 10 or 30 μM CHX (Fig. 3B), suggesting that de novo protein synthesis is required for the induction of TLR10 mRNA. The CHX doses used did not induce cytopathic effect or alter the expression of the housekeeping gene β-actin.
Soluble factors secreted by influenza A virus-infected human macrophages induce TLR10 expression.
As the expression of TLR10 required viral replication and de novo protein synthesis, we investigated whether soluble mediators (e.g., cytokines or other intermediates) released from influenza A virus-infected human macrophages could induce TLR10 expression in uninfected cells. We thus developed an in vitro cell model to investigate the effect of these soluble mediators on TLR10 expression. Culture supernatants of human macrophages infected with influenza A virus were collected at 6 h after infection, filtered through a 100-kDa pore filter (Millipore) to remove any infectious virus, and added to uninfected human macrophages. Real-time RT-PCR data showed that TLR10 expression was up-regulated by more than fourfold in uninfected cells stimulated with supernatants of H5N1-infected cells compared with mock-infected cells. Similarly, supernatants from H1N1-infected cells also induced TLR10 expression in uninfected cells, but to a lower extent (2.4-fold) (Fig. 3C). Because TNF-α is one of the cytokines that are up-regulated in human macrophages and secreted to culture supernatant in response to influenza A virus infection (13), we next investigated the effect of TNF-α on TLR10 expression. Different concentrations of TNF-α (5 × 101 and 5 × 103 pg/mL) were chosen to mimic the secretory level of this cytokine in response to H1N1 and H5N1 infections, respectively (13). Expression of TLR10 was up-regulated in the presence of TNF-α in a dose-dependent manner (Fig. 3D), suggesting that TNF-α is one possible mediator for TLR10 induction in human macrophages after influenza virus infection.
Cytokine induction by influenza A virus is mediated via TLR10.
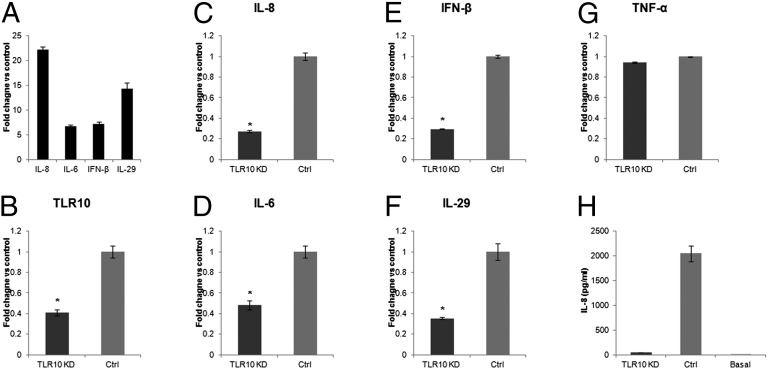
We next investigated whether the expression of cytokines typically up-regulated following influenza A virus infection (Fig. 4A) (14, 15) is TLR10 dependent. TLR10 shRNA knockdown (KD) in THP-1 cells (Fig. 4B) significantly reduced the expression of TLR10 mRNA by ∼60% compared with control cells. H1N1 virus-induced IL-8 and IL-6 mRNA expression was significantly suppressed by 73% and 52%, respectively, in TLR10 shRNA knockdown cells compared with control cells at 6 h after infection (Fig. 4 C and D). TLR10 knockdown also significantly suppressed IFN-β and IL-29 mRNA induction in virus-infected cells by 71% and 65%, respectively (Fig. 4 E and F). TNF-α mRNA expression was not affected by TLR10 knockdown (Fig. 4G). Secretory IL-8 protein was measured using ELISA, and in agreement with the mRNA data, H1N1 virus-induced IL-8 protein was markedly suppressed in TLR10 knockdown cells compared with control (Fig. 4H).
Fig. 4.
