Significance
Clostridium difficile is the leading cause of antibiotic-associated infection in hospitals worldwide. Disease symptoms are caused by toxins A and B, which form membrane-spanning pores that deliver associated cytotoxic enzyme domains into target cells leading to cellular death and tissue damage. Despite a wealth of information for the enzymatic domains that act once inside the cell, very little is known about the translocation pore and its role in disease pathogenesis. Here we describe the structural features of the pore and identify mutants that prevent pore formation and show that they are no longer toxic to host cells. These findings offer a glimpse into the elusive translocation pore and further provide the basis for a unique strategy to target toxins therapeutically.
Abstract
Disease associated with Clostridium difficile infection is caused by the actions of the homologous toxins TcdA and TcdB on colonic epithelial cells. Binding to target cells triggers toxin internalization into acidified vesicles, whereupon cryptic segments from within the 1,050-aa translocation domain unfurl and insert into the bounding membrane, creating a transmembrane passageway to the cytosol. Our current understanding of the mechanisms underlying pore formation and the subsequent translocation of the upstream cytotoxic domain to the cytosol is limited by the lack of information available regarding the identity and architecture of the transmembrane pore. Here, through systematic perturbation of conserved sites within predicted membrane-insertion elements of the translocation domain, we uncovered highly sensitive residues—clustered between amino acids 1,035 and 1,107—that when individually mutated, reduced cellular toxicity by as much as >1,000-fold. We demonstrate that defective variants are defined by impaired pore formation in planar lipid bilayers and biological membranes, resulting in an inability to intoxicate cells through either apoptotic or necrotic pathways. These findings along with the unexpected similarities uncovered between the pore-forming “hotspots” of TcdB and the well-characterized α-helical diphtheria toxin translocation domain provide insights into the structure and mechanism of formation of the translocation pore for this important class of pathogenic toxins.
The primary virulence determinants of pathogenic Clostridium difficile are two protein toxins, TcdA and TcdB, which are responsible for the symptoms associated with infection, including diarrhea and pseudomembranous colitis (1). TcdA and TcdB are large (i.e., 308 and 270 kDa, respectively) homologous toxins (sharing 48% sequence identity) that appear to intoxicate target cells using a strategy that is similar in principle to that described for a number of smaller A–B toxins, such as anthrax toxin (2) and diphtheria toxin (DT) (3). In addition to a cytotoxic enzymic A domain and receptor-binding B domain responsible for binding and translocating the A domain into cells, TcdA and TcdB are additionally equipped with an internal autoprocessing domain that proteolytically cleaves and releases the N-terminal glucosyltransferase domain in response to intracellular inositol hexakisphosphate (4).
The series of events leading to the delivery of the A domain into cells begins with toxin binding to an as yet unidentified receptor on target cells via the C-terminal receptor-binding domain (i.e., the B domain), which triggers toxin internalization into acidified vesicles via clathrin-mediated endocytosis (5). In the endosome, cryptic regions from within the large ∼1,000-aa translocation domain emerge and insert into the endosomal membrane, creating a pore that is believed to enable translocation of the N-terminal glucosyltransferase (i.e., the A domain) into the cytosol. Processed and released A chains enzymatically glucosylate and thereby inactivate intracellular Rho and Ras family GTPases (6, 7) leading first to cytopathic effects (i.e., cell rounding) (8), and later, cytotoxic effects (i.e., apoptosis and necrosis) (9, 10).
Like many other A–B toxins that mediate their own delivery into cells, high-resolution structures of the enzymic A domains (11–13) and the receptor-binding portion of the B domains of glucosylating toxin family members are known (14, 15), whereas the structure and mechanism of the pore-forming translocation domain remains poorly characterized. These interconnected processes have been proposed to be mediated by the central ∼1,000-aa D domain (i.e., amino acids 801–1,850); however, with the absence of any structural information for this domain in either the prepore or pore state, no framework exists for resolving the functional determinants for this large domain that govern pore formation and translocation. It is well established that in response to acidic pH, the D domain undergoes a conformational change that results in the formation of ion-conductive pores in both biological membranes and artificial lipid bilayers (16, 17).
It has been hypothesized that the cluster of 172 hydrophobic, highly conserved amino acids in the middle of the translocation domain (i.e., residues 958–1,130 in TcdA and 956–1,128 in TcdB) comprised some, if not all, of the segments that form the translocation pore (18). Demonstrating this, however, has been challenging, in large part due to difficulties associated with manipulating clostridial toxin genes at the genetic level. The recent availability of clones of both TcdA and TcdB in Bacillus megaterium expression plasmids, which enable the high-level production of stably folded toxin, has facilitated research in this direction (19, 20); however, studies specifically addressing the structure and function of the translocation domain have been limited to large-fragment deletions to probe function (21, 22).
