Why are humans susceptible to certain illnesses but not others? Diseases such as gout and hypertension have plagued us for centuries. The prevalence of gout in the United States has risen to 3.9% in recent years (1), and a staggering one in three adults have hypertension (2). It has often been assumed that some of the less than desirable aspects of our physiology are relics of our evolutionary past, but rarely have these ideas been subjected to experimental investigation, and many of these theories have been surprisingly difficult to prove (3). Although the idea of resurrecting ancient proteins using sequence information from extant organisms was first proposed half a century ago by Pauling and Zuckerkandl (4), it is only in the last decade or so that advances in phylogenetic likelihood and Bayesian models (5), combined with fast, inexpensive gene synthesis technology, have made this approach increasingly feasible. These methods have since been used with great success to investigate a variety of topics, including the evolution of protein regulatory complexes in developmental pathways (6), vertebrate steroid hormone receptors (7), paleoenvironments and the thermophilicity of early life (8), archosaur vision (9), vaccine development (10), yeast transcriptional circuits following gene duplication (11), and the evolution of complexity in coral fluorescent proteins (12). Evolutionary questions can be notoriously intractable to experimental investigation, but the recreation of ancestral proteins provides us a window to the past through which we can directly test hypotheses regarding our evolutionary history.
Resurrecting Ancestral Uricase Enzymes
This is the approach taken by Kratzer et al. (13) in PNAS to address the fascinating question of why certain primates have lost uricase, a key enzyme that metabolizes uric acid, leaving us vulnerable to diseases such as gout that are not prevalent in other mammals. Uricases are liver enzymes involved in purine metabolism that convert uric acid into a more water-soluble form that is easily excreted by the kidneys (14). It has long been known that the loss of uricase has resulted in high levels of uric acid in humans compared with other mammals (15), but the reasons for this loss remain puzzling. In humans, depending on diet and other factors, uric acid levels can reach a threshold where urate crystals precipitate in soft tissues, giving rise to the well-known joint pain associated with gout. Elevated uric acid levels have also been associated with a number of other common diseases in modern humans including chronic hypertension, cardiovascular disease, kidney and liver diseases, metabolic syndrome, diabetes, and obesity (14).
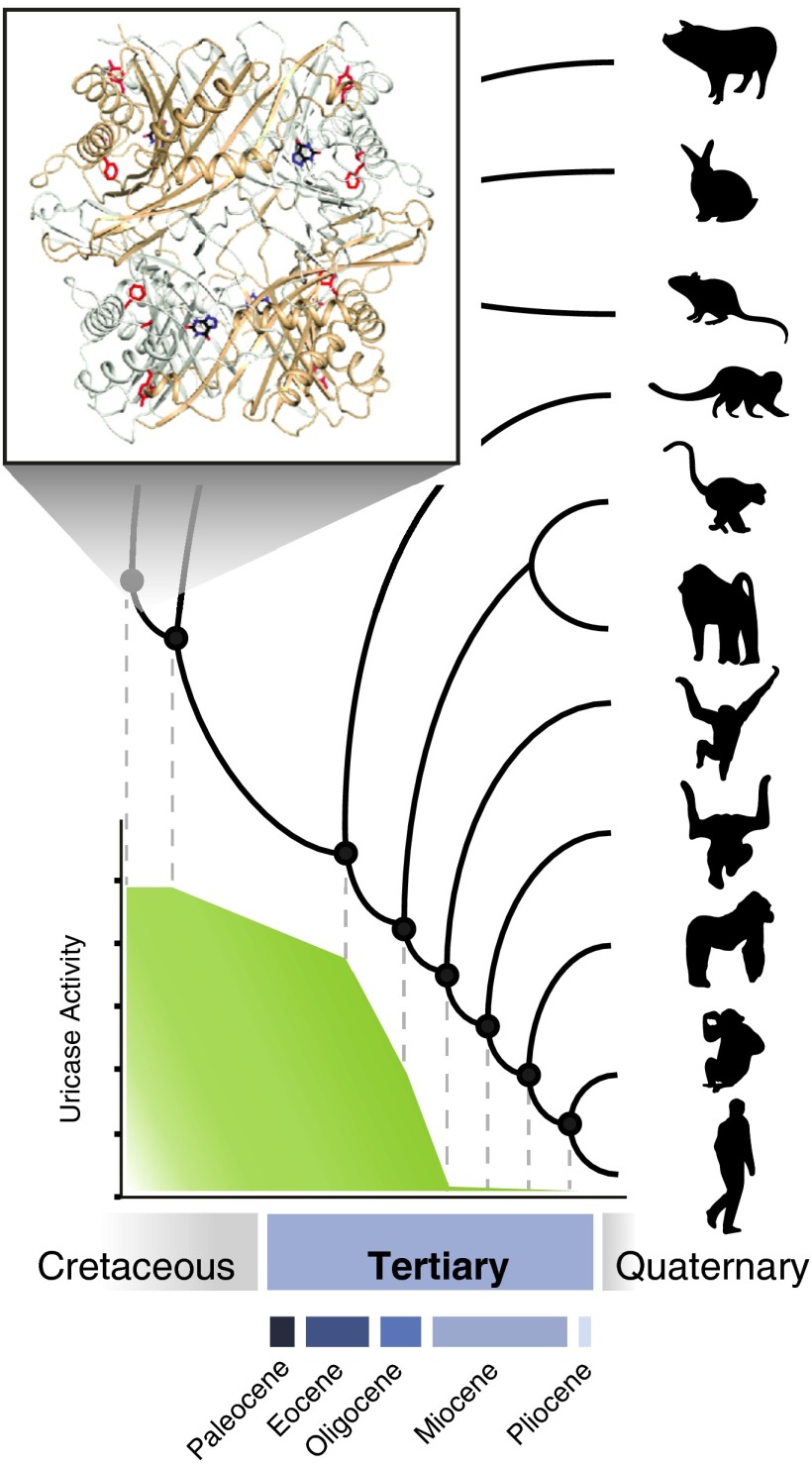
Why was such a key enzyme lost during the evolution of hominoid primates? In other mammals, this enzyme is crucial in controlling uric acid levels. Disrupting uricase function in mice results in mortality within the first 4 wk of life due to extreme uric acid levels in the blood (16). Kratzer et al. (13) used experimentally recreated ancient uricases to determine exactly when, and how, uricase function was lost in primates. They show notable, but surprisingly gradual, decreases in catalytic