Significance
Human susceptibility to gout is driven by the fact that we have a pseudogene for uricase that prevents a functional enzyme from being produced. Our inability to convert highly insoluble uric acid into a more soluble molecule makes us vulnerable to disease and other health complications. We have exploited ancestral sequence reconstruction to better understand how and why apes lost this functional enzyme. Our ancient proteins support one hypothesis that the progressive loss of uricase activity allowed our ancestors to readily accumulate fat via the metabolism of fructose from fruits. This adaptation may have provided our ancestors with an advantage when the energy-rich rainforests of Europe and Asia were displaced by temperate forests by the end of the Oligocene.
Keywords: hyperuricemia, pseudogene, evolution
Abstract
Uricase is an enzyme involved in purine catabolism and is found in all three domains of life. Curiously, uricase is not functional in some organisms despite its role in converting highly insoluble uric acid into 5-hydroxyisourate. Of particular interest is the observation that apes, including humans, cannot oxidize uric acid, and it appears that multiple, independent evolutionary events led to the silencing or pseudogenization of the uricase gene in ancestral apes. Various arguments have been made to suggest why natural selection would allow the accumulation of uric acid despite the physiological consequences of crystallized monosodium urate acutely causing liver/kidney damage or chronically causing gout. We have applied evolutionary models to understand the history of primate uricases by resurrecting ancestral mammalian intermediates before the pseudogenization events of this gene family. Resurrected proteins reveal that ancestral uricases have steadily decreased in activity since the last common ancestor of mammals gave rise to descendent primate lineages. We were also able to determine the 3D distribution of amino acid replacements as they accumulated during evolutionary history by crystallizing a mammalian uricase protein. Further, ancient and modern uricases were stably transfected into HepG2 liver cells to test one hypothesis that uricase pseudogenization allowed ancient frugivorous apes to rapidly convert fructose into fat. Finally, pharmacokinetics of an ancient uricase injected in rodents suggest that our integrated approach provides the foundation for an evolutionarily-engineered enzyme capable of treating gout and preventing tumor lysis syndrome in human patients.
Uric acid (monosodium urate) is a metabolic product of purine catabolism generated from the breakdown of nucleic acids. In most species, uric acid is metabolized to 5-hydroxyisourate by the enzyme uricase (urate oxidase) (1). Depending on the species, 5-hydroxyisourate is further degraded to allantoin, urea, or ammonia and excreted from the body (2–5). Uricase comprises the core of peroxisomes in terrestrial vertebrates and exists as an insoluble crystalloid (6). This is in contrast to most microbial and aquatic vertebrate species in which uricases are soluble and reside in either the cytoplasm (bacteria) or peroxisome (e.g., yeast). Crystal structures of bacterial and fungal uricases demonstrate that the active form of the protein exists as a homotetramer (dimer-of-dimers) with each of the four active sites located at the head-to-tail monomer–dimer interface (7, 8).
Uric acid is recognized as an antioxidant and it has been hypothesized to be an important antioxidant in the plasma (9), although its role in oxidative stress has been disputed (10). More recently, uric acid has been implicated in metabolic syndrome and fat accumulation (2, 11–13). Mammals possessing a functional uricase typically display serum uric acid levels in the 1- to 2-mg/dL range (0.06–0.12 mM). In contrast, uric acid levels are 3 to 10 times higher in apes (including humans) as a result of parallel nonsense mutations that caused a pseudogenization of the uricase gene during the early Miocene era (∼17 Ma) (14, 15). Despite being nonfunctional, cDNA sequencing confirmed that uricase mRNA is present in human liver cells and that these transcripts have two premature stop codons (15–17).
Homozygous deletion of functional uricase in mice results in death shortly after birth. These mice can be rescued if fed a xanthine oxidase inhibitor (allopurinol) to prevent the conversion of xanthine to uric acid (18). The inability of these mice to manage a sudden buildup of uric acid has supported one hypothesis that ancient apes slowly decreased uricase transcription via mutations in the uricase promoter before the pseudogenization events (14). However, an alternative hypothesis to prevent the sudden buildup of uric acid in ancient primates would be to slowly diminish the enzymatic activity of uricase before pseudogenization events. This alternative mechanism may have provided more robust control to slowly abolish uricase activity compared with mutations in gene expression that would be susceptible to compensatory changes that up-regulate expression. This mechanism may also provide greater control than modifying transcriptional expression, as uricases are highly resistant to proteasome degradation and thus protein turnover (19).
Results
To test this alternative hypothesis, we resurrected ancient uricase enzymes throughout mammalian history. Fig. 1 shows the biochemical properties of uricases at major ancestral mammalian divergences whose descendents ultimately trace to humans. The most ancient of the ancestral uricases (An19/22) had the highest enzymatic activity and is significantly more active than modern pig uricase (considered to be one of the more active modern mammalian uricases; P < 0.0001). Enzymatic assays for descendents of An19/22 reveals a progressive stepwise decrease in uricase activity from the last common ancestor of mammals (∼90 Ma) to the last common ancestor of apes (∼20 Ma). In support of this conclusion, An19/22 is significantly more catalytically efficient than An26 (P < 0.0001), and An26 is significantly more catalytically efficient than An27 (P < 0.0001). The activity of An30 demonstrates that uricase function was nearly enzymatically abolished before the pseudogenization events in ancestral apes.
Fig. 1.