In the present study, we set out initially with the goal of identifying the determinants of pore formation and translocation through a comprehensive mutagenesis study using the B. megaterium platform. We discovered very early in this pursuit that site-specific mutagenesis of the inherently AT-rich toxin sequence (i.e., G + C = 27%) using the B. megaterium system was laborious and inefficient. To address this, we generated a GC-enriched copy of TcdB (i.e., G + C = 45%) with codons optimized for Escherichia coli expression, which allowed us to perform high throughput probing of the translocation domain. We identify several single point mutations clustering to within the hydrophobic region of the delivery domain that result in major defects in pore formation and translocation. We report the unexpected similarity of the identified pore-forming region to that of the translocation domain of DT and use these data en bloc to propose an α-helical model for the translocation pore of TcdB and homologous pathogenic toxins.
Results
Patterns of Hydrophobicity and Secondary Structure Suggest a Helical Pore for TcdB.
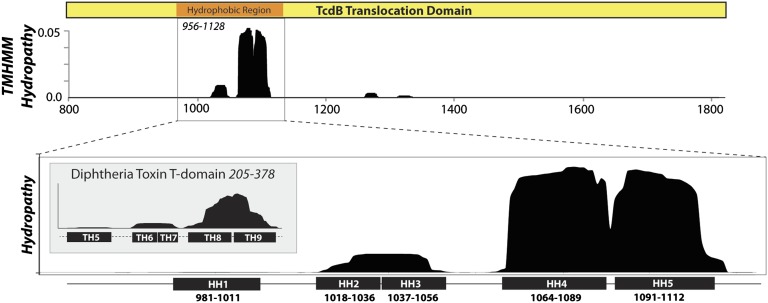
To begin to unravel the determinants of pore formation and translocation, we first analyzed the hydrophobicity, sequence conservation, and predicted secondary structure elements of the 1,050-aa translocation domain. Seven stretches of hydrophobicity were identified in TcdB: 985–1,005; 1,018–1,036; 1,037–1,056; 1,064–1,089; 1,091–1,112; 1,261–1,281; and 1,310–1,330 (Fig. 1A). We excluded the latter two regions because neither were predicted to be hydrophobic in the homolog from Clostridium novyi (TcnA) (Fig. S1). Notably, the former five hydrophobic segments fell within the “hydrophobic region” of the translocation domain that was predicted previously (i.e., 956–1,128). The length of the four hydrophobic segments that all were between 18 and 25 amino acids, combined with the absence of any alternating hydrophobic–hydrophilic “β-barrel” motifs in this region, suggest that the membrane-inserted form of these segments likely adopt an α-helical conformation. When the primary sequence of the hydrophobic region was analyzed using secondary structure propensity algorithms, five α-helical structural elements with four intervening disordered loops were predicted (Fig. S2).
Fig. 1.
Hydropathy analysis of the translocation domain. (Upper) Hydropathy analysis of the entire translocation domain of TcdB was performed using a membrane protein topology prediction method (TMHMM v2.0) that uses a hidden Markov model to predict transmembrane helices (36). Seven distinct peaks of hydrophobicity are evident: five within the previously described hydrophobic region (18), along with two smaller regions of hydrophobicity between 1,280 and 1,350, which were poorly conserved among homologous toxins and thus not pursued. (Lower) Hydropathy analysis of the 172-residue hydrophobic region showing predicted hydrophobic helices (HH1–HH5). (Inset) Hydropathy analysis of the 173-aa diphtheria toxin translocation domain with established α-helical segments predicted to comprise the translocation pore of DT (30, 31).
Positing an α-helical mode of membrane insertion, we evaluated the well-characterized α-helical diphtheria toxin (DT) translocation domain for comparison. The hydrophobic helices (TH5–6/7 and TH8–TH9) previously shown to be involved in pore formation and translocation of DT (23, 24) were correctly mapped by this analysis (Fig. 1B, Inset). Unexpectedly, we observed that the general pattern of hydrophobicity was strikingly similar for the 173-residue translocation and the 172-residue hydrophobic region of TcdB. Three peaks of similar length and amplitude were predicted in both toxins. We examine the functional link between DT translocation and the hydrophobic region of TcdB in greater detail below.
Validation of a GC-Enriched Toxin B Gene for Mutagenesis and Expression in E. coli.
The observation that the putative pore-forming hydrophobic regions of the large translocation domain were localized within the 172-aa window led us to investigate which specific amino acids in this region were involved in pore formation and translocation. To circumvent experimental barriers associated with generating many mutants to the AT-rich clostridial toxin gene (i.e., G + C = 27%), a copy of the 7,098 base pair TcdB gene was synthesized in which the G + C content was increased to 45%. This mutagenesis-competent copy of TcdB was then cloned into an E. coli expression plasmid to enable expression in a host that is more amenable for high throughput characterization than the existing B. megaterium expression system.
To validate the newly constructed GC-enhanced copy of TcdB, we characterized the structure and function of E. coli produced TcdB and compared it to benchmark standards. TcdB produced in E. coli was indistinguishable from TcdB produced in the well-validated B. megaterium system showing equal potency on Chinese hamster ovary (CHO) cells’ toxicity (Fig. S3). We next measured the cytotoxicity of both purified toxin and soluble clarified lysate from induced E. coli on CHO cells and compared this to uninduced controls, a glucosyltransferase (GTD) defective mutant (D270A), and WT TcdB produced in B. megaterium (Fig. S3). Purified WT toxins produced in either system yielded similar potency on CHO cells, whereas D270A was similarly inactive when produced in either system. Importantly, WT toxin produced in E. coli-clarified soluble lysate was equipotent with the purified toxins (after normalizing toxin concentration of toxin in crude lysates using densitometry). This set of experiments showed that E. coli-produced toxin is functional, and further, that there are no confounding contaminants in the E. coli preparations as evidenced by the complete lack of toxicity of the uninduced control on CHO cell viability (Fig. S3).
