This year marks the 50th anniversary of the publication of a landmark paper in PNAS in which Till et al. proposed a remarkable model of stem cell proliferation (1). Their idea, based on assessment of colony-forming statistics in light of a mathematical model of a stochastic birth–death process, was that individual stem cell dynamics are inherently random. This surprising proposal quickly ignited a heated, and long-running, debate over stochastic and instructive models of stem cell behavior (2). In the intervening half century, many models of stem cell dynamics have been proposed, yet the mechanisms by which stem cell numbers and activity are regulated are still not completely understood. In PNAS, Lei et al. contribute an original idea to the ongoing discussion (3). Drawing on notions from evolutionary theory, they propose a general mathematical framework that views regulation of stem cell population activity as an optimization problem, which achieves best solution when there is cross-talk between genetic and epigenetic feedback mechanisms.
Stem cells are present throughout development and adulthood and are characterized by their ability to self-renew and differentiate along multiple different cellular lineages. In the adult, stem cells are responsible for regulating tissue homeostasis and response to injury and typically reside in small numbers in tissue-specific locations known as niches that provide precisely regulated microenvironments to nurture stem cell activity (4). Although tissue-specific differences are apparent, a number of broad regulatory principles have become clear. Typically, adult stem cells divide relatively infrequently and regulate tissue homeostasis through a hierarchy of increasingly committed progenitor cells, which serve to increase cell numbers and ensure that an appropriate balance of cell types is robustly maintained over an entire lifetime. However, quiescent stem cells must also remain poised to initiate rapid tissue regeneration when needed, for instance subsequent to disease or damage. To achieve both robustness and sensitivity requires precise regulation of stem cell proliferation, differentiation, and apoptosis and continual adaptation of these functions to changes in environmental conditions. Elucidation and recapitulation of the feedback mechanisms by which this balance is achieved are major challenges in stem cell biology and regenerative medicine. Studies since the late 1970s have emphasized the importance of the niche in coordinating extrinsic and intrinsic regulatory mechanisms and maintaining appropriate stem cell proliferation (4). However, niches are themselves subject to continual turnover, and stem cells do not necessarily remain in their niche, even under homeostatic conditions (5). For example, trafficking of hematopoietic stem cells between the bloodstream and their bone marrow niche is necessary for development and healthy lifelong hematopoiesis and provides a means for stem cells to initiate a rapid systemic response to tissue damage (6). This trafficking is regulated by various different cytokines and is subject to numerous complex systemic feedback control mechanisms, including, for example, circadian oscillations (7), which ensure appropriate balance between circulating and quiescent hematopoietic stem cells in the body and continually optimize stem cell numbers and activity. However, although much is now known about the individual molecular and cellular components involved in these regulatory mechanisms (see ref. 6 for a recent review), the general strategies by which these diverse components combine to regulate stem cell activity at the systems level are largely unknown. However, deciphering such systemic optimization strategies is of central importance to understanding stem cell dynamics.
Against the backdrop of these challenges, Lei et al. outline a general mathematical framework that applies tools from optimization theory to understand stem cell dynamics (3). In their model, stem cell numbers are regulated by rates of proliferation, differentiation, and apoptosis that are continually tuned by both genetic and epigenetic feedback mechanisms to maximize population—not individual cell—performance. Key to this process is diversification of the stem cell population over a variety of different epigenetic states (taken in the broad sense to mean characteristics heritable through cell division, including molecular expression patterns, not associated with changes in DNA sequence) and association of different epigenetic states with different propensities for proliferation, differentiation, and apoptosis. Importantly, both total cell numbers and the distribution of epigenetic states within the population are regulated by system-level feedback mechanisms and coevolve to maximize tissue performance. This approach provides fresh perspective on some well-known phenomena. For instance, the authors argue that optimization strategies that use heterogeneous apoptosis rates, in which unfit cells have a higher propensity for apoptosis, support healthier long-term tissue homeostasis. This conclusion is not surprising. However, their reasoning is important. Rather than dissecting the molecular machinery associated with the cellular stress response (i.e., focusing on the cell as the regulated entity), they argue that heterogeneous apoptosis emerges naturally from evolutionary selection mechanisms that optimize long-term population function. Regulatory mechanisms do not control the fates of individual cells per se, but rather the overall structure of the population. Similarly, they reason that optimization strategies that use heterogeneous proliferation rates, which depend on both the total cell number and cell-cell variations in epigenetic status, provide a more robust and rapid response to tissue perturbation. Thus, regulated heterogeneity emerges spontaneously within stem cell populations as a consequence of optimization of population function by evolutionary selection principles. Variable populations perform better and should therefore be expected.