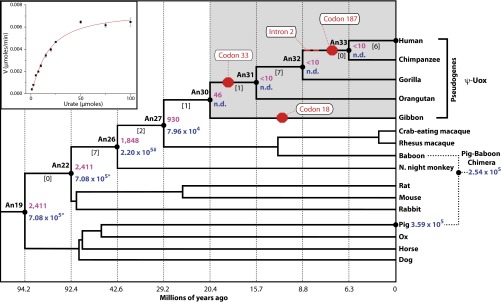
Phylogeny of the uricase gene family and enzymatic properties of inferred ancestral uricases. Shown is a phylogenetic tree depicting the evolutionary relationship among mammalian uricase genes. Three genetic lesions are attributed with the pseudogenization of the human uricase gene. A nonsense mutation at codon 33 is common to all great apes (orangutan, gorilla, chimpanzee, and human). An additional nonsense mutation is common for the chimpanzee and human sequences at codon 187. Further, a mutation is located at the splice-acceptor site in intron 2 for the chimpanzee and human sequences. A missense mutation at codon 18 of lesser apes is independent of the great ape uricase pseudogenization event. The ancestral nodes (An) on the tree are labeled in bold. The protein sequences at nodes 19 and 22 are identical and labeled as An19/22. In addition, the protein sequences at nodes 32 and 33 are identical and labeled as An32/33. The numbers in brackets between nodes represent the number of amino acid replacements that occur along each branch. The two stop codons in a full-length human uricase gene were replaced with arginine codons but this recombinant protein exhibited properties analogous to An32/33. Enzymatically characterized uricases are denoted by solid black circles. The specific activity is the amount of uric acid substrate oxidized per minute per milligram of total insoluble protein (shown in purple). Catalytic efficiency was also determined for modern and ancient uricases when possible (shown in blue). (Inset) Example kinetic reaction as monitored by following the oxidation of uric acid. (*kcat/KM for An19/22 is significantly greater than An26’s, P < 0.0001; #kcat/KM for An26 is significantly greater than An27’s, P < 0.0001.) Divergence times for ancestral nodes are provided along the x axis (49).
Such a progressive accumulation of slightly deleterious amino acid replacements is consistent with the observation that it is not possible to “reactivate” a functional human uricase enzyme by simply replacing the two premature stop codons present in the gene (Fig. 1). To better understand why the human uricase is not functional without the premature stop codons, the 22 amino acid replacements inferred to have occurred between node An19/22 and humans (numbers in brackets in Fig. 1) were individually assayed within the background of the An19/22 enzyme. Nearly all of the 22 amino acid replacements caused a substantial decrease in catalytic efficiency (kcat/Km; Fig. S1). Three of these replacements had striking effects on uricase purification and activity. Two amino acid replacements (S232L and Y240C) prevented uricase solubility; thus, catalytic efficiency could not be determined. Both these replacements presumably occurred after the pseudogenization events. Alternatively, the single replacement F222S caused a decrease in uricase efficiency of nearly two orders of magnitude, and this replacement is inferred to have occurred on the branch connecting the origins of apes to the last common ancestor of apes (i.e., before pseudogenization events).
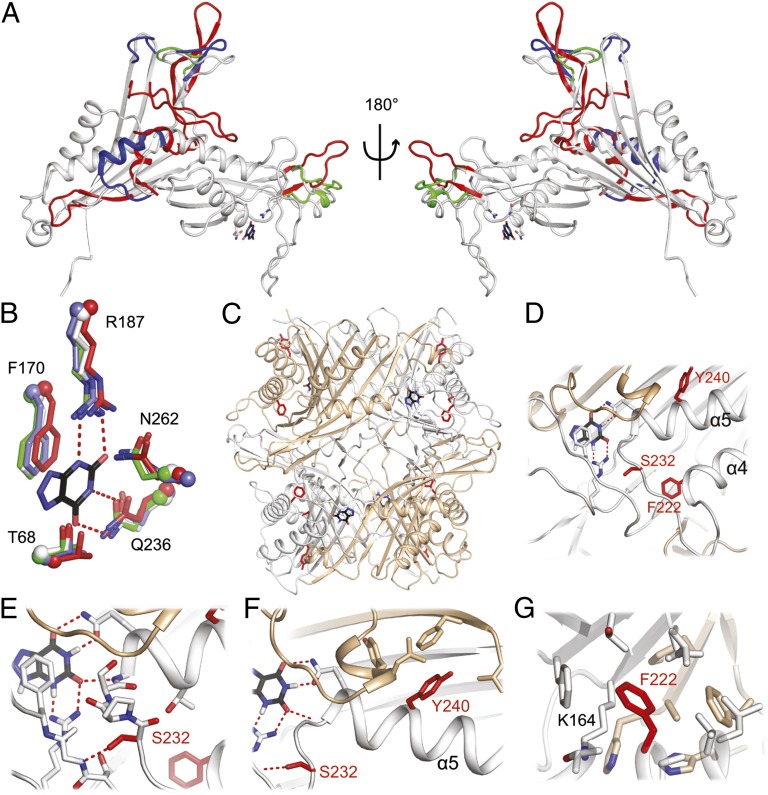
To better understand the effects of these amino acid replacements, as well as why it is not possible to reactivate the human uricase gene by simply replacing the premature stop codons, we determined the 3D crystal structure of apo An19/22. The mammalian structure reveals a conserved uricase fold organized as a homotetramer (dimer-of-dimers) with the active site located at the monomer–dimer interface. Mammalian uricase displays divergent structural features compared with microbial uricases, with large differences in the composition and conformation of loops adjacent to subunit interfaces (i.e., β4–β5, α3, β13–β14, α3) that are well separated from the active site (Fig. 2A and Fig. S2). Despite these differences, the oligomeric interfaces and active site residues are remarkably conserved (Fig. 2B) (7, 8). The three mutations with the largest deleterious effect on catalytic activity cluster on the α4–α5 element, which is critical for positioning the ligand during catalysis within the active site (Fig. 2 C and D) (20). The α4–α5 loop contributes three hydrogen bonds