High-Throughput Mapping of the Functional Determinants in the Translocation Domain.
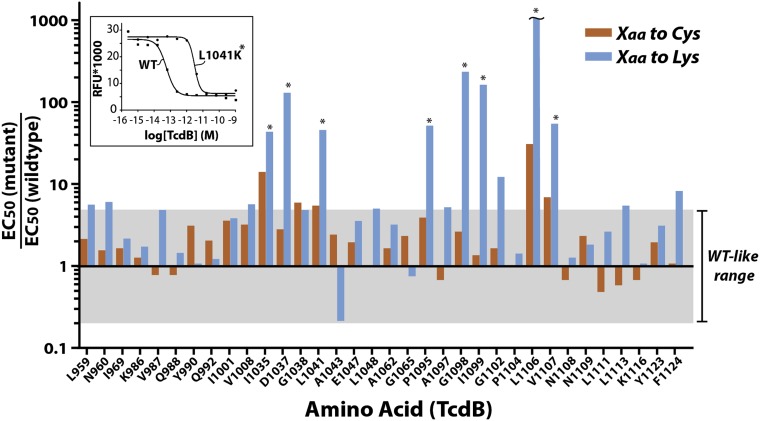
With a robust system to probe toxin function in place, we probed residues that were absolutely conserved among the large clostridial toxin (LCT) family members, reasoning that functionally important residues would be conserved in homologous toxins. We used a double-mutant strategy in which we mutated each of the residues to both a highly disruptive residue (lysine), and a more conservative residue (cysteine). The highly polar lysine side chain was selected to increase the probability of identifying a membrane-spanning segment; introducing the polar and charged Lys side chain into a marginally hydrophobic membrane-inserting segment could be expected to prevent insertion of this segment and thus pore formation. On the other hand, cysteine, like alanine, is a relatively benign substitution that can both help identify key functional residues and has the added downstream benefit of offering the possibility of attaching sulfhydryl probes in TcdB for structure/function studies.
The impact of each mutation on TcdB function was quantified by measuring the dose-dependent reduction in cell viability 48 h posttoxin addition relative to wild-type TcdB (Fig. 2). Of the nearly 90 mutants generated, only Y971 and L1048 were not expressed upon mutation, and thus were not evaluated further. Of the remaining mutants, we identified eight residues that when mutated, gave rise to a greater than 90% reduction in toxicity relative to WT (Fig. 2). As expected, for virtually all sites tested, mutation to lysine was more detrimental to function than mutation to cysteine. Four positions (i.e., D1037, G1098, I1099, and L1106) displayed a greater than 99% reduction in toxicity of which L1106K was the most defective, with an observed shift of over 1,000-fold relative to wild-type TcdB. Mutants displaying greater than a 10-fold reduction in toxicity were reexpressed in large scale and purified using Ni+-affinity chromatography and/or anion exchange chromatography and retested in triplicate. We observed excellent overall correlation in potency between the soluble lysates and purified toxins, which both confirmed our screening results and further validated our soluble lysate screening approach.
Fig. 2.
High throughput mapping of the functional determinants in the translocation domain of TcdB. Functional consequences of Cys and Lys substitutions in the hydrophobic region of TcdB. Mutant soluble lysates were titrated onto CHO cells (using threefold dilutions) in 96-well plates and incubated for 48 h at 37 °C (n = 4). In parallel, an aliquot of each mutant was used to measure the concentration using band densitometry after SDS/PAGE. Forty-eight hours later, cell viability of treated cells was quantitated by measuring PrestoBlue fluorescence using a SpectraMax M2 fluorescence microplate reader. (Inset) Sample titration curves of WT TcdB and L1041K mutant TcdB. Gray shading represents the wild-type–like range of activity (i.e., ±5-fold wild-type TcdB).
Pinpointing the Nature of the Defects in TcdB That Diminish Function.
Defective TcdB mutants were studied in detail to determine the mechanistic basis for their defects. To rule out the possibility that point mutations were causing gross and global misfolding of toxins—thus rendering them unable to intoxicate cells—we evaluated the state of folding for each mutant using the hydrophobic dye 2-p-toluidinylnaphthalene-6-sulphonate (TNS), which displays increased fluorescence when binding to hydrophobic patches of polypeptide (i.e., unfolded proteins). In addition to confirming that all mutants tested were folded, these studies show that the pH dependence of unfolding was preserved for mutant toxins (Fig. S4). To address this further, we evaluated the enzymatic activity of the GTD for each defective mutant. All mutants tested showed comparable activity to WT TcdB with specific activities that were within ±2-fold wild-type levels (Fig. S4), further suggesting that mutant toxins were otherwise folded.