Cytokines induced by influenza A virus are regulated via TLR10. (A) Induction of proinflammatory cytokines and IFNs in H1N1 virus infected THP-1 cells. (B) The knockdown efficiencies of TLR10 shRNA knockdown (TLR10 KD) in THP-1 cells assessed by RT-PCR. Relative expression of virus induced proinflammatory cytokine genes, (C) IL-8 and (D) IL-6 and type I and III IFNs, (E) IFN-β and (F) IL-29 in TLR10 shRNA knockdown cells compared with control at 6 h after H1N1 virus infection. (G) TNF-α gene expression was not affected by TLR10 knockdown. (H) Secretory IL-8 protein level in culture supernatant collected from TLR10 knockdown and control cells after virus infection determined using ELISA. Basal IL-8 protein level in culture supernatant before infection was included for comparison. Data shown are average of two independent experiments and error bars indicate SD of six measurements obtained from the two independent experiments. (*P < 0.05).
Similar to the H1N1 virus, H5N1 virus-induced proinflammatory cytokine and IFN expression was also markedly suppressed in TLR10 knockdown cells generated using the siRNA silencing approach (Fig. S4). In conclusion, the data here indicated that expression of proinflammatory cytokines as well as type I and type III IFNs induced by influenza A virus infection was, at least partially, mediated via TLR10.
We investigated the effect of TLR10 knockdown on virus replication. Influenza virus titer was comparable in TLR10 knockdown cells compared with control (Fig. S5).
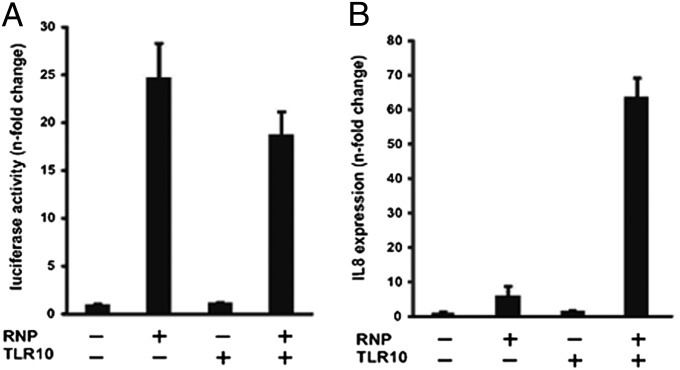
As influenza A virus-induced IL-8 was found to be the most suppressed cytokine observed in the TLR10 gene silencing experiments (Fig. 4 and Fig. S4), we then performed a virus RNP reconstitution assay to further determine whether the induction of IL-8 is TLR10 dependent. Viral RNP complex genes (PB2, PB1, PA, and NP) of influenza A virus were transfected into 293T cells to mimic viral replication. The plasmid pPOLI-NS-Luc that expresses vRNA-like luciferase RNA was used to monitor the activity of viral polymerases. If the RNP is functional, the vRNA produced from the pPOLI-NS-Luc plasmid will be transcribed by the viral polymerases leading to the synthesis of the luciferase protein used as a reporter (20). Human TLR10 expression plasmid was also cotransfected into 293T cells to examine the effect of its expression on viral replication as reflected in the luciferase activity. At 20 h posttransfection, the luciferase activity had increased 25-fold and 20-fold in RNP-transfected and RNP/TLR10-transfected cells, respectively (Fig. 5A). This increased luciferase activity confirmed that the RNP reconstitution assay is functional in both RNP-transfected and RNP/TLR10-transfected cells and that TLR10 expression has no influence on the polymerase activity of H5N1 virus. During the course of influenza virus infection, RNP and RNP-generated products are some of the PAMPs recognized by cellular sensors leading ultimately to the production of cytokines, including IL-8 (21). Therefore, we next determined the IL-8 mRNA expression in the same setting. IL-8 expression has only increased 5-fold in RNP-transfected cells, whereas its expression was increased 65-fold in RNP/TLR10-transfected cells at 20 h posttransfection (Fig. 5B). The data suggest that TLR10 substantially enhanced vRNP-induced activation of IL-8 expression. These results are in agreement with the effect of TLR10 gene silencing data, indicating that TLR10 can contribute to IL-8 induction during viral replication.
Fig. 5.