to the bound ligand via Gln236 and the backbone amide of Val235, and comprises one wall of the active site. The net effect of each individual mutation is to destabilize oligomerization and thus the formation of the active site. The S232L replacement eliminates an important hydrogen bond to the backbone of Gly169 destabilizing the positions of Phe170 and Gln236, which are critical for positioning the bound ligand within the active site. S232L also introduces a polar incompatibility with neighboring residues S168, R187, and T238 preventing proper packing of the β4–β5 and α4–α5 loop likely disrupting protein folding and dimerization. The Y240C replacement eliminates aromatic stacking and hydrophobic interactions across the monomer–dimer interface with a cluster of hydrophobic residues destabilizing the dimer interface and formation of the active site (Fig. 2F), confirming previous computational predictions (21). Phe222, located on α4, stabilizes the dimer–dimer interface via aromatic stacking and hydrophobic interactions and an intrasubunit cation–π interaction with the highly conserved Lys164 (Fig. 2E). Mutation to serine would eliminate these interactions and generate an energetic penalty by introducing a hydrophilic residue in this hydrophobic pocket. The remaining amino acid replacements are axially distributed around the structure and their effects on activity are correlated across the nodes of the phylogeny (Fig. S3).
Fig. 2.
Structural features of an ancient mammalian uricase. (A) Superposition of the Anc19/22 structure (gray) on uricases from Bacillus sp. (red; PDB ID code 1J2G), Aspergillus flavus (green; PDB ID code 2YZE), and Arthrobacter globiformis (blue; PDB ID code 1R56). Structurally conserved regions are shown in gray, and divergent regions are highlighted red, green, and blue, respectively. For reference, uric acid from holo A. flavus uricase (PDB ID code 3OBP) is modeled as black sticks; active site residues are shown as white, side-chain sticks. (B) Close-up view of the active site with side chains shown as sticks (N, blue; O, red) and the α-carbon shown as a sphere. (C) Global and close-up view of the tetramer showing the location of the three large-effect amino acid replacements in red. Monomers A and C are colored white, whereas B and D are colored tan. (D–G) Close-up views of S232, Y240, and F222, respectively.
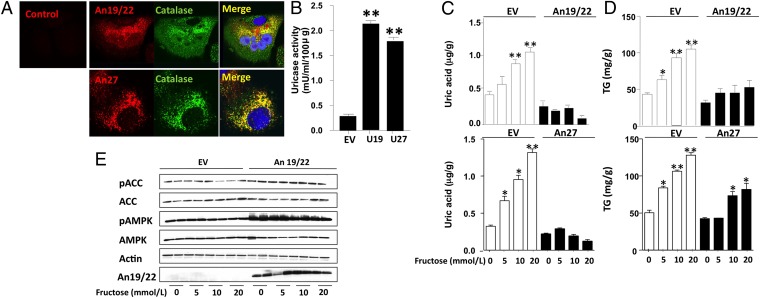
The progressive loss of uricase activity in the ape lineage may have provided a selective advantage (22). Although several theories have been proposed, a recent hypothesis is that the loss of uricase may have aided the accumulation of fat stores in response to fructose, a major nutrient in fruits that were a primary staple of ancestral simians (23). Fructose increases hepatic fat stores in part because of its ability to generate uric acid during metabolism (24), whereas the inhibition of uricase enhances fat accumulation in the liver in response to fructose (25). To better understand the role ancient uricases would have in fructose metabolism, human HepG2 cells were transfected with two ancestral uricases (An19/22 and An27; Fig. 3A). Localization studies documented that An19/22 and An27 colocalized with catalase, demonstrating peroxisomal location (Fig. 3A). Increased uricase activity was confirmed in lysates obtained from these uricase-expressing cells (Fig. 3B). Uricase-expressing cells exposed to fructose (5 mM for 72 h) showed less intracellular uric acid accumulation (Fig. 3C) and less triglyceride accumulation (Fig. 3D) compared with cells treated with an empty vector. In addition, cells expressing An19/22 (Fig. 3E) or An27 (Fig. S4) showed a tendency to have greater levels of phosphorylated AMP kinase, and its target protein ACC, in response to fructose compared with control cells. These results are consistent with earlier work showing that uric acid is an inhibitor of AMP kinase (26). These results also demonstrate that triglyceride accumulation is correlated with the enzymatic efficiency of uricases and that, whereas ancient uricases are capable of managing uric acid levels in our assay conditions, more derived versions such as recombinant human uricase are not able to manage uric acid.
Fig. 3.