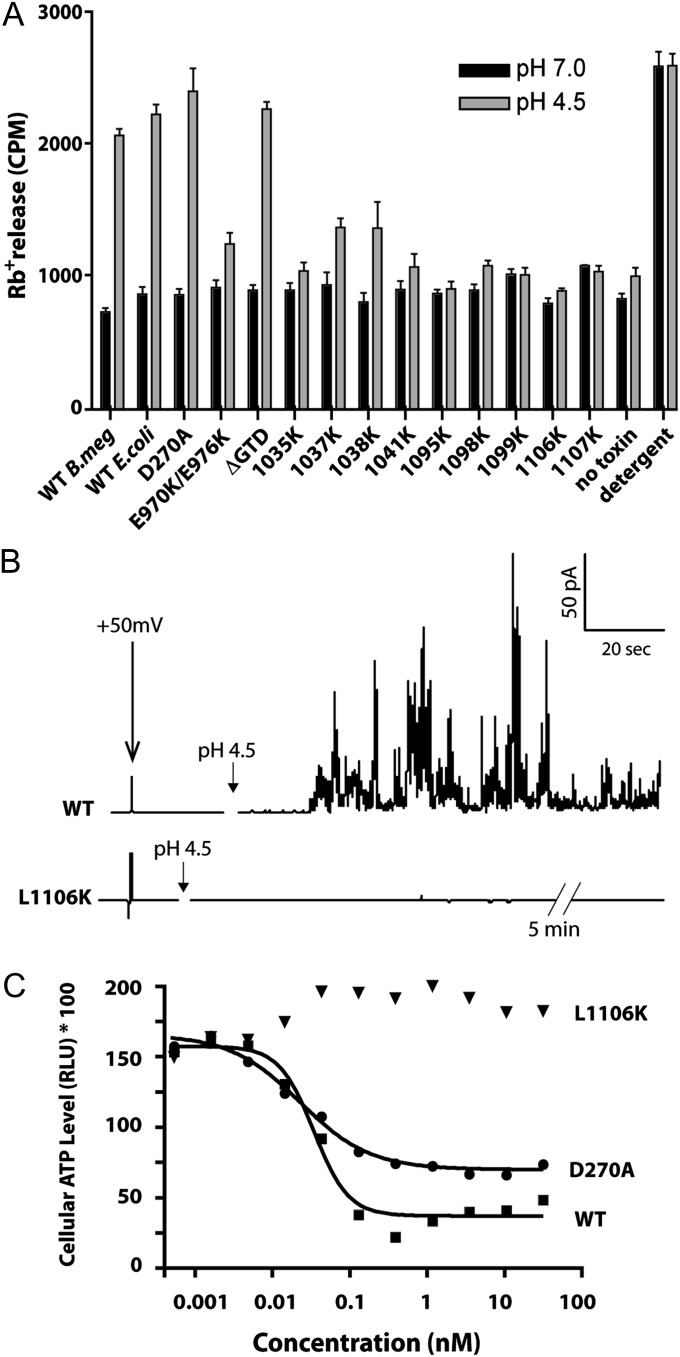
Defective mutants were then tested for translocation domain-specific functions. We first tested the ability of each of the defective mutants and control toxins to release 86rubidium ions from CHO-K1 cells upon binding to the cell surface and acidification of the medium to trigger insertion into the plasma membrane (Fig. 3A). Wild-type TcdB from all sources was able to form pores and release rubidium upon acidification to levels comparable to the detergent controls. As expected, the GTD inactive mutant D270A, and a construct in which the entire GTD was removed (i.e., ΔGTD) were also able to form pores at low pH. In contrast, all of the defective mutant toxins tested showed defects in pore formation, with most showing levels comparable to the no-toxin control. Interestingly, two mutant toxins, 1037K and 1038K, showed intermediate levels of pore formation, suggesting that pore formation was reduced, but not ablated under these conditions. Similarly, the previously identified mutant E970K/E976K, which to date is the only other mutant reported that affects pore formation, showed a partial, but not complete reduction in pore formation. In parallel, we tested pore formation in synthetic lipid bilayers using electrophysiological methods. As shown in Fig. 3B, Top, WT TcdB induced an increase in membrane activity at acidic pH (pH 4.5) with a holding potential of +50 mV (Δψ = +50 mV, cis-positive). The observed large and transient currents were reproducible and similar in character to previous planar lipid bilayer experiments (21). By contrast, despite several attempts (n = 8), with the use of higher concentrations and extended traces, we were unable to detect any channel activity for the L1106K mutant, consistent with an inability to form ion-conductive pores (Fig. 3B).
Fig. 3.
Characterization of defective purified TcdB mutants. (A) Pore formation on biological membranes. Pore formation of purified mutant toxins was tested on CHO cells preloaded with 86Rb+. Pore formation was induced by acidification of the external medium (control pH 7.0; black bars, pH 4.5; gray bars)—see Materials and Methods for details of assay (n = 5). (B) Pore formation on planar lipid bilayers. Approximately 100 pM of WT or L1106K toxin was added to the cis-chamber of planar DiPhPC/n-decane membranes in buffer containing 1 M KCl. After 2 min the pH of the cis-chamber was dropped to pH 4.5 using a defined concentration of dilute HCl. Measurements were performed with +50 mV (Ψ = +50 mV, cis-positive) at room temperature. (C) Effect of pore formation on enzyme-independent cytotoxicity. The high-dose acute cytotoxicity of purified WT TcdB, a glucosyltransferase-defective mutant (D270A), and a pore-formation defective mutant (L1106K). Constructs were tested on human IMR-90 fibroblasts under necrosis-like conditions as described previously (10).