The notion that apparently functionally homogeneous stem cell populations are intrinsically variable either in their expression patterns or responses to directive stimuli has received much recent experimental attention. For instance, cell-cell variations in the expression of a range of important transcription factors have been observed to confer sensitivity to differentiation inducing stimuli without marking definite commitment in clonal populations of both pluripotent and adult stem cells (8). As a mechanism for regulating stem cell dynamics, such “nongenetic” variability is appealing because it allows the population as a whole to remain
Lei et al. outline a general mathematical framework that applies tools from optimization theory to understand stem cell dynamics.
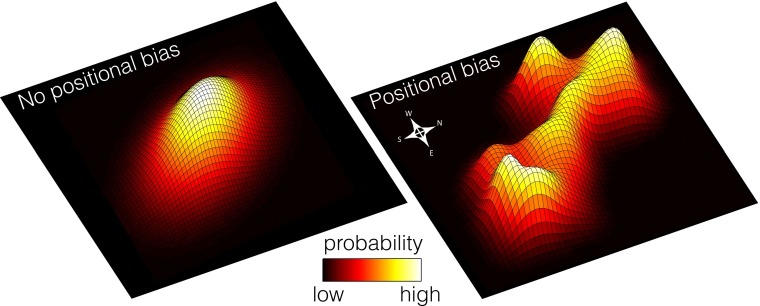
primed to respond quickly to a range of different stimuli while remaining robust to cell loss (Fig. 1) (9, 10). However, the mechanisms by which such diversity is regulated have yet to be fully determined, and demonstrations of the powerful regenerative potency of individual stem cells appear to argue against a purely system-level perspective (11, 12). Consequently, the extent to which observed cell-cell variation is functionally significant, rather than due to deficiencies in current stem cell selection and culture procedures, is unclear (13). Although these issues have yet to be fully resolved, it seems that stem cell dynamics are regulated at both the population and individual cell level by interplay between cell intrinsic and system-level feedback mechanisms (14, 15). Dissecting this interplay is a major challenge for the coming years that will require both reductionist and system-level approaches.
Fig. 1.
(Left) Model 1: Stem cell populations are homogeneous, cell identity is determined, and regulation is exerted at the single cell level. (Right) Model 2: Stem cell populations are inherently heterogeneous, different molecular states (positions) confer different functional biases to individual cells, and regulation is exerted at the population level. Both panels show population expression distributions over a hypothetical 2D expression space.
Fifty years ago, Till et al. saw that a mathematical model previously used to describe cosmic ray showers also explained certain aspects of stem cell dynamics (1). This observation opened up a whole new perspective on stem cell biology that is still being explored today. By adapting ideas from evolutionary theory and population biology, Lei et al. offer another approach to this complex problem (3). Such cross-fertilization of ideas has been central to progress in stem cell research in the past and may be necessary for progress in the future.
Acknowledgments
This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/L000512/1.
Footnotes
The author declares no conflict of interest.
See companion article on page E880.
References
- 1.Till JE, McCulloch EA, Siminovitch L. A stochastic model of stem cell proliferation, based upon the growth of spleen colony-forming cells. Proc Natl Acad Sci USA. 1964;51(1):29–36. doi: 10.1073/pnas.51.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackett N, Gordon M. “Stochastic”: 40 years of use and abuse. Blood. 1999;93(9):3148–3149. [PubMed] [Google Scholar]
- 3.Lei J, Levin SA, Nie Q. Mathematical model of adult stem cell regeneration with cross-talk between genetic and epigenetic regulation. Proc Natl Acad Sci USA. 2014;111:E880–E887. doi: 10.1073/pnas.1324267111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311(5769):1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 5.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441(7097):1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 6.Mazo IB, Massberg S, von Andrian UH. Hematopoietic stem and progenitor cell trafficking. Trends Immunol. 2011;32(10):493–503. doi: 10.1016/j.it.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452(7186):442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 8.Graf T, Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell. 2008;3(5):480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacArthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat Rev Mol Cell Biol. 2009;10(10):672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273(5272):242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 12.Wang ZD, Jaenisch R. At most three ES cells contribute to the somatic lineages of chimeric mice and of mice produced by ES-tetraploid complementation. Dev Biol. 2004;275(1):192–201. doi: 10.1016/j.ydbio.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 13.Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat Rev Mol Cell Biol. 2013;14(6):357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacArthur BD, Lemischka IR. Statistical mechanics of pluripotency. Cell. 2013;154(3):484–489. doi: 10.1016/j.cell.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145(6):851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]