Expression and function of ancestral uricases in human HepG2 cells. (A) Colocalization studies between An19/22 (red, Upper) and An27 (red, Lower) with the peroxisome marker catalase (green). Merged images show colocalization (yellow). Nuclear staining with DAPI (blue). (B) Intracellular uricase activity in control and An19/22- and An27-expressing cells. (C) Intracellular uric acid levels in HepG2 control cells (empty vector; EV) and cells stably expressing An19/22 or An27 and exposed to increasing levels of fructose (from 0 to 20 mM). (D) Intracellular triglyceride levels in HepG2 control cells (EV) and cells stably expressing An19/22 or An27 and exposed to increasing levels of fructose (from 0 to 20 mM). (*P < 0.05 and **P < 0.01 vs. no fructose.) (E) Representative Western blot of lysates from HepG2 cells stably expressing an empty vector control or An19/22. ACC, Acetyl-Coa carboxylase; AMPK, AMP kinase; P, phosphorylated protein.
The buildup of serum urate in the blood (hyperuricemia) can have severe consequences for human health. As such, the medical community has a strong interest in developing a recombinant “human-like” uricase to treat gout, prevent tumor lysis syndrome, and control hyperuricemia in general (27). The difficulties in developing a therapeutic uricase are many-fold, but mostly rely on a compromise between activity, stability, and immunoreactivity in human patients. A recently Food and Drug Administration-approved uricase consists of a combination of pig and baboon enzymes [pig–baboon chimera (PBC)], whereby the pig portion provides a high catalytic efficiency and the substantially less active baboon portion may help to avoid an immune response in humans (Fig. 1) (28). However, this recombinant enzyme has a t1/2 on the order of minutes in animal models and thus requires PEG modification to increase its t1/2 to an acceptable level (pegloticase) (28, 29). Based on the observations that An19/22 is significantly more catalytically efficient than pig uricase and yet has greater sequence identity to human uricase than PBC (93% vs. 90%, respectively; Fig. S5), we tested the pharmacokinetics of native An19/22 in healthy rats. Monitoring plasma uricase activity revealed that the t1/2 of An19/22 is nearly two orders of magnitude (100-fold) more stable than native (non-PEGylated) PBC uricase (Fig. S6). These results suggest that An19/22 uricase has potential therapeutic value as a non-PEGylated protein, which is important because human patients develop antibodies to the PEG moiety on pegloticase (30).
Discussion
We have demonstrated that ancient mammalian uricases have accumulated amino acid replacements that have in turn progressively diminished the enzyme’s activity before proposed pseudogenization events in the lesser and great ape lineages. Interestingly, this progressive decrease in uricase activity is not reflected in the branch-based nonsynonymous-to-synonymous (dN/dS) metric used to detect changes in selective constraints acting on protein-coding genes; however, a neutral signal is clearly evident among the ape pseudogenes (Fig. S7). Further, a sites-based model of adaptive evolution did not detect any sites under positive selection. This progressive loss of uricase activity was initiated in the Eocene, with complete loss of activity occurring by the early Miocene (∼15–20 Ma). This geologic window is signified by a period of progressive cooling that substantially affected the diet of early hominoids as inferred from fossilized dental records, especially for those that migrated around Eurasia (23). The Miocene period hosted the diversification of our ape ancestors in Europe and Asia (e.g., Kenyapithecus and Dryopithecus) and uricase loss could have helped these frugivorous organisms “follow the fruit” back to Africa as the rainforests receded to the equatorial zone during this time (23). The loss of uricase may have provided a survival advantage by amplifying the effects of fructose to enhance fat stores (24), and by the ability of uric acid to stimulate foraging (31), while also increasing blood pressure in response to salt (13). Thus, the loss of uricase may represent the first example of a “thrifty gene” [as hypothesized by Neel (32)] to explain the current epidemic of obesity and diabetes, except that it is the loss of a gene (33), and not the acquisition of a new gene, that has increased our susceptibility to these conditions. We anticipate that future resurrection studies on enzymes upstream and downstream of uricase in purine catabolism will provide greater details on the evolution of flux through a metabolic pathway experiencing functional adaptation (3, 34). For instance, the resurrection of ancestral xanthine oxidoreductases will reveal when primates experienced the lowering of activity displayed by modern forms of the enzyme, and whether this correlates with the decrease in ancestral uricase activity.
In total, we have shown that ancient mutations decrease uricase activity over evolutionary time, and this progressive decrease is correlated with physiological function and (potential) adaptation. Our integrated approach of combining ancestral sequence reconstruction, structural biology, and cell biology provides unique insights into the evolutionary history of uricases and primate evolution, and also provides the framework necessary for the biomedical field to engineer (or reactivate) a recombinant human uricase for therapeutic purposes. We have shown that one of our ancestral uricases has orders-of-magnitude greater bioavailability than state-of-the-art therapeutic uricases. We anticipate that ancient enzymes will play a greater role in therapeutic and industrial applications in the near future, with uricase paving the way.
Methods
cDNA, Ancestral Sequence Resurrection, and in Vitro Assays.
Transcripts of the human uricase pseudogene were verified by using fetal liver cDNA (Clontech) as template for a series of PCR reactions (Promega). Forward and reverse primers were designed against the published sequence of human uricase gene (15). All PCR reactions were performed in a total volume of 50 μL consisting of 1 μL of template cDNA (1 ng), 2 μL 10 μM each of forward and reverse primers, 0.25 μL GoTaq polymerase enzyme (1.25 U), 10 μL 5× GoTaq buffer, and 1 μL deoxyribonucleotide mix (10 mM), brought to volume with dH2O. Products were examined via 1.5% to 2% (wt/vol) agarose gel electrophoresis, and bands were purified by using a gel extraction kit (Qiagen) and subjected to DNA sequence analysis (Fig. S8).