Examining the Role of Pore Formation on TcdB-Induced Necrosis.
The experimental conditions that we used in this study to measure TcdB cytotoxicity depend on functional autoprocessing and GTD functions. Recently, Lacy and coworkers identified a second, alternative mode of cytotoxicity for TcdB that was found to be independent of autoprocessing and glucosyltransferase functions (10). At higher doses of TcdB—which are hypothesized to be possible during infection—cells undergo a rapid necrotic-like cell death via an NADPH oxidase pathway, characterized by a rapid depletion of ATP (25). We took advantage of the pore-defective mutants identified here to examine the importance of pore formation on this alternative mode of TcdB-mediated cell death. Whereas both WT TcdB and the glucosyltransferase inactive D270A mutant equally induced a rapid depletion of ATP at 100 pM, we saw no reduction in cellular ATP for L1106K up to 100 nM, indicating that pore formation is indeed required for the TcdB-mediated necrosis (Fig. 3C).
Mapping the Determinants of Pore Formation onto a Model of the TcdB Pore.
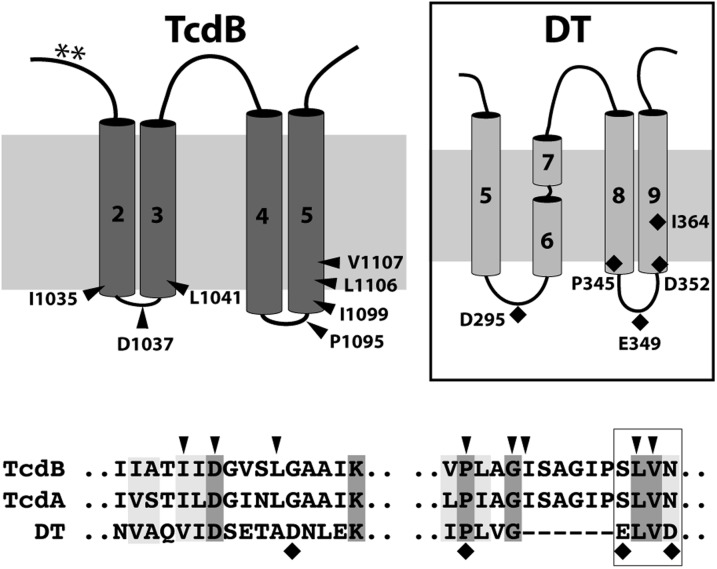
To place these functional data onto a structural framework, we considered a model of the membrane-inserted form of TcdB based on the predicted hydrophobicity and secondary structure of the hydrophobic region and using knowledge of DT to help orient the model. Mapping the residues that we identified as being sensitive to mutagenesis onto the TcdB model revealed two “hotspots” that conspicuously localized to the distal side of the membrane (Fig. 4). In forming the pore, these residues would be expected to traverse the greatest distance into the membrane relative to the proximal side of the membrane where the prepore residues before insertion. Intriguingly, when we mapped the four residues in the DT translocation domain previously shown to reduce DT-mediated toxicity by more than 100-fold, we observed a similar phenomenon; defective mutants localize to the distal end of the pore. In support of this model, we found shared residues within the hotspots that appear to be functionally important in both toxins. Asp-1,037 in TcdB and Asp-295 in DT, although offset slightly in primary sequence alignments, are positioned in the loop between the first two membrane-spanning helices and mutations to lysine in both cases as shown here for TcdB and previously with DT (26) resulting in a >100-fold shift relative to WT toxin. In the loop region intervening the second helical hairpin hotspot, Pro-345 in TcdB is aligned with Pro-1,095 in DT. Pro-345 in DT was previously shown to prevent pore formation and membrane insertion, resulting in a 99% reduction in toxicity to Vero cells (27). Our mutagenesis studies show that the P1095K mutation also prevents pore formation, similarly giving rise to a ∼95% reduction in toxicity (Fig. 2). Finally, at the heart of the membrane-insertion region are the well-studied pore-formation/translocation defective DT mutants E349K and D352K, which were shown previously to reduce functional toxicity in Vero cells by >100-fold due to an impairment in membrane insertion of the TH8–TH9 helical hairpin (26, 28). Sandwiched between these two residues are L350 and V351 in DT, corresponding to the two defective mutants, L1106 and V1107 uncovered here in TcdB.
Fig. 4.
Mapping the functional determinants of pore formation and translocation onto a working model of the TcdB translocation pore and the model of the “double-dagger” DT pore proposed originally by Choe et al. (30) and later supported by Wang and London (31). Four hydrophobic segments of ∼20 amino acids are proposed to span the lipid bilayer in the pore state as two helical hairpins—similar to DT. Functionally important residues affecting membrane-insertion/pore-formation map to the cytosolic face of the membrane in TcdB and DT. (Lower) A ClustalX alignment of TcdB, TcdA, and DT shows regions of conservation in the two pore-formation hotspots with defective mutants indicated as arrowheads (for TcdB) or diamonds (for DT). Residues within the boxed region (i.e., S1105-N1108 for TcdB, S1107-N1110 for TcdA, and E349-D352 for DT) are highly susceptible to mutagenesis.
