Significance
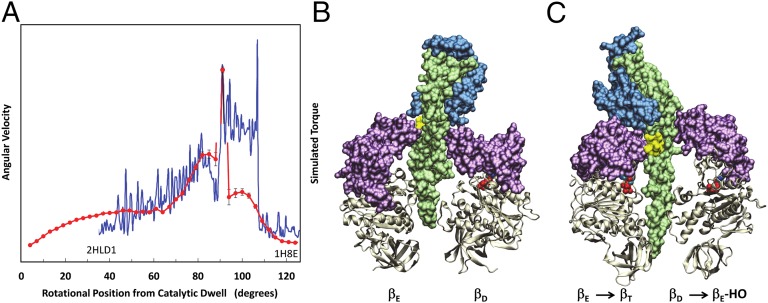
We present a description of the angular velocity of the power stroke as a function of rotational position for the F1-ATPase molecular motor. The angular velocity of this motor, which rotates in 120° power strokes separated by catalytic dwells, is part of the FoF1 ATP synthase in oxidative phosphorylation, and has been thought not to vary. This paper reports the unexpected discovery that a series of angular accelerations and decelerations occur, providing direct evidence that angular velocity depends on substrate binding affinity. The data support a model in which rotation is powered by Van der Waals repulsive forces during the final 85° of rotation, consistent with a transition from F1 structures 2HLD1 and 1H8E (Protein Data Bank).
Keywords: FoF1, torque, rotary motor, ATP hydrolysis
Abstract
F1-ATPase, the catalytic complex of the ATP synthase, is a molecular motor that can consume ATP to drive rotation of the γ-subunit inside the ring of three αβ-subunit heterodimers in 120° power strokes. To elucidate the mechanism of ATPase-powered rotation, we determined the angular velocity as a function of rotational position from single-molecule data collected at 200,000 frames per second with unprecedented signal-to-noise. Power stroke rotation is more complex than previously understood. This paper reports the unexpected discovery that a series of angular accelerations and decelerations occur during the power stroke. The decreases in angular velocity that occurred with the lower-affinity substrate ITP, which could not be explained by an increase in substrate-binding dwells, provides direct evidence that rotation depends on substrate binding affinity. The presence of elevated ADP concentrations not only increased dwells at 35° from the catalytic dwell consistent with competitive product inhibition but also decreased the angular velocity from 85° to 120°, indicating that ADP can remain bound to the catalytic site where product release occurs for the duration of the power stroke. The angular velocity profile also supports a model in which rotation is powered by Van der Waals repulsive forces during the final 85° of rotation, consistent with a transition from F1 structures 2HLD1 and 1H8E (Protein Data Bank).
The purified F1-ATPase is a molecular motor that can hydrolyze ATP to drive counterclockwise (CCW) rotation of the γ-subunit within the (αβ)3-ring (Fig. 1A). In most living organisms, the Fo component of the FoF1 complex uses energy derived from a proton-motive force across a membrane to power F1-dependent synthesis of ATP from ADP and Pi. Consumption of an ATP at each F1 catalytic site, primarily composed of a β-subunit, correlates with a 120° rotational power stroke of the γ-subunit separated by a catalytic dwell with an 8-ms duration in Escherichia coli enzyme (1). A second “ATP-binding” dwell can occur after the γ-subunit has rotated ∼30° to 40° from the catalytic at limiting substrate concentrations (2, 3). Thus, three successive catalytic events that include power strokes and dwells are required to complete one revolution of the γ-subunit. Once bound to F1, ATP is retained for 240° (3) such that the ADP and Pi generated are released two catalytic events later.
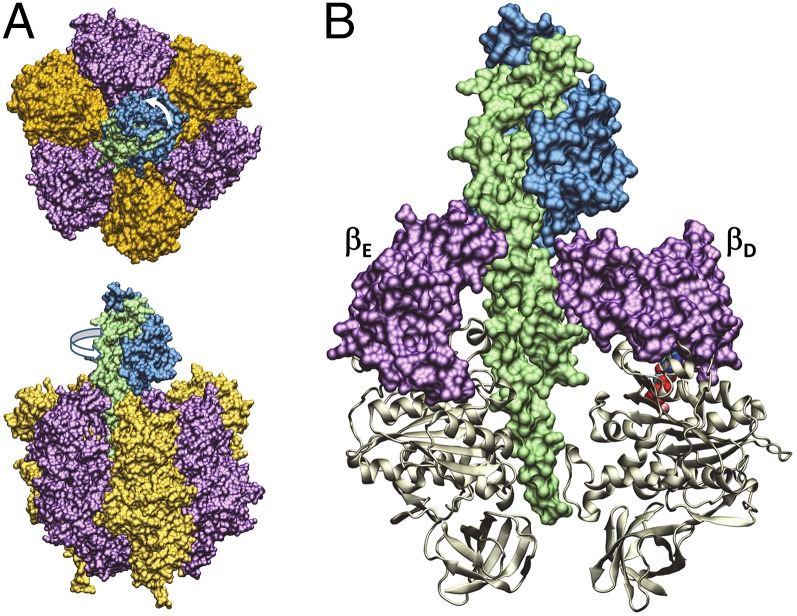
Fig. 1.
Structural components of the F1-ATPase molecular motor. (A) Top (from membrane) and side views of F1 composed of the ring of α (orange) and β (purple) subunits surrounding the γ-subunit rotor (blue and green). (B) Open (βE) and closed (βD) conformations of the catalytic site composed of the catalytic domain (tan ribbon) and the lever domain (purple) relative to the γ-subunit coiled-coil (green) and foot (blue) domains. In the Gibbons et al. (32) F1 structure (PDB ID code 1E79) used here, the γ-subunit foot domain is rotated 7° CCW from the structure determined by Menz et al. (4).