efficiency in these resurrected ancient enzymes, until activity was completely abolished in the ancestor of the great apes (Fig. 1). Site-directed mutagenesis was used to determine which amino acid substitutions were responsible for the decreases in activity. This study also provides a crystal structure of an ancestral mammalian uricase, which indicates that ancient mutations conferring reduced enzyme activity cluster near, but not in, the active site. Expression of ancient uricases in modern human liver cells in culture revealed not only colocalization of the ancestral enzyme with catalase, a peroxisomal marker, but also showed reduced uric acid and triglyceride accumulation in response to fructose. In other words, the ancient proteins are able to function in a modern cell in a manner similar to extant mammals with functional uricases, demonstrating that the enzymatic pathway is still intact despite millions of years of nonfunctional hominoid uricases. Finally, to assess the possibilities of using ancestral uricases for therapeutic applications, the authors test its pharmacokinetics in healthy rats, finding a greatly increased half-life relative to the native (non-PEGylated) form of a recently US Food and Drug Administration-approved pig-baboon chimeric uricase.
Fig. 1.
Schematic representation of a mammalian phylogeny, showing the experimentally recreated uricase ancestors for which enzymatic activity was measured by Kratzer et al. (13). (Inset) Ancestral mammalian uricase crystal structure (13). Approximate geologic timescale is indicated below.
The results of this paper are remarkable in a number of respects. Human uricase falls into a class of genes called unitary (or unprocessed) pseudogenes; these are previously functional genes, with no close paralogs, that have been subsequently disabled (17). This is in contrast to pseudogenes generated by gene duplication or retrotransposition, which are substantially more common in humans. The losses of single copy nuclear genes that perform essential functions and the circumstances under which they can occur are often puzzling. Current theories postulate particular conditions that might render a gene redundant, such as the naturally ascorbic acid-rich diets of guinea pig and simian ancestors, which may have allowed for pseudogenization of the gene that synthesizes vitamin C (18). However, alternate theories have suggested that pseudogenization need not always be the result of neutral processes. Instead, the loss of gene function could be advantageous and therefore the target of positive selection (19). Regardless of why pseudogenes originate, few studies have directly addressed the evolutionary history of their functional decline; in the absence of evidence, it might be tempting to simply assume an abrupt loss of function. However, the authors demonstrate that for primate uricases, this is clearly not the case (13). Their results indicate marked but gradual decreases in uricase function tens of millions of years before pseudogenization likely occurred, with specific activity all but abolished by the time the gene is lost. This is both unexpected and intriguing, particularly in light of current theories of primate origins and evolution (20).