Discussion
Research over the past decade has provided tremendous insight into the structure and function of the pathogenic toxins of C. difficile. Despite these significant advances, there remains a large gap in our understanding of the underlying structural and functional features that explain how the central translocation domain mediates the critical step of delivering the cytotoxic glucosyltransferase domains across the endosomal membrane into the cytosol. In this study we provide a comprehensive analysis of the functional determinants of the translocation domain and offer a model of the TcdB pore. Using a synthetic GC-enriched copy of TcdB, we identified eight residues, between residues 1,035 and 1,107 that when mutated resulted in a greater than 90% decrease in function. One of these pore-formation defective mutants, L1106K, reduced TcdB toxicity by greater than 1,000-fold relative to WT toxin. These key functional determinants are conserved in the members of the large clostridial toxin family as well as among the different variants of TcdB (Fig. S5). That these studies were conducted under conditions where autoprocessing and glucosyltransferase functions are required for cytotoxicity, argues that formation of this pore is required for translocation of the glucosyltransferase effector into cells.
Recently, Genisyuerek et al. generated a series of internal-, amino-, and carboxyl-terminal truncations of the translocation domain to delineate the determinants of pore formation and translocation for TcdB (21). Consistent with our findings and our model, they concluded that residues 1–1,500 (with a receptor-binding domain) encompassed the translocation machinery enabling functional toxicity. Importantly, however, they also showed that a small fragment (i.e., 830–1,025) was able to form channels in planar lipid bilayers (21). Because this fragment only partially overlaps with the residues in our model and excludes the most hydrophobic helices within the delivery domain, this finding would appear to contradict our model; however, it is important to note, as the authors of this study also suggest, that the appearance of ion channel activity in a synthetic lipid bilayer with fragments of a toxin, does not necessarily indicate the existence of the minimal translocation domain. Distinctions between the minimal channel forming region and the minimal translocation domain have been made previously for DT. A C-terminal fragment of DT, containing only half of TH8 and all of TH9 formed channels in lipid bilayers that had similar electrophysiological properties as those formed by wild-type toxin (29), despite lacking the necessary determinants for translocation.
Taken together, the data presented here support a model for the TcdB pore that is similar in principle to the double-hairpin model, as has been suggested previously for DT (30, 31). As with DT, we found that introducing a positive charge in regions that must travel farthest across the apolar bilayer during insertion to be the most detrimental to function. Whether the four transmembrane segments proposed require further oligomerization for functional translocation is not known at present. Furthermore, how the hydrophobic helices are concealed within the large 1,050-aa translocation domain prepore structure before insertion is unknown and awaits a high-resolution structure of this elusive domain.
Our observation that single point mutants within the translocation domain of TcdB could prevent pore formation and translocation additionally highlights the importance of this domain in pathogenesis. Several recent studies have shown that under certain conditions, the glucosyltransferase domain (10), the cysteine protease domain (10, 32), and even the receptor-binding domain (33) are dispensable for function. Our panel of pore-defective mutants identified here, uniquely allowed us to test the importance of translocation domain function in toxin function. We found that pore-formation defective mutants are nontoxic under all toxic paradigms tested, including cytopathic function and cytotoxic function both at low (GTD dependent) and high concentrations (GTD independent) (10). Along with providing a mechanistic basis for understanding how TcdB may elicit cytotoxicity in the absence of enzyme activity, these findings also offer a unique avenue for therapeutic intervention to halt the progress of these pathogenic toxins with small molecules or monoclonal antibodies. Agents that prevent the prepore-to-pore conversion, or block proper insertion/assembly of the pore can be expected, based on our findings, to fully inhibit toxin pathogenesis. Furthermore, given the magnitude of the reduction in toxicity combined with the subtly of a single point mutation on holotoxin folding (as demonstrated), the defective single-point mutants described herein offer ideal candidates for vaccine and/or monoclonal antibody development to neutralize these potent toxins. As these mutants have thus far only been tested in vitro on cultured mammalian cells, confirmation of the exciting potential therapeutic applications of these mutants awaits future in vivo validation studies in a disease-relevant model of infection.
Materials and Methods
Expression and Purification of Recombinant TcdB from B. megaterium.
Recombinant TcdB wild type was a B. megaterium expression vector pHis1522 encoding the strain VPI10463 obtained from Hangping Feng (University of Maryland, Baltimore). Proteins were expressed and purified as previously described (20).
Expression and Purification of Recombinant Codon-Optimized TcdB Constructs.
Codon-optimized TcdB sequence was synthesized (GenScript) to increase GC percentage to 45%. The codon-optimized gene was cloned into an E. coli expression vector pET28a and transformed into E. coli BL21 DE3 competent cells and expressed as C-terminal His-tagged proteins. See SI Materials and Methods for details.
Mutagenesis of TcdB Mutants.