The γ-subunit is composed of coiled-coil and globular “foot” domains where the former extends through the core of the (αβ)3-ring (Fig. 1B). The β-subunits contain a catalytic domain and a C-terminal “lever” domain that is extended or open when the catalytic site is devoid of nucleotide and contracted (closed) when nucleotide is bound. In most F1 crystal structures (4, 5), the coiled-coil faces the β-subunit with the open lever (βE) whereas the foot domain extends over the lever domains of catalytic sites that usually contain bound ADP (βD) and ATP (βT). Although crystal structures provide excellent pictures of the subunit conformations at one rotational position, the rotational movement of the γ-subunit between these static structures and the mechanism in which ATP fuels this movement occurs remain major unresolved questions.
Consensus is currently lacking regarding the relationship of nucleotide occupancy at the three catalytic sites to the catalytic dwell and ATP-binding dwell despite the intense scrutiny this question has received since Boyer and coworkers (6, 7) showed that F1 operates via an alternating site mechanism. The catalytic dwell includes ATP hydrolysis and is believed to be terminated by the release of phosphate (3). Some single-molecule experiments support a mechanism whereby ATP binding and ADP release are concurrent during the ATP-binding dwell (3). As a result, only two catalytic sites are occupied the majority of the time such that three-site occupancy occurs transiently during the ATP-binding dwell. However, these results are inconsistent with the F1 structure that contains transition state analogs and has three-site nucleotide occupancy (4). Nucleotide binding studies also strongly support a mechanism in which all three sites must be occupied (8, 9) and are consistent with other single-molecule studies that support alternative three-site mechanisms (10–12). At this time there is no consistent evidence that correlates any of the crystal structures to the prevailing rotational mechanism.
The β-subunit lever domain is positioned to push against the γ-foot and the γ–coiled-coil as it opens and closes, respectively (Fig. 1B). The asymmetry of the γ-subunit at these interfaces resembles a camshaft that would be consistent with CCW directionality in response to lever movement. The energy for a 120° power stroke has been proposed to derive from the binding affinity of ATP that is used as ATP binding-induced closing of the β-lever (13) and is supported by experiments in which the lever was truncated (14).
Based on single-molecule measurements, it was concluded that F1 is nearly 100% efficient (15). A necessary outcome of this conclusion is that the 120° power strokes must occur at a constant angular velocity (13). Although a number of simulation studies have modeled rotation of the F1-ATPase γ-subunit (16–19), only one of these (19) has provided a result showing that the angular velocity should vary during a power stroke. The claim of 100% efficiency (15) that serves as the energetic basis of this power stroke mechanism is unwarranted because the magnitude of the viscous flow coupling to the surface was unknown owing to technical limitations, and the authors erroneously used the value of ΔG° in lieu of ΔG in their calculation. The technical handicap was subsequently overcome by Junge and coworkers (20, 21), who relied on elastic probe curvature instead of rotation speed to calculate the average torque of the power stroke. Similar average torque values were subsequently obtained using rotation under limiting drag conditions, when the drag on the probe was measured directly (22).
Based on 100% enzyme efficiency, it was difficult to explain how the energy from ATP binding was able to power 120° of rotation when the catalytic dwell interrupts rotation 80° after ATP binds. It was hypothesized that the remaining binding energy needed to power the final 40° of rotation until the next ATP binds is stored as elastic energy in the closed β-lever, which upon product release pushes on the γ-foot as it opens (13). However, to date, experimental evidence that tests these hypotheses is lacking owing to the inability to measure the rotary motion under conditions where the angular velocity is limited by the internal mechanism of the motor.
Here we have resolved the angular velocity as a function of the rotational position using an assay that provides 10-μs time resolution. The results clearly show that the angular velocity is not constant during a power stroke, but undergoes a series of accelerations and decelerations as a function of rotational position. The slower angular velocity observed with the lower-affinity substrate ITP provides direct evidence that substrate binding affinity provides energy to power rotation. The correlation of the angular velocity profile of the final 85° of rotation presented here to the profile resulting from the simulations of Pu and Karplus (19) strongly supports a model in which ATP binding-dependent closure of the lever applies force to the γ-subunit. This also provides evidence that associates the F1 structures of Kabaleeswaran et al. (23) and Menz et al. (4) with the protein conformations at 35° and 120° because they were used as the reference structures for the simulations of Pu and Karplus (19). The data also show that elevated ADP concentrations increase dwells at ∼35° and decrease the angular velocity between 85° and 120°. This indicates that ADP can remain bound subsequent to the ATP-binding dwell consistent with a three-site mechanism.
Results
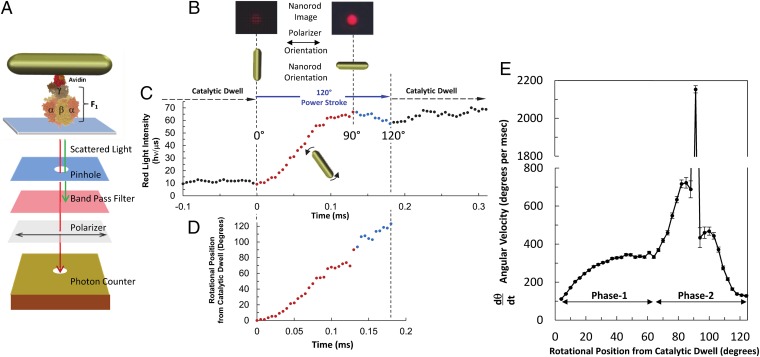
We observed the rotational position of individual 75- × 35-nm gold nanorods attached to the γ-subunit of single F1-ATPase molecules from E. coli using dark-field microscopy (Fig. 2A). A nanorod typically appears as a stationary spot with a diameter that is half the wavelength of the scattered light when viewed through a filter to eliminate all but the red light scattered from it. Upon rotation, the light intensity changes in a sinusoidal manner as a function of its rotary position relative to the direction of a polarizer positioned in the light path (Movie S1). To measure the angular velocity of the 120° F1-ATPase power strokes, the polarizing filter was aligned with each gold rod (Fig. 2B) such that the scattered light intensity was near a minimum for one of the catalytic dwells.