Ancestral sequence reconstruction was performed following our previous protocols (35, 36). Modern uricase sequences from 17 taxa were retrieved from public databases and were aligned by using MUSCLE. An evolutionary tree was inferred by using MrBayes (37). The following parameters were incorporated during the analysis: the generalized time reversible DNA substitution model, a proportion of the nucleotide sites were invariable whereas the remaining sites were drawn from a γ-distribution. To search tree space, two independent Metropolis-coupled Markov chain Monte Carlo (MCMCMC) simulations, four chains each, were performed for 1,000,000 generations with parameter sampling every 100 generations. The first 100 samples were discarded during the burn-in phase of the MCMCMC analysis. The resulting gene tree recapitulated the species tree for these organisms.
Ancestral sequences and dN/dS were inferred by using PAML version 4.1 (38). Ancestral sequences were inferred by using DNA-based and amino acid-based models. All nonsense mutations present in an ancestral sequence were converted to the sense codon present in homologs (an otherwise universally conserved arginine for both stop codons 33 and 137). The DNA-based analysis incorporated the same parameters as described earlier. The amino acid-based analysis incorporated a γ-distribution and the WAG replacement matrix. These various analyses generated identical ancestral sequences. The inferred ancestral sequences displayed high posterior probabilities (Fig. S9) and were robust to alternative models used during the inference process (35, 39, 40). For instance, an alternative topology that places primates and rodents as sister clades with Laurasiatherians as an outgroup was also used to infer ancestral sequences (41). No ancestral sequence differences were observed for the corresponding nodes between the two topologies. Premature codons were substituted for the highly conserved arginines present in uricase homologs. Many CGA (arginine) codons in ape uricases have been converted to TGA (stop) codons, likely as a result of methylation and subsequent deamination at the CG dinucleotides after selective constraints were lifted from the gene. Although it has been hypothesized that the common ancestor of great apes had a premature stop codon at position 33, this hypothesis lacks full support because there are multiple instances of homoplasy (CGA to TGA) at other positions in the ape uricases (14). Codon-based analyses were performed to calculate dN/dS ratios by using branch-based (free-ratio) and sites-based models. These analyses incorporated a γ-distribution, codon frequencies F3 × 4, and a transition/transversion ratio. For the sites-based method, M1/M2 and M7/M8 analyses were not significantly different according to a likelihood ratio test. Further, Bayes empirical Bayes did not identify any positively selected sites with a P value >95%.
Uricase encoding genes were codon-optimized for expression in Escherichia coli (Epoch Laboratories). These genes were cloned into the pET-21a vector (Novagen) using NdeI and XhoI sites. E. coli Tuner (DE3; EMD Millipore) cells were freshly transformed with uricase-containing vectors. A single colony was used to inoculate a 5-mL overnight culture. This overnight culture was used to seed 1 L Luria broth with 100 μg/mL carbenicillin and 100 μg/mL chloramphenicol. Cells were grown to an OD600 between 0.6 and 0.8 at which point they were induced with 1 mM IPTG. Expression was carried out overnight (16–20 h) at 37 °C with shaking at 250 rpm. Cells were then ruptured by sonication and the inclusion bodies (IBs) were washed five times with PBS buffer. The IBs were resuspended in a 0.1 M to 1 M Na2CO3 buffer at a pH between 10 and 11 and 1 mM PMSF. The carbonate extraction supernatant contained active uricase as confirmed by the Amplex Red Uricase/Uric Acid Activity Kit (Invitrogen).
Soluble uricases were purified using FPLC (GE Healthcare) at 4 °C and followed the two-step purification scheme reported for the PBC uricase (29). Anion exchange chromatography was performed on a HiLoad 16/10 Q-Sepharose HP column (GE Healthcare) preequilibrated with 0.075 M Na2CO3, pH 10.2. After sample application, contaminants were washed from the column by using salt gradient from 0.15 M NaCl to 1.5 M NaCl in 10 mM NaHCO3, pH 8.5. A three-column volume wash with 10 mM NaHCO3, pH 8.5, 1.5 M NaCl ensured that all major contaminants had been eluted from the column media. This was followed by a three-column volume wash with 10 mM NaHCO3, pH 8.5. In preparation for the elution of purified uricase, the pH was raised with one column volume of Na2CO3, pH 11. Uricase was then eluted by a 10-column volume salt gradient from 0 to 0.6 M NaCl in 0.1 M Na2CO3, pH 11. Uricase was collected and concentrated by using 9-kDa molecular weight cutoff spin concentrators (Pierce). The final purification step was performed by size exclusion chromatography on a HiLoad 16/60 Superdex 200 prep-grade column preequilibrated with 0.1 M Na2CO3, pH 10.2 (GE Life Sciences). Tetrameric uricase was eluted with 0.1 M Na2CO3, pH 10.2, at ∼140 kDa and concentrated by centrifugation with spin concentrators.