What is the reason for such a long, slow decline in uricase activity spanning more than 50 million years of early primate evolution? In their study, the authors (13) suggest a link between the last step of uricase pseudogenization and changing climatic conditions in the late Miocene. They hypothesize that climatic cooling during this period may have coincided with periods of food scarcity, selecting for loss of uricase function, thereby allowing more efficient metabolism of fructose and maintenance of fat stores. Although this might help to explain the final stages of gene loss, it does not address the long history of gradual decline in uricase function, which according to their results commenced much earlier in primate evolution. One of the more popular theories of primate origins suggests that the rise of the angiosperms in the late Cretaceous was an important component of early primate diversification, an effect that has also been noted in other fruit-eating birds and mammals (20). That this period in primate evolution could be coincident with a gradual decline in uricase function is fascinating and might be suggestive of more general physiological adaptations in response to changes in climate and vegetation (21), but these may have been occurring in primates over broader evolutionary time scales than previously assumed. It seems likely that uricase loss is the combined result of many factors that will be challenging to pinpoint and tease apart. However, it is interesting to note that this period in early primate evolution is also roughly coincident with major shifts in sensory abilities such as increased color vision (trichromacy) (22) and decreases in olfaction and pheromonal receptors (23, 24).
Search for Thrifty Genes
In 1962, Neel (25) proposed that a thrifty genotype provided a selective advantage to hunter-gatherer ancestors subject to a feast
Kratzer et al. used experimentally recreated ancient uricases to determine exactly when, and how, uricase function was lost in primates.
or famine lifestyle that may have set us on a course for increased risk of diabetes and obesity in our present-day times of plenty. Although intuitively appealing, conclusive genetic evidence for positive selection on genes that increase the risk of diabetes in human evolution, a key prediction based on Neel’s hypothesis, has been surprisingly difficult to demonstrate (3). The results of Kratzer et al. (13) do provide convincing experimental evidence for notable declines in uricase activity that could be consistent with ancient physiological adaptations for more efficient processing of fructose and to increase fat stores in times of scarcity (21). It is also evident that loss of uricase activity is an important factor in the high rates of gout and hypertension prevalent today and may be a risk factor for diabetes and obesity as well (14). However, in the absence of any statistical evidence for positive selection (their study did not find elevated nonsynonymous to synonymous substitution rates in lineages leading to primates), their results are also consistent with a “drifty” gene hypothesis in which uricase function is lost merely as a result of random genetic drift (26). Could this simply be due to a lack of statistical power? Future studies are needed to clarify this issue.
In sum, the recent study by Kratzer et al. (13) elegantly highlights the power of experimental ancestral reconstruction approaches in probing ancient evolutionary events. Our evolutionary history may hold not only answers to why humans are susceptible to certain diseases but may also provide the key to their treatment, particularly in the case of evolutionary losses of gene function. Resurrecting ancient proteins with a view toward potential therapeutics is a currently unexploited and thoroughly intriguing possibility. We are all prisoners of our history, but perhaps we can find better solutions for the future by learning from the past.
Acknowledgments
I thank Alex Van Nynatten for assistance in preparing the figure. Grants from the Human Science Frontier Research Program and the Natural Sciences and Engineering Research Council of Canada support my work on ancestral reconstruction and molecular evolution.
Footnotes
The author declares no conflict of interest.
See companion article on page 3763.