Single point mutations were made in the TcdB codon-optimized sequence using QuikChange lightning multimutagenesis kit (Agilent Technologies). Sequenced plasmids with confirmed mutations were transformed and expressed using the same conditions as wild type.
Small-Scale Expression of TcdB Mutants.
Plasmids expressing TcdB mutants were transformed into E. coli BL21 DE3 cells. Overnight cultures were prepared in a 24-well block (BD Biosciences) in 5 mL of Luria Broth. Cells were harvested by centrifugation and resuspended in buffer (20 mM Tris, 500 mM NaCl pH 8.0 and protease inhibitor; Sigma) and lysed by lysozyme (Bioshop) according to the manufacturer’s instructions, followed by centrifugation at 4,000 × g for 20 min. Supernatants were collected. The concentration of each full-length mutant protein in the lysates was determined by densitometry (Image Lab 3.0).
Cell Viability Assay.
TcdB variants were added to CHO-K1 cells at a serial dilution of 1/3 starting at 1 nM. Cell viability was assessed after 48 h by PrestoBlue Cell Viability Reagent (Life Technologies). Fluorescence was read on a Spectramax M5 plate reader (Molecular Devices).
Rubidium Release Assay.
86Rb+ release assay was performed as previously reported (21). Briefly, CHO-K1 cells were seeded in 96-well plates supplemented with 1 μCi/mL 86Rb+ (PerkinElmer) at a density of 1 × 104 cells per well. 86Rb+ released was determined by liquid scintillation counting with TopCount NXT (PerkinElmer). See SI Materials and Methods for more details.
TNS Fluorescence Assay.
pH-induced conformational changes of TcdB were assessed as described previously (34). Assay plates were read in Spectramax M5 plate reader (Molecular Devices).
Black Lipid Bilayer Experiments.
Lipid bilayer experiments were performed essentially as described previously (35). Both cis- and trans-compartments contained 1 mL of solutions containing 1 M KCl, 10 mM Tris pH 7.4. Pore formation was initiated by adding appropriate amounts of 2 M HCl to the cis-compartment to lower the pH to 4.5.
CellTiterGlo ATP Assay.
Cell death assay was performed as previously described (10). Assay plates were read in Spectramax M5 plate reader (Molecular Devices).
Supplementary Material
Acknowledgments
This work was supported by a National Sciences and Engineering Research Council of Canada grant (RGPIN 418405) and a Canadian Institutes of Health Research grant (286650) (to R.A.M.). D.B.L. was supported by AI095755 from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400680111/-/DCSupplemental.
References
- 1.Kelly CP, LaMont JT. Clostridium difficile—more difficult than ever. N Engl J Med. 2008;359(18):1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 2.Collier RJ. Membrane translocation by anthrax toxin. Mol Aspects Med. 2009;30(6):413–422. doi: 10.1016/j.mam.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collier RJ. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon. 2001;39(11):1793–1803. doi: 10.1016/s0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 4.Reineke J, et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446(7134):415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- 5.Papatheodorou P, Zamboglou C, Genisyuerek S, Guttenberg G, Aktories K. Clostridial glucosylating toxins enter cells via clathrin-mediated endocytosis. PLoS ONE. 2010;5(5):e10673. doi: 10.1371/journal.pone.0010673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Just I, Selzer J, von Eichel-Streiber C, Aktories K. The low molecular mass GTP-binding protein Rho is affected by toxin A from Clostridium difficile. J Clin Invest. 1995;95(3):1026–1031. doi: 10.1172/JCI117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genth H, Aktories K, Just I. Monoglucosylation of RhoA at threonine 37 blocks cytosol-membrane cycling. J Biol Chem. 1999;274(41):29050–29056. doi: 10.1074/jbc.274.41.29050. [DOI] [PubMed] [Google Scholar]
- 8.Donta I, et al. Effect of beta-adrenergic blockade on physiologic growth in the Wistar rat. Res Commun Chem Pathol Pharmacol. 1982;37(1):147–150. [PubMed] [Google Scholar]
- 9.Hippenstiel S, et al. Rho protein inactivation induced apoptosis of cultured human endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2002;283(4):L830–L838. doi: 10.1152/ajplung.00467.2001. [DOI] [PubMed] [Google Scholar]
- 10.Chumbler NM, et al. Clostridium difficile toxin B causes epithelial cell necrosis through an autoprocessing-independent mechanism. PLoS Pathog. 2012;8(12):e1003072. doi: 10.1371/journal.ppat.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinert DJ, Jank T, Aktories K, Schulz GE. Structural basis for the function of Clostridium difficile toxin B. J Mol Biol. 2005;351(5):973–981. doi: 10.1016/j.jmb.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 12.Pruitt RN, et al. Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J Biol Chem. 2012;287(11):8013–8020. doi: 10.1074/jbc.M111.298414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler MO, Jank T, Aktories K, Schulz GE. Conformational changes and reaction of clostridial glycosylating toxins. J Mol Biol. 2008;377(5):1346–1356. doi: 10.1016/j.jmb.2007.12.065. [DOI] [PubMed] [Google Scholar]