The enzymatic activity of purified tetrameric uricase was determined spectrophotometrically by monitoring the decrease of uric acid by following the absorbance at 293 nm (A293). A freshly prepared 1 mM uric acid stock in 0.1 M sodium phosphate buffer, pH 7.4, was diluted with 0.1 M sodium phosphate buffer, pH 7.4, to prepare a range of uric acid concentrations. Specifically, the assays were performed at the following uric acid concentrations: 1 μM, 2.5 μM, 5 μM, 10 μM, 15 μM, 20 μM, 25 μM, 50 μM, 75 μM, and 100 μM. The averages from triplicate runs of initial velocities at each uric acid concentration were used to plot a hyperbolic regression curve to determine Km, Vmax, and kcat/Km of the purified uricases (Hyper32 Kinetics Application). The concentration of purified tetrameric uricase was determined by the Quick Bradford Assay (Bio-Rad). Finally, the kcat was determined by dividing Vmax by the concentration of tetrameric uricase used in the kinetics experiments. We applied the Gaussian error propagation approach to generate SEs for our kcat/Km values and then applied an unpaired Student t test (two-tailed) to determine significance. Unfortunately, not all uricases could be solubilized in the range of tested conditions. Thus, specific activity for all modern and ancient uricases was determined as a reference point and defined as the amount of uric acid substrate oxidized per minute per milligram of total insoluble protein.
Three-Dimensional Structure Determination.
A variant of An19/22 (R147K, E220K) was crystallized by vapor diffusion under conditions optimized from initial crystal screening trials using the commercially available Nucleix Suite (Qiagen). Before crystallization, purified protein was concentrated to 10 mg/mL in 10 mM sodium carbonate (pH 10.5) and combined with twice its volume of reservoir solution, 25% (wt/vol) 2-meththyl-2,4-pentanediol (MPD), 40 mM MgCl2, and 50 mM sodium cacodylate (pH 6). Box-shaped crystals of nearly 200 µm in each dimension grew in 3 to 5 d at 20 °C. The concentration of MPD was raised to 40% for harvesting and cryoprotection.
Diffraction data for apo uricase were collected under cryogenic conditions at Southeast Regional Collaborative Access Team 22ID at Argonne National Synchrotron Light Source (Argonne, IL; Table S1). Indexing, integration, and scaling of crystallographic reflections was performed by using HKL2000 software, version 0.98.699a (42). Initial phases were determined by molecular replacement by using a search model generated by the Phyre2 homology-based protein structure prediction server (43) using the crystal structure of the Aspergillus flavus urate oxidase [Protein Data Bank (PDB) ID code 1R56] as the template (20). The structure was refined in the PHENIX crystallographic software package (44) and model building was performed in the COOT molecular graphics program (45). The R factors for the final model are 20.7% and 25.9% for R and Rfree, respectively. A total of 88.9% of residues are Ramachandran-favored for the uricase with <2.5% outliers. PyMOL was used to visualize the structures and generate images (46).
In Vivo Assays.
The established human hepatocarcinoma cell line HepG2 was purchased from the American Type Culture Collection. The cells were cultured in RPMI medium (no. 17850; Gibco) supplemented with 10% FBS (Gibco) and 100 U/mL penicillin and 100 µg/mL streptomycin (Gibco) in a humidified incubator with 5% CO2 at 37 °C.
Transduction and generation of stable clones expressing uricases was performed as previously described (26). The coding sequences of the various uricases were inserted into the pLVXIRES Puro vector by direct cloning using EcoRI and NotI sites. Clones were confirmed by sequencing analysis. Seven micrograms of vector at 1 mg/mL was combined with the Lenti-X HTX packaging system (Clontech) and HEK-293T cells maintained in tetracycline-free medium were transfected with the components. Medium was collected at 48 h and efficient production of lentiviral particles was assessed with the Lenti-X Gostix (Clontech). HepG2 cells were exposed to the virus and stable clones generated with puromycin (10 µg/mL). Controls refer to puromycin selected clones transduced with lentiviral particles containing an empty pLVXIRES Puro vector. In experiments involving fructose, cells were incubated with these compounds at different concentrations (from 0 to 20 mM) for 72 h with fresh changes of medium twice daily.
Protein lysates were prepared from confluent cell cultures and rat livers by using MAP Kinase lysis buffer as previously described (47). Sample protein content was determined by the BCA Protein Assay (Pierce). A total of 40 μg of total protein was loaded per lane for SDS‐PAGE (10% wt/vol) analysis and then transferred to PVDF membranes. Membranes were incubated with primary antibodies (uricase 1:200; Santa Cruz Biotechnology; ACC, ACL, FAS, pACC, AMPK, pAMPK, and actin 1:1,000; Cell Signaling) and visualized using a horseradish peroxidase secondary antibody (1:2,000) and the HRP Immun-Star detection kit (Bio‐Rad). Chemiluminescence was recorded with an Image Station 440CF and results analyzed with 1D Image Software (Kodak Digital Science).
Cell preparation, staining, and imaging were performed as previously described (48). Briefly, HepG2 cells were grown to confluence in eight-well glass slides (no. 177402; Nunc) and fixed with paraformaldehyde 4% in PBS solution for 20 min. Excess paraformaldehyde was removed by incubating fixed cell to 20 mM glycone in PBS solution. Cells were then permeabilized with 0.3% Triton X-100 in PBS solution and incubated overnight with uricase antibody (Santa Cruz Biotechnology). Cells were rinsed and incubated with a goat anti-rabbit Alexa Fluor 568-conjugated secondary antibody (Molecular Probes). Preparations were imaged with a 63× oil immersion objective by using a laser scanning confocal microscope (model LSM510; Zeiss). Data were analyzed by using LSM Image Analyzer Postacquisition software (Zeiss).