Expression of proinflammatory cytokine IL-8 was mediated via TLR10 during influenza A virus replication. (A) Dual-luciferase assay: 293T cells were transfected with pPOLI-NS-Luc reporter and pRL-CMV control plasmid, together with the plasmids including the expression plasmids of PB2, PB1, PA and NP (RNP) and human TLR10 expression plasmid (TLR10). Cells were harvested at 20 h after transfection. Luciferase activity shown was the Firefly luciferase activity normalized to Renilla luciferase activity. (B) RT-PCR analysis of proinflammatory cytokines IL-8 mRNA expression. Similar to A, 293T cells were harvested at 20 h after transfection. Data shown are representative of two independent experiments and error bars indicate SD of technical triplicates.
Discussion
Since its initial identification in 2001 (22), little is known about the role of TLR10. In the present study, we demonstrated a previously unidentified role of TLR10 in innate immune responses elicited by influenza virus infection.
Basal expression of TLR10 was detectable in human primary macrophages and the related cell line THP-1. Our present data show the induction of this receptor by H5N1 virus infection, and to a lesser extent by human H1N1 virus, suggesting TLR10 may have a role related to virus host defense and pathogenicity. The difference between H5N1 and H1N1 viruses in TLR10 induction is far more pronounced following low MOI infection, which is likely to be more akin to the conditions pertaining to natural infection in vivo.
We investigated the mechanism of TLR10 induction after influenza virus infection. Our findings suggested that induction of TLR10 requires virus replication and is dependent on de novo protein synthesis following infection. Culture supernatants of virus-infected human macrophages modestly up-regulated TLR10 expression in uninfected cells in a paracrine manner (Fig. 3C), as did TNF-α (Fig. 3D), but such up-regulation was markedly less than that observed with replicating virus (Fig. 3A). The relative roles of virus replication, secretory mediators (e.g., TNF-α) in supernatants and synergy between these in TLR10 induction needs further investigation.
Although the function of TLR10 was previously unknown, we now identify its involvement in cytokine expression in influenza virus-infected cells. TLR10 contributes to the induction of both antiviral IFNs such as type I and type III IFNs as well as proinflammatory cytokines such as IL-8 and IL-6. Cytokine dysregulation has been proposed to be one of the mechanisms contributing to the pathogenesis of H5N1 disease (13–15, 23). The significantly higher level of TLR10 induction following H5N1 infection compared with H1N1 infection also raises the possibility that TLR10 may play a role in immunopathology and virus pathogenesis by amplifying proinflammatory cytokine cascades. Our previous data has also demonstrated a role of RIG-I and to a lesser extent, TLR3, in regulating influenza A virus-induced cytokines (24). These pathways may act in synergy as RIG-I, TLR3, and TLR10 are all up-regulated upon H5N1 infection. The cross-talk and balance of different PRR signaling pathways in response to virus infection will thus be an interesting area for future study.
TLR10 was first cloned in 2001 (22), yet little is known about this receptor. It remains the only member of the human TLR family without a defined agonist, signaling pathway, or function. Recent studies have suggested that TLR10 may be responsible for sensing of bacterial lipopeptides (12, 25). Here we report that TLR10 acts as an innate immune sensing receptor for influenza virus infection. Importantly, both TLR10 and viral RNP complex were required for the activation of the expression of the proinflammatory cytokine gene IL-8. Because viral RNP can be sensed by RIG-I to induce IFN (21), it will be of great interest to determine whether and how TLR10 might affect IFN induction. The involvement of RIG-I in IL-8 induction by TLR10 and viral RNP also merits further investigation. Nevertheless, our findings suggest that viral RNP might act through TLR10 to activate cytokine production. In other words, component(s) of the viral RNP complex or its products could be a ligand for TLR10 to trigger signaling for downstream cytokine expression. Further studies are needed to define which viral component is recognized by TLR10, i.e., viral protein, viral RNA, or the whole RNP complex.