Fig. 2.
Single-molecule measurements of rotation using gold nanorods as a probe. (A) Schematic model of experimental setup. (B) Scattered light intensity when the long axis of a nanorod is perpendicular and parallel to the direction of polarization. (C) Scattered red light intensity collected at 200 kHz from a nanorod attached to the γ-subunit of F1 with 1 mM MgATP during a period that includes the end of one catalytic dwell through a 120° power stroke until the start of the next catalytic dwell. (D) Rotational position for 0° to 90° of the power stroke was determined from minimum to maximum intensity values of C using Eq. 1 and for 90° to 120° (blue) using Eq. 2. (E) Average angular velocity of F1 ATPase-driven γ-subunit rotation during a power stroke binned for every 3° of rotation from the end of the catalytic dwell.
Fig. 2C shows an example of the raw data collected during a single 120° power stroke, and the conversion of the same data from light intensity to rotational position is shown in Fig. 2D. At the end of the catalytic dwell the light intensity during the 120° of power stroke rotation increased through the maximum, which occurred when the nanorod had rotated 90°. After collecting 5 s of data from each molecule at 200 kHz with a single-photon counting avalanche photodiode, the power strokes that met the minimum-to-maximum-intensity criteria were collected and the rotational position as a function of time for each power stroke was determined from the arcsine of the intensity (Fig. 2D) as described in Methods.
The power stroke angular velocity (ω) as a function of rotational position was obtained by dθ/dt for each power stroke, then the angular velocities of all of the power strokes acquired were binned and averaged for every 3° of rotation to obtain the data of Fig. 2E. The average angular velocity of the γ-subunit power strokes as a function of the rotational position from the end of the catalytic dwell was not constant in the presence of saturating MgATP, but accelerated in two phases. In phase 1 following the catalytic dwell, the angular acceleration decreased steadily to reach a relatively constant angular velocity of ∼330°⋅ms−1 between 50° and 60°. The power stroke accelerated again (phase 2) to ∼710°⋅ms−1 at ∼83°. Subsequent angular decelerations were briefly interrupted by accelerations at 93° and 100°, with the former rotating at an estimated 2,100°⋅ms−1 during a 3° period. The resolution of the angular velocity changes, which in Fig. 2E are the average of 2,397 power strokes from nine F1-ATPase molecules, is evidence that the phase shift between power strokes is minimal.
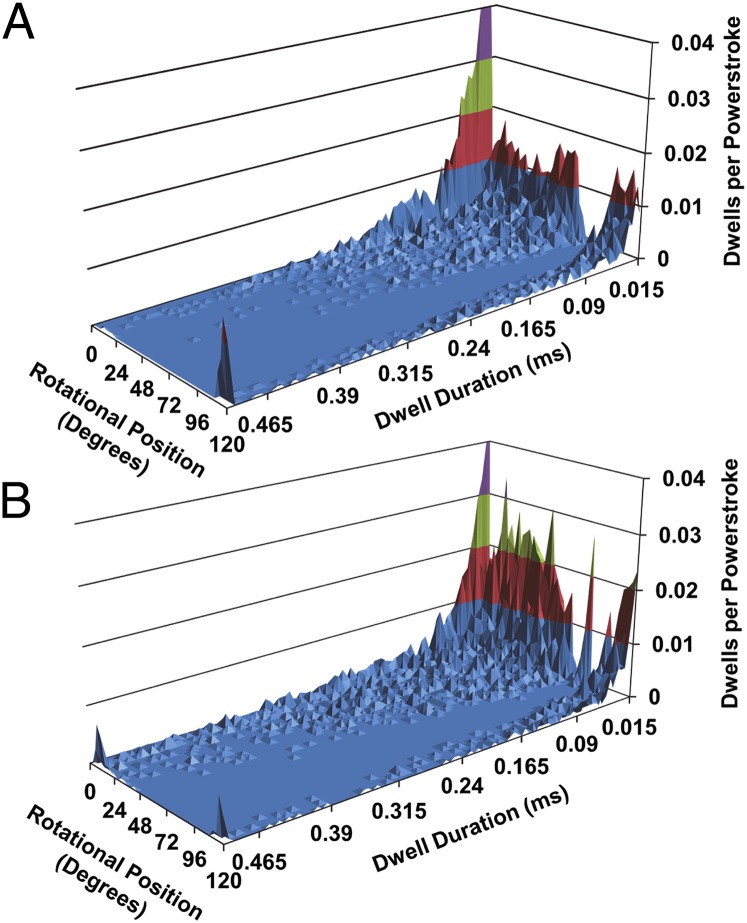
For some individual power strokes the angular velocity at one or more rotational positions was zero. Fig. 3A shows the location and duration of these pauses for every 3° of rotational position. The most common occurrence of pauses (four per 100 power strokes) was observed during the first 3° of rotation from the end of the catalytic dwell. This likely represents the end of the catalytic dwell for power strokes that were slightly phase-shifted during alignment with the polarizer. Most pauses were <20 μs, which was not much longer than the 10-μs time resolution limit defined by the data acquisition speed. Pauses were almost nonexistent between 80° and 110° when the angular velocity was at its highest.