Rodent Assays.
Sprague–Dawley rats (Charles River Laboratories) were obtained and acclimated after delivery for at least 1 wk before testing. For each uricase tested, five age- and weight-matched male rats served as biological replicates. The uricase preparations were kept on ice and then immediately diluted in room temperature PBS solution, pH 7.4, to a concentration of 0.2 mg/mL. One milliliter of a 0.2-μm filter-sterilized uricase preparation (Pall) was i.v. injected. Postinjection blood samples were collected at 5 min and every 60 min thereafter for a minimum of 5 h. Each sample containing 100 μL of fresh blood was immediately transferred to lithium heparin coated Microtainer blood collection tubes (BD) and mixed by rocking at room temperature and then centrifuged for 5 min at 2,350 × g to separate plasma from whole blood. Samples were stored on ice until assayed as described earlier.
Supplementary Material
Acknowledgments
We thank Betül Kaçar, Valery Adorno-Cruz, and Laura O’Farrell for their assistance. This work was supported by Technological Innovation: Generating Economic Results (TI:GER), Graduate Assistance in Areas of National Need (GAANN), and Translational Research Institute for Biomedical Engineering and Science (TRIBES) programs at the Georgia Institute of Technology (J.T.K.), the Georgia Research Alliance (E.A.G.), and start-up funds from the Georgia Institute of Technology (E.A.G.), University of Colorado (R.J.J.), and Emory University (E.A.O.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
Footnotes
Conflict of interest statement: E.A.G. declares financial interest because a patent related to this research has been filed by the Georgia Institute of Technology, and this technology is licensed to General Genomics, LLC, which is a start-up founded by E.A.G.. R.J.J. is an inventor on several patents and patent applications related to lowering uric acid as a means for treating hypertension, diabetic nephropathy, and renal disease, and is a founder of XORT, Inc., which is a start-up company that has licensed these applications.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4MB8).
See Commentary on page 3657.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320393111/-/DCSupplemental.
References
- 1.Kahn K, Tipton PA. Spectroscopic characterization of intermediates in the urate oxidase reaction. Biochemistry. 1998;37(33):11651–11659. doi: 10.1021/bi980446g. [DOI] [PubMed] [Google Scholar]
- 2.Johnson RJ, et al. Lessons from comparative physiology: Could uric acid represent a physiologic alarm signal gone awry in western society? J Comp Physiol B. 2009;179(1):67–76. doi: 10.1007/s00360-008-0291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keebaugh AC, Thomas JW. The evolutionary fate of the genes encoding the purine catabolic enzymes in hominoids, birds, and reptiles. Mol Biol Evol. 2010;27(6):1359–1369. doi: 10.1093/molbev/msq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi S, Fujiwara S, Noguchi T. Evolution of urate-degrading enzymes in animal peroxisomes. Cell Biochem Biophys. 2000;32(Spring):123–129. doi: 10.1385/cbb:32:1-3:123. [DOI] [PubMed] [Google Scholar]
- 5.Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat Chem Biol. 2006;2(3):144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- 6.Hruban Z, Swift H. Uricase: Localization in hepatic microbodies. Science. 1964;146(3649):1316–1318. doi: 10.1126/science.146.3649.1316. [DOI] [PubMed] [Google Scholar]
- 7.Juan EC, et al. Structures of Arthrobacter globiformis urate oxidase-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2008;D64(pt 8):815–822. doi: 10.1107/S0907444908013590. [DOI] [PubMed] [Google Scholar]
- 8.Colloc’h N, et al. Crystal structure of the protein drug urate oxidase-inhibitor complex at 2.05 A resolution. Nat Struct Biol. 1997;4(11):947–952. doi: 10.1038/nsb1197-947. [DOI] [PubMed] [Google Scholar]
- 9.Ames BN, Cathcart R, Schwiers E, Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: A hypothesis. Proc Natl Acad Sci USA. 1981;78(11):6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hershfield MS, et al. Treating gout with pegloticase, a PEGylated urate oxidase, provides insight into the importance of uric acid as an antioxidant in vivo. Proc Natl Acad Sci USA. 2010;107(32):14351–14356. doi: 10.1073/pnas.1001072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson RJ, et al. Hypothesis: could excessive fructose intake and uric acid cause type 2 diabetes? Endocr Rev. 2009;30(1):96–116. doi: 10.1210/er.2008-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RJ, et al. The planetary biology of ascorbate and uric acid and their relationship with the epidemic of obesity and cardiovascular disease. Med Hypotheses. 2008;71(1):22–31. doi: 10.1016/j.mehy.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe S, et al. Uric acid, hominoid evolution, and the pathogenesis of salt-sensitivity. Hypertension. 2002;40(3):355–360. doi: 10.1161/01.hyp.0000028589.66335.aa. [DOI] [PubMed] [Google Scholar]
- 14.Oda M, Satta Y, Takenaka O, Takahata N. Loss of urate oxidase activity in hominoids and its evolutionary implications. Mol Biol Evol. 2002;19(5):640–653. doi: 10.1093/oxfordjournals.molbev.a004123. [DOI] [PubMed] [Google Scholar]
- 15.Wu XW, Muzny DM, Lee CC, Caskey CT. Two independent mutational events in the loss of urate oxidase during hominoid evolution. J Mol Evol. 1992;34(1):78–84. doi: 10.1007/BF00163854. [DOI] [PubMed] [Google Scholar]