Genetic polymorphisms of this receptor have been recently reported to be related to a number of diseases, including inflammatory disease such as Crohn’s disease (7) and respiratory disease such as asthma (8) making the study of TLR10 an extremely important area to explore in general and in relation to the respiratory tract.
In this investigation of the role of TLR10 in influenza virus infection, we used primary human peripheral blood monocyte-derived macrophages as one cell type for our experimental studies. It is noted that alveolar macrophages and epithelial cells are likely to be the first targets of an invading pathogen and the role of TLR10 in these cells need to be investigated in more detail in future. However, previous studies have shown that, compared with human alveolar macrophages, primary human peripheral blood monocyte-derived macrophages are more permissive to, and are more active in expressing cytokines in response to influenza virus infection in vitro (26). Thus, peripheral blood monocyte-derived macrophages remain relevant to pathogenesis because blood monocytes are quickly recruited and differentiated into macrophages in the infected lung. They are likely to be involved in amplification of cytokine cascades, which may be relevant to protection as well as to immune pathology.
The absence of functional TLR10 in mice implies that the role of this receptor cannot be investigated in conventional experimental mouse infection studies of influenza (or other viruses) using mice. The work reported here provides important data that point to a role for TLR10 in influenza virus infection and open a new field of study. It is likely that TLR10 plays a role in sensing and responding to a range of virus infections other than influenza.
Materials and Methods
Viruses.
H5N1 clade 1 virus A/Vietnam/3212/04 and the human seasonal influenza H1N1 virus A/HK/54/98 were passaged in Madin-Darby canine kidney (MDCK) cells and titrated in these cells by tissue culture infectious dose 50 (TCID50) assays.
Cells.
Human peripheral blood monocytes were separated from buffy coats obtained from healthy blood donors provided by the Hong Kong Red Cross Blood Transfusion Service. The mononuclear cells were separated by density gradient centrifugation (Ficoll-Paque; Pharmacia Biotech) and purified by adherence (14). All protocols using human donor cells were approved by the ethics committee of the University of Hong Kong. The cells were allowed to differentiate for 14 d in vitro in RPMI-1640 (Life Technologies) with 5% (vol/vol) autologous human serum before virus infection experiments. Human monocytic cell line THP-1, alveolar epithelial cell line A549, and the human kidney epithelial cell line 293T were obtained from ATCC. Cells were cultured and maintained according to manufacturer instructions. Phorbol-12-myristate-13-acetate activation of THP-1 cells was not used, as we wished to examine the impact of influenza virus infection on nonactivated cells.
Virus Infection.
Cells were infected with influenza A viruses (or UV-inactivated viruses) at MOI of 2 or 0.001, as indicated. After 30 min of virus adsorption, the virus inoculum was removed, the cells were washed with warm culture medium, and incubated in macrophage serum-free medium, RPMI-1640 or DMEM (Life Technologies) supplemented with 0.6 mg/L penicillin and 60 mg/L streptomycin.
Drug Treatment.
Cells were pretreated with protein synthesis inhibitor (CHX) (Invitrogen) for 45 min before infection. Cells were treated with TNF-α for 3 h before cell lysate collection. The respective drugs were maintained in the culture medium throughout the experiments.
shRNA-Mediated Gene Silencing.
TLR10 knockdown THP-1 cell lines were constructed by stable expression of shRNA designed to knockdown TLR10 gene expression. Five different MISSION shRNA plasmids (Sigma-Aldrich) containing hairpin TLR10 shRNA sequence were used for generating TLR10 knockdown cells. Each of the plasmids was cotransfected with ViraPower lentiviral packaging mix (Life Technologies) into 293T cells to produce TLR10 shRNA lentiviral particles. THP-1 cells were infected with the lentiviral particles followed by antibiotic selection in culture medium containing 20 μg/mL puromycin (Sigma-Aldrich), resulting in five THP-1 cell lines with different shRNA sequences inserted into the genome. Knockdown efficiency of the TLR10 gene expression was determined by RT-PCR using mRNA extracted from parental and puromycin-selected THP-1 cells. The two cell lines that showed the highest knockdown efficiency were used to confirm the specificity of gene silencing of TLR10 in independent experiments.