References
- 1.Zhu YY, Pandya BJ, Choi HK. Prevalence of gout and hyperuricemia in the US general population: The National Health and Nutrition Examination Survey 2007-2008. Arthritis Rheum. 2011;63(10):3136–3141. doi: 10.1002/art.30520. [DOI] [PubMed] [Google Scholar]
- 2.Go AS, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics—2013 update: A report from the American Heart Association. Circulation. 2013;127(1):e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayub Q, et al. Revisiting the thrifty gene hypothesis via 65 loci associated with susceptibility to type 2 diabetes. Am J Hum Genet. 2014;94(2):176–185. doi: 10.1016/j.ajhg.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauling L, Zuckerkandl E. Chemical paleogenetics: Molecular ‘restoration studies’ of extinct forms of life. Acta Chem Scand. 1963;17:9–16. [Google Scholar]
- 5. Cannarozzi GM, Schneider A, eds (2012) Codon Evolution: Mechanisms and Models (Oxford Univ Press, Oxford)
- 6.Brayer KJ, Lynch VJ, Wagner GP. Evolution of a derived protein-protein interaction between HoxA11 and Foxo1a in mammals caused by changes in intramolecular regulation. Proc Natl Acad Sci USA. 2011;108(32):E414–E420. doi: 10.1073/pnas.1100990108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461(7263):515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akanuma S, et al. Experimental evidence for the thermophilicity of ancestral life. Proc Natl Acad Sci USA. 2013;110(27):11067–11072. doi: 10.1073/pnas.1308215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang BSW, Jönsson K, Kazmi MA, Donoghue MJ, Sakmar TP. Recreating a functional ancestral archosaur visual pigment. Mol Biol Evol. 2002;19(9):1483–1489. doi: 10.1093/oxfordjournals.molbev.a004211. [DOI] [PubMed] [Google Scholar]
- 10.Ducatez MF, et al. Feasibility of reconstructed ancestral H5N1 influenza viruses for cross-clade protective vaccine development. Proc Natl Acad Sci USA. 2011;108(1):349–354. doi: 10.1073/pnas.1012457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker CR, Hanson-Smith V, Johnson AD. Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science. 2013;342(6154):104–108. doi: 10.1126/science.1240810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ugalde JA, Chang BS, Matz MV. Evolution of coral pigments recreated. Science. 2004;305(5689):1433. doi: 10.1126/science.1099597. [DOI] [PubMed] [Google Scholar]
- 13. Kratzer JT, et al. (2014) Evolutionary history and metabolic insights of ancient mammalian uricases. Proc Natl Acad Sci USA 111:3763–3768. [DOI] [PMC free article] [PubMed]
- 14.Gustafsson D, Unwin R. The pathophysiology of hyperuricaemia and its possible relationship to cardiovascular disease, morbidity and mortality. BMC Nephrol. 2013;14:164. doi: 10.1186/1471-2369-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keilin J. The biological significance of uric acid and guanine excretion. Biol Rev Camb Philos Soc. 1959;34(3):265–296. [Google Scholar]
- 16.Wu XW, et al. Hyperuricemia and urate nephropathy in urate oxidase-deficient mice. Proc Natl Acad Sci USA. 1994;91(2):742–746. doi: 10.1073/pnas.91.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M. Identification and analysis of unitary pseudogenes: Historic and contemporary gene losses in humans and other primates. Genome Biol. 2010;11(3):R26. doi: 10.1186/gb-2010-11-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drouin G, Godin JR, Pagé B. The genetics of vitamin C loss in vertebrates. Curr Genomics. 2011;12(5):371–378. doi: 10.2174/138920211796429736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson MV. When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64(1):18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sussman RW, Tab Rasmussen D, Raven PH. Rethinking primate origins again. Am J Primatol. 2013;75(2):95–106. doi: 10.1002/ajp.22096. [DOI] [PubMed] [Google Scholar]
- 21.Johnson RJ, et al. Lessons from comparative physiology: Could uric acid represent a physiologic alarm signal gone awry in western society? J Comp Physiol B. 2009;179(1):67–76. doi: 10.1007/s00360-008-0291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt DM, et al. Molecular evolution of trichromacy in primates. Vision Res. 1998;38(21):3299–3306. doi: 10.1016/s0042-6989(97)00443-4. [DOI] [PubMed] [Google Scholar]
- 23.Gilad Y, Przeworski M, Lancet D. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2(1):E5. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang JZ, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci USA. 2003;100(14):8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neel JV. Diabetes mellitus: A “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14(4):353–362. [PMC free article] [PubMed] [Google Scholar]
- 26.Speakman JR. Thrifty genes for obesity, an attractive but flawed idea, and an alternative perspective: The 'drifty gene' hypothesis. Int J Obes. 2008;32(11):1611–1617. doi: 10.1038/ijo.2008.161. [DOI] [PubMed] [Google Scholar]