- 16.Wu XW, Lee CC, Muzny DM, Caskey CT. Urate oxidase: Primary structure and evolutionary implications. Proc Natl Acad Sci USA. 1989;86(23):9412–9416. doi: 10.1073/pnas.86.23.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeldandi AV, et al. Molecular evolution of the urate oxidase-encoding gene in hominoid primates: Nonsense mutations. Gene. 1991;109(2):281–284. doi: 10.1016/0378-1119(91)90622-i. [DOI] [PubMed] [Google Scholar]
- 18.Kelly SJ, et al. Diabetes insipidus in uricase-deficient mice: A model for evaluating therapy with poly(ethylene glycol)-modified uricase. J Am Soc Nephrol. 2001;12(5):1001–1009. doi: 10.1681/ASN.V1251001. [DOI] [PubMed] [Google Scholar]
- 19.Yokota S, Kamijo K, Oda T. Degradation of overexpressed wild-type and mutant uricase proteins in cultured cells. J Histochem Cytochem. 1999;47(9):1133–1140. doi: 10.1177/002215549904700905. [DOI] [PubMed] [Google Scholar]
- 20.Retailleau P, et al. Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: A reassignment of the active-site binding mode. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 3):453–462. doi: 10.1107/S0907444903029718. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, et al. Structure-based characterization of canine-human chimeric uricases and its evolutionary implications. Biochimie. 2012;94(6):1412–1420. doi: 10.1016/j.biochi.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Johnson RJ, Lanaspa MA, Gaucher EA. Uric acid: A danger signal from the RNA world that may have a role in the epidemic of obesity, metabolic syndrome, and cardiorenal disease: Evolutionary considerations. Semin Nephrol. 2011;31(5):394–399. doi: 10.1016/j.semnephrol.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson RJ, Andrews P. Fructose, uricase, and the back-to-Africa hypothesis. Evol Anthropol. 2010;19(6):250–257. [Google Scholar]
- 24.Lanaspa MA, et al. Uric acid induces hepatic steatosis by generation of mitochondrial oxidative stress: Potential role in fructose-dependent and -independent fatty liver. J Biol Chem. 2012;287(48):40732–40744. doi: 10.1074/jbc.M112.399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tapia E, et al. Synergistic effect of both treatments on renal alterations. Am J Physiol Renal Physiol. 2013;304(6):F727–F736. doi: 10.1152/ajprenal.00485.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanaspa MA, et al. Counteracting roles of AMP deaminase and AMP kinase in the development of fatty liver. PLoS ONE. 2012;7(11):e48801. doi: 10.1371/journal.pone.0048801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Yuan Y, Zhan CG, Liao F. Uricases as therapeutic agents to treat refractory gout: Current states and future directions. Drug Dev Res. 2012;73(2):66–72. doi: 10.1002/ddr.20493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams LD, Hershfield MS, Kelly SJ, Saifer MG, Sherman MR. 2011. US Patent 8,067,553.
- 29.Hershfield MS, Kelly SJ. 2006. US Patent 7,056,713.
- 30.Ganson NJ, Kelly SJ, Scarlett E, Sundy JS, Hershfield MS. Control of hyperuricemia in subjects with refractory gout, and induction of antibody against poly(ethylene glycol) (PEG), in a phase I trial of subcutaneous PEGylated urate oxidase. Arthritis Res Ther. 2006;8(1):R12. doi: 10.1186/ar1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutin AR, et al. Impulsivity is associated with uric acid: Evidence from humans and mice. Biol Psychiatry. 2014;75(1):31–37. doi: 10.1016/j.biopsych.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neel JV. Diabetes mellitus: A “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 33.Olson MV. When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64(1):18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abadeh S, Killacky J, Benboubetra M, Harrison R. Purification and partial characterization of xanthine oxidase from human milk. Biochim Biophys Acta. 1992;1117(1):25–32. doi: 10.1016/0304-4165(92)90157-p. [DOI] [PubMed] [Google Scholar]
- 35.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451(7179):704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 36.Gaucher EA, Thomson JM, Burgan MF, Benner SA. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature. 2003;425(6955):285–288. doi: 10.1038/nature01977. [DOI] [PubMed] [Google Scholar]
- 37.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294(5550):2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 39.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian β-lactamases. J Am Chem Soc. 2013;135(8):2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 40.Hanson-Smith V, Kolaczkowski B, Thornton JW. Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol Biol Evol. 2010;27(9):1988–1999. doi: 10.1093/molbev/msq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu FG, et al. Molecular and morphological supertrees for eutherian (placental) mammals. Science. 2001;291(5509):1786–1789. doi: 10.1126/science.1056346. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Schrodinger LLC . The PyMOL Molecular Graphics System, version 1.3r1. New York: Schrodinger LLC; 2010. [Google Scholar]
- 47.Lanaspa MA, et al. ZAC1 is up-regulated by hypertonicity and decreases sorbitol dehydrogenase expression, allowing accumulation of sorbitol in kidney cells. J Biol Chem. 2009;284(30):19974–19981. doi: 10.1074/jbc.M109.001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andres-Hernando A, et al. Effects of 2-bromoethanamine on TonEBP expression and its possible role in induction of renal papillary necrosis in mice. Toxicol Sci. 2010;118(2):510–520. doi: 10.1093/toxsci/kfq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hedges SB, Dudley J, Kumar S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22(23):2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.