Construction of TLR10-Overexpressed THP-1 Cells.
TLR10 gene sequence was cloned into pLenti6.2/V5 DEST vector (Life Technologies) for the construction of a TLR10 overexpression cell line. Lentiviral particles that incorporated TLR10 RNA were produced as described above using pLenti6.2/TLR10. THP-1 cells were infected with the TLR10 lentivirus and selection was carried out using culture medium containing 10 μg/mL blasticidin (Life Technologies). mRNA was extracted from the blasticidin-resistance cells and overexpression of TLR10 was confirmed using RT-PCR.
Small Interfering RNA (siRNA)-Mediated Gene Silencing.
Accell siRNAs against human TLR10 and control siRNAs were transiently transfected into THP-1 cells using siRNA delivery medium according to the manufacturer’s protocol (Accell; Thermo Fisher Scientific). Three days after transfection, the cells were infected with influenza A viruses and cell lysates collected as indicated. Three individual and pooled siRNAs targeting TLR10 were used to confirm the specificity of gene silencing of TLR10 in independent experiments.
RT-PCR.
Total RNA from influenza virus-infected cells and viral RNA from culture supernatants of virus-infected cells were isolated using RNeasy Mini kit (Qiagen) and QIAamp Viral RNA Mini kit (Qiagen), respectively. cDNA was synthesized from RNA using poly(dT) primers or uni-12 primers (5′-AGCAAAAGCAGG-3′) and SuperScript III reverse transcriptase (Invitrogen). The PCR primers used in this study are shown in Table S1. Transcript expression was monitored using a SYBR Fast qPCR master mix kit (Kapa Biosystems) with specific primers. The fluorescence signals were measured in real time with a 7500 Fast Real-Time PCR System (Applied Biosystems). The specificity of the SYBR Green PCR signal was confirmed by melting curve analysis. The threshold cycle (CT) was defined as the fractional cycle number at which the fluorescence reached 10 times the SD of the baseline (from cycles 2–10). The ratio change in target gene relative to the β-actin control gene was determined by the 2-ΔΔCT1 method, as described previously (14).
Immunofluorescence Staining.
Cells were seeded on coverslips and infected with influenza virus at MOI of 2 for 18 h. For immunofluorescence staining, the cells were fixed with 4% (vol/vol) paraformaldehyde for 10 min and then permeabilized with 0.2% Triton X-100 in PBS for 30 min at room temperature (RT). After washing with PBS, cells were incubated with 10% (vol/vol) normal blocking serum in PBS for 20 min, followed by incubation with primary anti-human mouse TLR10 antibody (Imgenex) for 1.5 h at RT. After washing, the cells were incubated with FITC-conjugated anti-mouse IgG (Imgenex). Cell nuclei were counterstained with 4′6-diamidino-2-phenylindole (Sigma-Aldrich).
Western Blotting.
Western blotting was performed using the whole cell lysate. Equivalent amounts of protein from cell lysates were heat denatured in sample buffer [62.5 mM Tris⋅HCl, 30% (vol/vol) glycerol, 2% (wt/vol) SDS, 5% (vol/vol) 2-mercaptoethanol, 0.01% bromophenol blue] and separated by electrophoresis on 6% (wt/vol) SDS/PAGE and then transferred to polyvinylidene difluoride membranes. The membrane was blocked in PBS-0.1% Tween 20 containing 5% (vol/vol) skim milk for 1.5 h and immunoblotted with TLR10 antibody (Imgenex). A Full-Range Rainbow molecular weight marker (GE Healthcare) was used to determine the size of TLR10. Bound antibody was detected by incubation with HRP-conjugated IgG antibody (GE Healthcare) and ECL Plus solution (GE Healthcare).
Flow Cytometry.