Fig. 3.
Abundance of pauses as a function of duration and rotational position from the catalytic dwell in the presence of (A) 2 mM ATP and 1 mM Mg2+ or (B) 2 mM ATP, 3 mM Mg2+, and 4 mM ADP.
In the presence of 2 mM MgADP, the abundance and duration of dwells during phase 1 increased (Fig. 3B). The fractional change in dwell abundance owing to 2 mM MgADP increased during phase 1 with a maximal increase of 36% at a rotational position of ∼30°, which is consistent with previous observations (3). Fig. 4A shows the effects of 2mM MgADP on the average angular velocity as a function of rotational position. The fractional changes in angular velocity in the presence versus the absence of MgADP are shown in Fig. 4B (blue) to enable a direct comparison with the changes in dwells. To determine the contribution of the increased occurrence of MgADP-dependent dwells on the average angular velocity, the fractional changes in dwell abundance in the presence (Fig. 3B) versus the absence of MgADP (Fig. 3A) as a function of rotational position is shown in Fig. 4B (green).
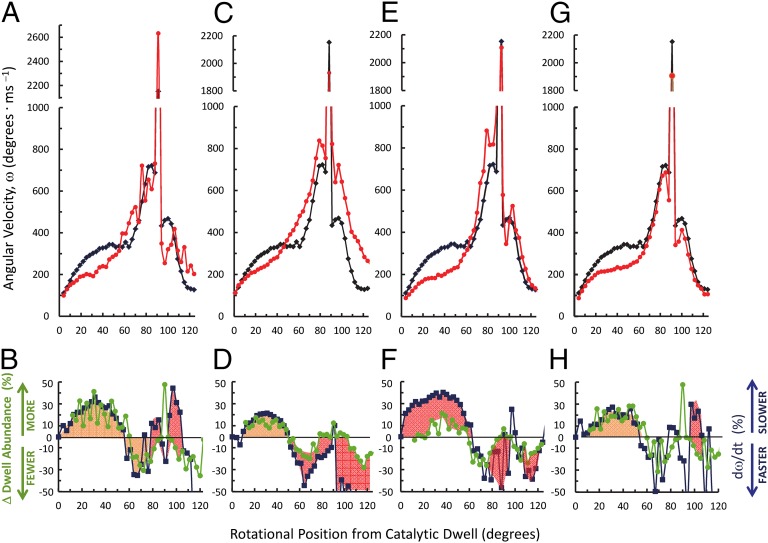
Fig. 4.
Dependence of power stroke angular velocity as a function of rotational position on substrate and product. Red lines show angular velocity in the presence of (A) 2 mM ATP, 3 mM Mg2+, and 4 mM ADP; (C) 0.3 mM ATP and 0.6 mM Mg2+; (E) 2 mM ITP and 1 mM Mg2+; or (G) 2 mM ITP, 3 mM Mg2+, and 4 mM ADP versus (black) 1 mM MgATP as in Fig. 2E. Percent changes in angular velocity (blue) and dwell abundance (green) resulting from (B) increased MgADP from A, (D) limiting MgATP from C, (F) 1 mM MgITP from E, and (H) MgITP with increased MgADP from G. Changes in dwell abundance were determined from the difference of Fig. 3 B and A for B and from analogous plots for D, F, and H. Equal percent changes in angular velocities and dwell abundances are shown as tan, and inequalities are orange in B, D, F, and H.
During phase 1 the fractional decreases in angular velocity mapped closely to the fractional increase in dwells, with the former having a maximal decrease of 41% at ∼30°. This indicates that the decrease in angular velocity during this phase was primarily the result of the abundance of dwells. A decline in dwells also resulted in an apparent increase in angular velocity between 50° and 80°. However, during much of phase 2, MgADP-dependent decreases in angular velocity were observed that could not be explained by increases in dwell abundance.
When MgATP was rate-limiting for ATP hydrolysis (Fig. 4 C and D), fractional decreases in phase 1 angular velocity by as much as 21% at 30° were observed. The magnitudes of these changes were approximately the same as the fractional increase in dwells during the first 45° of rotation. Because limiting substrate is known to cause ATP-binding dwells that occur with a duration inversely proportional to the time required for ATP to bind to the empty catalytic site (3), the decrease in angular velocity during phase 1 can be attributed to an increase in ATP-binding dwells. During phase 2, the power stroke angular velocity increased by as much as twofold (at ∼100°) compared with what was observed when MgATP was saturating, which could not be explained by changes in dwell abundance.
The contribution of nucleotide binding affinity to the angular velocity of the γ-subunit was determined by examining ITP-dependent rotation. The dissociation constants for IDP for each of the three catalytic sites of F1 was measured using tryptophan fluorescence quenching (Table S1), which had been used to determine the affinities of the enzyme for ATP, ITP, and ADP (8, 9). Compared with ATP, the comparative decreases in affinities for ITP varied by site between 41-fold and 330-fold, whereas the decreases of IDP versus ADP were 38-fold to 730-fold.
The average angular velocity powered by 1 mM MgITP (Fig. 4E) was slower during phase 1 by as much as 40% compared with that of MgATP. The decreased affinity of F1 for ITP compared with ATP was also reflected in a change in the kcat/Km of ATP hydrolysis (Fig. S1). As a result, 1 mM ITP was rate-limiting for hydrolysis, which resulted in a fractional increase of ITP-binding dwells during this rotational phase (Fig. 4F). However, the fractional decrease in the MgITP-dependent angular velocity was at least twofold greater than was accounted for by the increase in ITP-binding dwells. This indicates that substrate affinity to the empty catalytic site contributes significantly to the angular velocity during this phase of the power stroke.