- 14.Ho JG, Greco A, Rupnik M, Ng KK. Crystal structure of receptor-binding C-terminal repeats from Clostridium difficile toxin A. Proc Natl Acad Sci USA. 2005;102(51):18373–18378. doi: 10.1073/pnas.0506391102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murase T, et al. Structural basis for antibody recognition in the receptor-binding domains of toxins A and B from Clostridium difficile. J Biol Chem. 2014;289(4):2331–2343. doi: 10.1074/jbc.M113.505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qa'Dan M, Spyres LM, Ballard JD. pH-induced conformational changes in Clostridium difficile toxin B. Infect Immun. 2000;68(5):2470–2474. doi: 10.1128/iai.68.5.2470-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barth H, et al. Low pH-induced formation of ion channels by clostridium difficile toxin B in target cells. J Biol Chem. 2001;276(14):10670–10676. doi: 10.1074/jbc.M009445200. [DOI] [PubMed] [Google Scholar]
- 18.von Eichel-Streiber C, Laufenberg-Feldmann R, Sartingen S, Schulze J, Sauerborn M. Comparative sequence analysis of the Clostridium difficile toxins A and B. Mol Gen Genet. 1992;233(1-2):260–268. doi: 10.1007/BF00587587. [DOI] [PubMed] [Google Scholar]
- 19.Burger S, et al. Expression of recombinant Clostridium difficile toxin A using the Bacillus megaterium system. Biochem Biophys Res Commun. 2003;307(3):584–588. doi: 10.1016/s0006-291x(03)01234-8. [DOI] [PubMed] [Google Scholar]
- 20.Yang G, et al. Expression of recombinant Clostridium difficile toxin A and B in Bacillus megaterium. BMC Microbiol. 2008;8:192. doi: 10.1186/1471-2180-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genisyuerek S, et al. Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol Microbiol. 2011;79(6):1643–1654. doi: 10.1111/j.1365-2958.2011.07549.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. A segment of 97 amino acids within the translocation domain of Clostridium difficile toxin B is essential for toxicity. PLoS ONE. 2013;8(3):e58634. doi: 10.1371/journal.pone.0058634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Identification of shallow and deep membrane-penetrating forms of diphtheria toxin T domain that are regulated by protein concentration and bilayer width. J Biol Chem. 1997;272(40):25091–25098. doi: 10.1074/jbc.272.40.25091. [DOI] [PubMed] [Google Scholar]
- 24.Ren J, et al. Interaction of diphtheria toxin T domain with molten globule-like proteins and its implications for translocation. Science. 1999;284(5416):955–957. doi: 10.1126/science.284.5416.955. [DOI] [PubMed] [Google Scholar]
- 25.Farrow MA, et al. Clostridium difficile toxin B-induced necrosis is mediated by the host epithelial cell NADPH oxidase complex. Proc Natl Acad Sci USA. 2013;110(46):18674–18679. doi: 10.1073/pnas.1313658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silverman JA, Mindell JA, Finkelstein A, Shen WH, Collier RJ. Mutational analysis of the helical hairpin region of diphtheria toxin transmembrane domain. J Biol Chem. 1994;269(36):22524–22532. [PubMed] [Google Scholar]
- 27.Johnson VG, Nicholls PJ, Habig WH, Youle RJ. The role of proline 345 in diphtheria toxin translocation. J Biol Chem. 1993;268(5):3514–3519. [PubMed] [Google Scholar]
- 28.Kaul P, et al. Roles of Glu 349 and Asp 352 in membrane insertion and translocation by diphtheria toxin. Protein Sci. 1996;5(4):687–692. doi: 10.1002/pro.5560050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deleers M, Beugnier N, Falmagne P, Cabiaux V, Ruysschaert JM. Localization in diphtheria toxin fragment B of a region that induces pore formation in planar lipid bilayers at low pH. FEBS Lett. 1983;160(1-2):82–86. doi: 10.1016/0014-5793(83)80941-7. [DOI] [PubMed] [Google Scholar]
- 30.Choe S, et al. The crystal structure of diphtheria toxin. Nature. 1992;357(6375):216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, London E. The membrane topography of the diphtheria toxin T domain linked to the a chain reveals a transient transmembrane hairpin and potential translocation mechanisms. Biochemistry. 2009;48(43):10446–10456. doi: 10.1021/bi9014665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreimeyer I, et al. Autoproteolytic cleavage mediates cytotoxicity of Clostridium difficile toxin A. Naunyn Schmiedebergs Arch Pharmacol. 2011;383(3):253–262. doi: 10.1007/s00210-010-0574-x. [DOI] [PubMed] [Google Scholar]
- 33.Olling A, et al. The repetitive oligopeptide sequences modulate cytopathic potency but are not crucial for cellular uptake of Clostridium difficile toxin A. PLoS ONE. 2011;6(3):e17623. doi: 10.1371/journal.pone.0017623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanis JM, Barua S, Ballard JD. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog. 2010;6(8):e1001061. doi: 10.1371/journal.ppat.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melnyk RA, Collier RJ. A loop network within the anthrax toxin pore positions the phenylalanine clamp in an active conformation. Proc Natl Acad Sci USA. 2006;103(26):9802–9807. doi: 10.1073/pnas.0604000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.