Cells were washed in PBS and incubated with an optimal concentration of Fixable Viability Dye eFluor 780 (eBioscience) before fixation with 2% (vol/vol) paraformaldehyde. Nonspecific binding was reduced by incubation with 3% (vol/vol) FBS, 3% (vol/vol) normal human serum, 3% (vol/vol) normal mouse serum, 1 mM EDTA, 0.1% sodium azide in PBS (blocking buffer) for 10 min at room temperature, in the presence (i.e., permeabilized cells) or absence (i.e., intact cells) of 0.5% saponin. Samples were then placed on ice for additional 20-min incubation. Finally, cells were incubated for 45 min on ice with 5 μg/mL of anti-human mouse TLR10-FITC antibody or isotype control diluted in blocking buffer, with or without saponin. Cells were then washed and data were collected from ≥30,000 viable cells on a LSRII flow cytometer (BD biosciences) and postacquisition analyses were performed with FlowJo software (TreeStar).
ELISA.
Cytokine proteins in culture supernatants were quantified by ELISA (R&D Systems) according to the manufacturer’s protocol. The detection limits for IL-8 detection using the ELISA kit is 7.5–2,000 pg/mL.
Virus-Free Supernatant Stimulation Assay.
To investigate the paracrine effects of virus-infected macrophages on TLR10 expression, the supernatant of influenza H5N1-infected (H5S), H1N1-infected (H1S), and mock-infected (MS) macrophages (at MOI of 2) were collected 6 h after infection. Supernatants were filtered using a 100-kDa pore filter (Millipore) to remove any virus and added to fresh (uninfected) macrophages. The macrophages were harvested at 3 h post supernatant challenge and RNA was extracted with RNeasy Mini kits (Qiagen) to study TLR10 expression. The expression of influenza M gene in uninfected cells was measured to ensure that no virus was present in culture supernatants.
RNP Reconstitution Assay.
Expression vectors of PB2, PB1, PA, and NP genes and pPOLI-NS-Luc reporter genes were provided by K. P. Mok (University of Hong Kong, Hong Kong, China). Briefly, the PB2, PB1, PA, and NP cDNAs of a highly pathogenic H5N1 virus, A/Vietnam/1203/04, were synthesized by reverse transcription of viral RNA and subcloned into the pHW2000 vector as described (27). In the RNP assay, pPOLI-NS-Luc was the reporter plasmid used for the determination of the viral polymerase activity (20). Expression of vRNA-like negative-sense luciferase RNA was driven by the polymerase I promoter. Sense luciferase RNA was synthesized/ replicated by viral RNPs and the activity of viral polymerases was quantitated by the luciferase activity. For the RNP assay, 293T cells were transfected with pPOLI-NS-Luc (100 ng), the control plasmid: pRL-CMV (10 ng; Promega), the expression plasmids of PB2, PB1, PA, and NP genes (100 ng each) and/or TLR10 expression plasmid using TransIT-Express Reagent (Mirus Bio). Dual luciferase assay was performed according to the manufacturer’s instruction (Promega). Expression of proinflammatory cytokine gene IL-8 was determined using real-time PCR.
Statistical Analysis.
Student t test was used for data analysis in this study and results were considered as statistically significant when P values were <0.05.
Supplementary Material
Acknowledgments
We thank Li Ping-Hung for his technical support and Dr. Mok Ka-Pun for providing the viral gene expression vectors. This work was supported by a grant from the Research Grant Council of Hong Kong Special Administrative Region (HKU-776110M), research fundings from the National Institute of Allergy and Infectious Diseases (Contract HHSN266200700005C), and Area of Excellence Scheme of the University Grants Committee (AoE/M-12/96).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324266111/-/DCSupplemental.