During phase 2, the MgITP-dependent power stroke angular velocity was higher than that of MgATP (Fig. 4 E and F). This increase in angular velocity was not the result of changes in the abundance of dwells. In the presence of MgADP (Fig. 4 G and H), the decrease in angular velocity observed with MgITP during phase 1 was dominated by an increase in dwells, and the angular velocity during phase 2 was suppressed. This indicates that the rotation rate during this phase is limited by bound product.
Discussion
Analysis of light scattered from a gold nanorod attached to the γ-subunit of the F1-ATPase provided unprecedented resolution of angular velocity as a function of time under conditions in which the rate-limiting steps result from the intrinsic enzyme mechanism rather than from drag on an optical probe. We found that the angular velocity is not constant but, following the catalytic dwell, rotation accelerates in two phases separated by 60°. Subsequent angular decelerations are briefly interrupted by accelerations at 93° and 100°. These results are consistent with a rotary mechanism that operates at an efficiency that is less than 100% of the energy available from hydrolysis.
The data here are presented in the form of angular velocity rather than torque. Torque (T) varies as a function of angular velocity (ω) by the relationship T = ωΓ when the angular velocity is limited by the viscous load (Γ). We had previously determined that the viscous load on F1 attached to nanorods under the same conditions as those presented here did not limit the average angular velocity of ∼360°⋅ms−1 measured during the first 90° of rotation (22, 24, 25). Because the phase-2 angular velocity sometimes substantially exceeds that average, more experiments are necessary to establish the point at which viscous load becomes rate-limiting. Nevertheless, under the conditions presented here, torque will vary proportionally with angular velocity.
The slower phase-1 angular velocity observed with the lower-affinity substrate ITP (Fig. 4) provides direct evidence that energy to power rotation is derived, at least in part, from substrate binding affinity. Based on coarse-grained plastic network modeling that showed variations in torque during a power stroke, Pu and Karplus (19) proposed that conformational changes of the catalytic β-subunits act on the γ-subunit through repulsive Van der Waals interactions to drive rotation. In the coarse-grained model, torque was calculated using a simple functional form for potential energy coupling of the action of the levers on the coiled-coil during the targeted molecular dynamics (TMD) trajectories. The superposition of torque derived from this simulation (19) on the angular velocity as a function of the rotational position is shown in Fig. 5A. The correlation of the simulation from the ATP-binding dwell at 35° to the catalytic dwell at 120° with the angular velocity profile presented here during the same part of the power stroke strongly supports the conclusion that ATP binding-dependent closure of the lever applies force to γ-subunit as summarized in Fig. 6 B and D.
Fig. 5.
(A) Superposition of the average angular velocity of F1 ATPase-driven γ-subunit rotation during a power stroke as a function of rotation angle from the catalytic dwell (red) with the torque profile during an 85° substep of rotation (blue) calculated from the simulation of Pu and Karplus (19). The simulation was obtained using the F1-ATPase structures (B) of Kabaleeswaran et al. (23) (PDB ID code 2HLD1 where the γ-subunit foot domain is rotated 29° CCW) and (C) Menz et al. (4) (PDB ID code 1H8E) as reference structures for rotational positions of 35° and 120° from the end of the catalytic dwell, respectively. Color designations of domains are the same as in Fig. 1. Yellow residues correspond to γ232–238 in the γ-subunit coiled-coil.
Fig. 6.
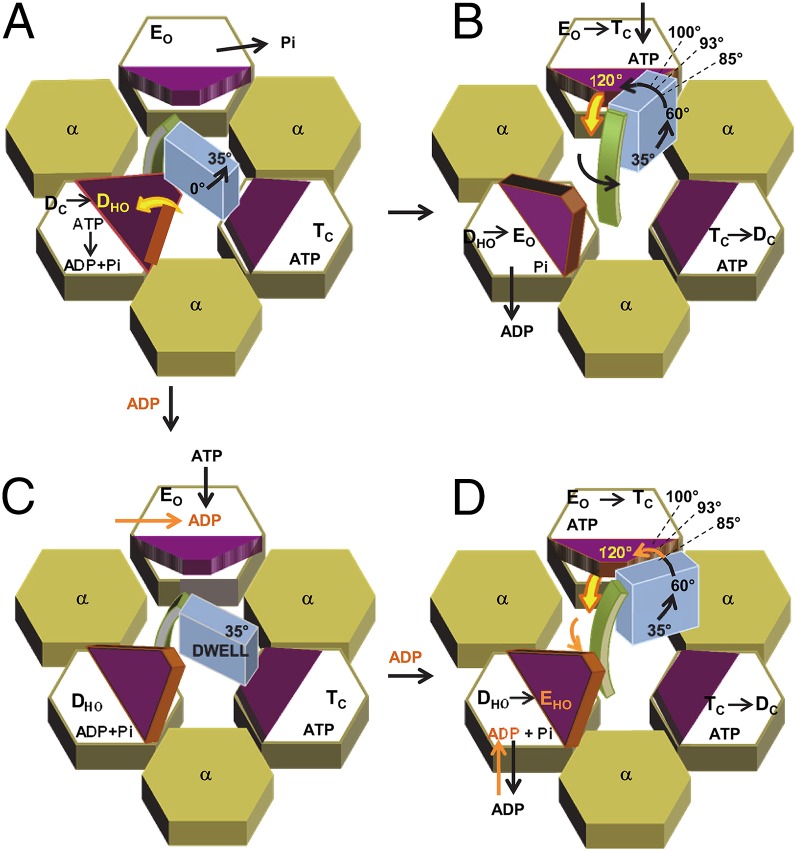
Reaction scheme of the angular velocity of F1-ATPase γ-subunit rotation. The γ-subunit foot (blue) and coiled-coil (green) domains are surrounded by the ring of alternating α-subunits (tan) and β-subunits, in which the lever domain of the latter is denoted in purple-brown. The β-subunits D, E, and T refer to βD, βE, and βT, respectively, with lever conformations of closed (C), open (O), or half-open (HO). (A) Initial rotation from the end of the catalytic dwell (0°) before the binding of ATP. Hydrolysis of ATP raises the β-lever from underneath the foot. (B) Rotation dependent upon ATP-binding to βE at low ADP concentrations. Rapid release of ADP from βD raises the lever so as not to interfere with the coiled-coil that is rotated in response to the ATP binding-dependent closing of the βE-lever. (C) High ADP concentration-dependent dwell resulting from competitive product ADP inhibition of ATP binding to βE. (D) High ADP concentration-dependent decrease in angular velocity between 85° and 120° owing to the retention of ADP at βD that retains the HO lever conformation, causing steric interference with rotation of the coiled-coil.