References
- 1.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449(7164):819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 4.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101(15):5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Hasan U, et al. Human TLR10 is a functional receptor, expressed by B cells and plasmacytoid dendritic cells, which activates gene transcription through MyD88. J Immunol. 2005;174(5):2942–2950. doi: 10.4049/jimmunol.174.5.2942. [DOI] [PubMed] [Google Scholar]
- 7.Morgan AR, Lam WJ, Han DY, Fraser AG, Ferguson LR. Genetic variation within TLR10 is associated with Crohn’s disease in a New Zealand population. Hum Immunol. 2012;73(4):416–420. doi: 10.1016/j.humimm.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Lazarus R, et al. TOLL-like receptor 10 genetic variation is associated with asthma in two independent samples. Am J Respir Crit Care Med. 2004;170(6):594–600. doi: 10.1164/rccm.200404-491OC. [DOI] [PubMed] [Google Scholar]
- 9.Guirado M, et al. Association between C13ORF31, NOD2, RIPK2 and TLR10 polymorphisms and urothelial bladder cancer. Hum Immunol. 2012;73(6):668–672. doi: 10.1016/j.humimm.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Zhou XX, et al. Sequence variants in toll-like receptor 10 are associated with nasopharyngeal carcinoma risk. Cancer Epidemiol Biomarkers Prev. 2006;15(5):862–866. doi: 10.1158/1055-9965.EPI-05-0874. [DOI] [PubMed] [Google Scholar]
- 11.Kim D, et al. Reactive oxygen species enhance TLR10 expression in the human monocytic cell line THP-1. Int J Mol Sci. 2010;11(10):3769–3782. doi: 10.3390/ijms11103769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan Y, et al. Human TLRs 10 and 1 share common mechanisms of innate immune sensing but not signaling. J Immunol. 2010;184(9):5094–5103. doi: 10.4049/jimmunol.0901888. [DOI] [PubMed] [Google Scholar]
- 13.Cheung CY, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: A mechanism for the unusual severity of human disease? Lancet. 2002;360(9348):1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 14.Lee SMY, et al. Hyperinduction of cyclooxygenase-2-mediated proinflammatory cascade: A mechanism for the pathogenesis of avian influenza H5N1 infection. J Infect Dis. 2008;198(4):525–535. doi: 10.1086/590499. [DOI] [PubMed] [Google Scholar]
- 15.Lee SMY, et al. Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages. PLoS ONE. 2009;4(12):e8072. doi: 10.1371/journal.pone.0008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SMY, Yen HL. Targeting the host or the virus: Current and novel concepts for antiviral approaches against influenza virus infection. Antiviral Res. 2012;96(3):391–404. doi: 10.1016/j.antiviral.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koyama S, et al. Differential role of TLR- and RLR-signaling in the immune responses to influenza A virus infection and vaccination. J Immunol. 2007;179(7):4711–4720. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 18.Le Goffic R, et al. Cutting edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol. 2007;178(6):3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 19.Hornung V, et al. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168(9):4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 20.Li OT, et al. Full factorial analysis of mammalian and avian influenza polymerase subunits suggests a role of an efficient polymerase for virus adaptation. PLoS ONE. 2009;4(5):e5658. doi: 10.1371/journal.pone.0005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140(3):397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Chuang T, Ulevitch RJ. Identification of hTLR10: A novel human Toll-like receptor preferentially expressed in immune cells. Biochim Biophys Acta. 2001;1518(1-2):157–161. doi: 10.1016/s0167-4781(00)00289-x. [DOI] [PubMed] [Google Scholar]
- 23.de Jong MD, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hui KP, et al. H5N1 influenza virus-induced mediators upregulate RIG-I in uninfected cells by paracrine effects contributing to amplified cytokine cascades. J Infect Dis. 2011;204(12):1866–1878. doi: 10.1093/infdis/jir665. [DOI] [PubMed] [Google Scholar]
- 25.Mikacenic C, Reiner AP, Holden TD, Nickerson DA, Wurfel MM. Variation in the TLR10/TLR1/TLR6 locus is the major genetic determinant of interindividual difference in TLR1/2-mediated responses. Genes Immun. 2013;14(1):52–57. doi: 10.1038/gene.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu WC, et al. Viral replication and innate host responses in primary human alveolar epithelial cells and alveolar macrophages infected with influenza H5N1 and H1N1 viruses. J Virol. 2011;85(14):6844–6855. doi: 10.1128/JVI.02200-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mok KP, et al. Viral genetic determinants of H5N1 influenza viruses that contribute to cytokine dysregulation. J Infect Dis. 2009;200(7):1104–1112. doi: 10.1086/605606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.