Based on the correlation in Fig. 5A, we concur with Pu and Karplus (19) that the angular velocity is largely the result of repulsive van der Waals interactions because amino acid side chains were not included in the simulation. This hypothesis is supported by experiments that show that the acidic nature of the well-conserved DELSEED residues in the lever is not essential for torque generation (26). The simulated torque profile (19) in Fig. 5 was calculated from the TMD trajectories as a function of rotational position using the coordinates of F1 structures from yeast (23) and bovine (4). At any one rotational position, the torque on the ith residue of the γ-stalk resulted from the cross-product of the position vector and the force vector acting on residue i, coarse-grained to its Cα atom. The observed changes in angular velocity are then primarily the result of the mechanical advantage of the lever acting on the γ-subunit as a cam shaft. This conclusion is supported by experiments that torque is decreased when the lever is shortened (14). In this regard, the spike in angular velocity at ∼93° from the catalytic dwell presented here is consistent with forces of the lever acting against the equivalent of bovine γ-subunit residues 232–238 (Fig. 5 B and C) based on the simulation (19). This relationship can be tested experimentally by determining the effects of mutations that substitute less bulky side chains at these positions.
The angular velocity profile presented here also provides evidence that associates the F1 crystal structures of Kabaleeswaren et al. (23) and Menz et al. (4) to the protein conformations at γ-subunit rotation angles of 35° and 120°, respectively, from the catalytic dwell because they were used as reference structures for these rotary positions in the Pu and Karplus simulation (19). The use of the Abrahams et al. (5) structure in lieu of that of Kabaleeswaren et al. (23) led to a γ-subunit rotation of −10° in the simulation (19), which is consistent with the differences in rotary positions of the γ-subunit foot domains between these structures. Two different rotational positions of the γ-subunit foot domain were resolved in Kabaleeswaren et al. (23). Specifically, the structure known by PDB ID code 2HLD1 (Fig. 5B) in which the foot domain is rotated 29° relative to that of Menz et al. (4) (Fig. 5C) was used in the in the Pu and Karplus (19) simulation.
The data of Fig. 4 C and D show that ATP can bind to the empty site of F1 over a ∼40° span of γ-subunit rotation starting from within a few degrees of rotation from the catalytic dwell (phase 1). This suggests that the energy derived from substrate binding is used to drive γ-subunit rotation from the initial rotational binding position onward such that the angular velocity at any subsequent angle is largely determined by the mechanical efficiency of the lever acting on the coiled-coil at that point. The increase in affinity for ATP as a function of rotary position during this same ∼40° span of γ-subunit rotation (27) is also likely to contribute to the increase in angular velocity during the power stroke. It is noteworthy that the affinities for ATP and ADP at the catalytic sites (8, 9) determined by tryptophan fluorescence quenching (Table S1) also correlate well to the affinities measured as a function of rotation angle (27).
Rotation of the γ-subunit from the end of the catalytic dwell before the binding of ATP is summarized in Fig. 6A. During the catalytic dwell ATP hydrolysis is believed to occur at the βD-catalytic site and subsequent release of Pi from the βE-site ends the dwell and initiates rotation (3). In most F1 crystal structures the βD-lever is in the down position and is partially beneath the foot domain of the γ-subunit such that the lever seems poised to push up against the foot in a cam-type manner as this catalytic site adopts a more open conformation consistent with empty catalytic sites. Based on the assumption of 100% efficiency, it was proposed that the γ-subunit rotation from the catalytic dwell to the point of substrate binding was powered by the release of elastic energy stored from ATP binding before the catalytic dwell (13). Because the data presented here clearly show that the enzyme is not 100% efficient, elastic energy storage is no longer a requirement of this mechanism. Our data do not eliminate the possibility that stored elastic energy contributes to the initial rotation from the catalytic dwell. Alternatively, simulations show that ATP hydrolysis at the βD-site is sufficient to raise the lever to a half-open (HO) position in a manner that can power this initial rotation (19). A comparable βHO conformation has been observed in the catalytic site with bound ADP of the F1 structure of Menz et al. (4). Thus, when ATP binds to the empty site early in the power stroke, the observed angular velocity is anticipated to have contributions from that initiated at the end of the catalytic dwell as well as that resulting from ATP binding.
The data of Figs. 3B and 4B show that elevated ADP results in an increase in dwells at about the same position as ATP-binding dwells, as observed previously (3). The increase in these dwells (the primary ADP effect) is explained by competitive product inhibition by ADP versus ATP for binding to the βE-catalytic site where ATP hydrolysis at the βD-site rotates the γ-subunit by ∼35° to form the DOH conformation (Fig. 6C). Once ATP is able to displace ADP from the βE-site, the βE-lever closes, applying force to the coiled-coil to complete the power stroke.
The data presented here also show that increased ADP concentrations have a secondary effect that decreases the phase-2 angular velocity between 85° and 120° from the catalytic dwell, as summarized in Fig. 6D. This indicates that ADP can remain bound subsequent to the ATP-binding dwell consistent with a three-site mechanism but decreases the enzyme efficiency in so doing. To explain the decrease in angular velocity observed between 85° and 120°, we propose that elevated ADP concentrations also decrease the rate of dissociation of ADP from the βD-site such that it remains in the HO conformation. In this conformation, the lever provides steric hindrance to the rotating coiled-coil, slowing rotation. The nucleotide occupancy under these conditions is then similar to several three-site mechanisms (10–12), and is also consistent with the nucleotide occupancy and conformation of the Menz et al. (4) F1 structure that coincides with the fit of the torque simulation and the angular velocity as rotation approaches 120° (Fig. 5C). In support of this mechanism, Adachi et al. (27) recently showed that there is no angular dependence for dissociation of ADP. It is noteworthy that dissociation of ADP is also thought to contribute some of the energy for rotation (3, 28). At low ADP concentrations, ADP dissociation can occur rapidly enough to be concurrent with ATP binding, as proposed by Adachi et al. (3). Under these conditions, the βD-lever has opened in time so as not to interfere with the angular velocity of the γ-subunit (Fig. 6B).
Methods
Purification of F1 from E. coli was accomplished as described previously (29) and stored at 0.1 mg/mL at −80 °C before use.
Single-molecule rotation assays were performed by attaching the biotinylated F1-ATPase to a coverslip, followed by assembly of the avidin-coated nanorod to the biotin on the γ-subunit as described (25). Light scattered from a rotating nanorod for single-molecule experiments was collected at 200 kHz for analysis using a single-photon counting avalanche photodiode as described by Ishmukhametov et al. (30).
The changes in scattered light intensities resulting from F1ATPase-dependent nanorod rotation were analyzed by custom software written in MatLab 6.5. An initial program established the minimum and maximum intensity values for each dataset and selected power stroke events in which the intensity changed from within 5% of the minimum at the end of a catalytic dwell to 5% of the maximum values (1). This aligned the power strokes in a manner that minimized the phase shift, ϕ, when averaging the data from many power strokes. This also set the same minimum and maximum intensity values, I(t), for any one molecule so the normalized value satisfies 0 ≤ I(t) ≤ 1, which is needed to take note of the relation between the rotational position, θ(t) and the intensity I(t). If ϕ = 0, then θ = 0 when I = 0. We let t0 be such that I(t0) = 0 is the first time at which the intensity is zero, then θ(t0) = −ϕ(t0). Moreover, we let tmax be the first time at which I(t0) = Imax, that is, I is increasing for t0 < t < tmax, with 0 < I(t) < 1. Because the scattered light intensity varies in a sinusoidal manner of the rotational position of the nanorod relative to the direction of polarization (1, 31), the rotational position during the power stroke was determined from
which is valid for the first 90° of the 120° power stroke (−π/2 < θ + ϕ < π/2). To obtain the rotational position from the scattered light intensity for the final 30° of the power stroke, we used
The calculated rotational position versus time for each power stroke from Eqs. 1 and 2 was used to determine the average angular velocity and the occurrence of pauses as a function of rotational position.
The angular velocity versus rotational position of each power stroke was determined by the slope of neighboring data points plotted versus time. Average velocities of power strokes were calculated for every 3° of rotational position from the end of the catalytic dwell.
The pauses within each power stroke were identified where the change in rotational position was less than 3° for a minimum of three data points (0.015 ms), and the pause duration was determined. The number of dwells for all power strokes examined for a given condition were binned according to their rotational position and duration and normalized by dividing each bin by the total number of power strokes.
Supplementary Material
Acknowledgments
We thank Dr. Rosemary Renaut for insightful discussions. This project was supported by National Institutes of Health Grant R01GM097510 (to W.D.F.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317784111/-/DCSupplemental.
References
- 1.Spetzler D, et al. Microsecond time scale rotation measurements of single F1-ATPase molecules. Biochemistry. 2006;45(10):3117–3124. doi: 10.1021/bi052363n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr, Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410(6831):898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 3.Adachi K, et al. Coupling of rotation and catalysis in F(1)-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130(2):309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: Implications for the mechanism of rotary catalysis. Cell. 2001;106(3):331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370(6491):621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Kayalar C, Rosing J, Boyer PD. An alternating site sequence for oxidative phosphorylation suggested by measurement of substrate binding patterns and exchange reaction inhibitions. J Biol Chem. 1977;252(8):2486–2491. [PubMed] [Google Scholar]
- 7.Boyer PD. Catalytic site occupancy during ATP synthase catalysis. FEBS Lett. 2002;512(1–3):29–32. doi: 10.1016/s0014-5793(02)02293-7. [DOI] [PubMed] [Google Scholar]
- 8.Senior AE, Nadanaciva S, Weber J. The molecular mechanism of ATP synthesis by F1Fo-ATP synthase. Biochim Biophys Acta. 2002;1553(3):188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 9.Weber J, Senior AE. Bi-site catalysis in F1-ATPase: Does it exist? J Biol Chem. 2001;276(38):35422–35428. doi: 10.1074/jbc.M104946200. [DOI] [PubMed] [Google Scholar]
- 10.Ariga T, Muneyuki E, Yoshida M. F1-ATPase rotates by an asymmetric, sequential mechanism using all three catalytic subunits. Nat Struct Mol Biol. 2007;14(9):841–846. doi: 10.1038/nsmb1296. [DOI] [PubMed] [Google Scholar]
- 11.Mao HZ, Weber J. Identification of the betaTP site in the x-ray structure of F1-ATPase as the high-affinity catalytic site. Proc Natl Acad Sci USA. 2007;104(47):18478–18483. doi: 10.1073/pnas.0709322104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sielaff H, Rennekamp H, Engelbrecht S, Junge W. Functional halt positions of rotary FOF1-ATPase correlated with crystal structures. Biophys J. 2008;95(10):4979–4987. doi: 10.1529/biophysj.108.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oster G, Wang H. Reverse engineering a protein: The mechanochemistry of ATP synthase. Biochim Biophys Acta. 2000;1458(2-3):482–510. doi: 10.1016/s0005-2728(00)00096-7. [DOI] [PubMed] [Google Scholar]
- 14.Usukura E, et al. Torque generation and utilization in motor enzyme FoF1-ATP synthase: Half-torque F1 with short-sized pushrod helix and reduced ATP synthesis by half-torque FoF1. J Biol Chem. 2012;287(3):1884–1891. doi: 10.1074/jbc.M111.305938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasuda R, Noji H, Kinosita K, Jr, Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell. 1998;93(7):1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 16.Dittrich M, Hayashi S, Schulten K. ATP hydrolysis in the betaTP and betaDP catalytic sites of F1-ATPase. Biophys J. 2004;87(5):2954–2967. doi: 10.1529/biophysj.104.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee S, Warshel A. Electrostatic origin of the mechanochemical rotary mechanism and the catalytic dwell of F1-ATPase. Proc Natl Acad Sci USA. 2011;108(51):20550–20555. doi: 10.1073/pnas.1117024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga N, Takada S. Folding-based molecular simulations reveal mechanisms of the rotary motor F1-ATPase. Proc Natl Acad Sci USA. 2006;103(14):5367–5372. doi: 10.1073/pnas.0509642103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pu JZ, Karplus M. How subunit coupling produces the gamma-subunit rotary motion in F1-ATPase. Proc Natl Acad Sci USA. 2008;105(4):1192–1197. doi: 10.1073/pnas.0708746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pänke O, Cherepanov DA, Gumbiowski K, Engelbrecht S, Junge W. Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: Angular torque profile of the enzyme. Biophys J. 2001;81(3):1220–1233. doi: 10.1016/S0006-3495(01)75780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary F(O)F(1)-ATPase. Nature. 2009;459(7245):364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 22.Hornung T, Ishmukhametov R, Spetzler D, Martin J, Frasch WD. Determination of torque generation from the power stroke of Escherichia coli F1-ATPase. Biochim Biophys Acta. 2008;1777(7-8):579–582. doi: 10.1016/j.bbabio.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabaleeswaran V, Puri N, Walker JE, Leslie AG, Mueller DM. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 2006;25(22):5433–5442. doi: 10.1038/sj.emboj.7601410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornung T, Martin J, Spetzler D, Ishmukhametov R, Frasch WD. Microsecond resolution of single-molecule rotation catalyzed by molecular motors. Methods Mol Biol. 2011;778:273–289. doi: 10.1007/978-1-61779-261-8_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spetzler D, et al. Single molecule measurements of F1-ATPase reveal an interdependence between the power stroke and the dwell duration. Biochemistry. 2009;48(33):7979–7985. doi: 10.1021/bi9008215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara KY, et al. The role of the DELSEED motif of the beta subunit in rotation of F1-ATPase. J Biol Chem. 2000;275(19):14260–14263. doi: 10.1074/jbc.275.19.14260. [DOI] [PubMed] [Google Scholar]
- 27.Adachi K, Oiwa K, Yoshida M, Nishizaka T, Kinosita K., Jr Controlled rotation of the F₁-ATPase reveals differential and continuous binding changes for ATP synthesis. Nat Commun. 2012;3:1022. doi: 10.1038/ncomms2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao YQ, Yang W, Marcus RA, Karplus M. A model for the cooperative free energy transduction and kinetics of ATP hydrolysis by F1-ATPase. Proc Natl Acad Sci USA. 2003;100(20):11339–11344. doi: 10.1073/pnas.1334188100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.York J, et al. Abundance of Escherichia coli F1-ATPase molecules observed to rotate via single-molecule microscopy with gold nanorod probes. J Bioenerg Biomembr. 2007;39(5-6):435–439. doi: 10.1007/s10863-007-9114-x. [DOI] [PubMed] [Google Scholar]
- 30.Ishmukhametov R, Hornung T, Spetzler D, Frasch WD. Direct observation of stepped proteolipid ring rotation in E. coli FoF₁-ATP synthase. EMBO J. 2010;29(23):3911–3923. doi: 10.1038/emboj.2010.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sönnichsen C, Alivisatos AP. Gold nanorods as novel nonbleaching plasmon-based orientation sensors for polarized single-particle microscopy. Nano Lett. 2005;5(2):301–304. doi: 10.1021/nl048089k. [DOI] [PubMed] [Google Scholar]
- 32.Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 Å resolution. Nat Struct Biol. 2000;7(11):1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.