Significance
Noncommunicable diseases, including cardiovascular disease and cancer, are growing worldwide but are challenging to diagnose because biomarkers that can accurately detect them in patients are lacking. Here, we designed nanoscale agents that are administered to reveal the presence of diseased tissues by producing a biomarker in the urine that can be detected using paper strips similar to a home pregnancy test. Using mouse models, we show that we can detect diseases as diverse as solid cancer and blood clots using only a single injection of our diagnostic followed by urine analysis on paper. This platform does not require expensive instruments, invasive procedures, or trained medical personnel, and may allow low-cost diagnosis of diseases at the point of care in resource-limited settings.
Keywords: protease nanosensor, urine biomarker, image-free diagnostic, engineered disease reporter, global health
Abstract
With noncommunicable diseases (NCDs) now constituting the majority of global mortality, there is a growing need for low-cost, noninvasive methods to diagnose and treat this class of diseases, especially in resource-limited settings. Molecular biomarkers combined with low-cost point-of-care assays constitute a potential solution for diagnosing NCDs, but the dearth of naturally occurring, predictive markers limits this approach. Here, we describe the design of exogenous agents that serve as synthetic biomarkers for NCDs by producing urinary signals that can be quantified by a companion paper test. These synthetic biomarkers are composed of nanoparticles conjugated to ligand-encoded reporters via protease-sensitive peptide substrates. Upon delivery, the nanoparticles passively target diseased sites, such as solid tumors or blood clots, where up-regulated proteases cleave the peptide substrates and release reporters that are cleared into urine. The reporters are engineered for detection by sandwich immunoassays, and we demonstrate their quantification directly from unmodified urine; furthermore, capture antibody specificity allows the probes to be multiplexed in vivo and quantified simultaneously by ELISA or paper lateral flow assay (LFA). We tailor synthetic biomarkers specific to colorectal cancer, a representative solid tumor, and thrombosis, a common cardiovascular disorder, and demonstrate urinary detection of these diseases in mouse models by paper diagnostic. Together, the LFA and injectable synthetic biomarkers, which could be tailored for multiple diseases, form a generalized diagnostic platform for NCDs that can be applied in almost any setting without expensive equipment or trained medical personnel.
In the last several decades, global health challenges have dramatically shifted, with substantial reductions in the burden of infectious diseases (e.g., HIV, tuberculosis, and malaria) and simultaneous growth in the prevalence of noncommunicable diseases (NCDs) such as stroke, heart disease, and cancer, which constitute an increasing majority of global mortality (1, 2). Strikingly, NCDs disproportionately affect the developing world: low- and middle-income countries bear nearly 80% of the world’s NCD burden (3), and cardiovascular diseases and cancer have been the first and second, respectively, highest causes of mortality in the developing world since 2001 (4). Diagnosing NCDs in remote and/or poor settings is difficult without access to costly imaging modalities [e.g., computed tomography (CT)/MRI], well-equipped clinical laboratories (e.g., for histopathology), and trained medical personnel. Consequently, developing diagnostics for NCDs that are cost effective and can be easily implemented remains an important goal in global health. One promising approach is to detect disease biomarkers from readily accessible bodily fluids with point-of-care (POC) devices that are inexpensive, noninvasive, and do not require trained medical personnel. Despite widespread interest, the lack of predictive, validated biomarkers significantly limits the types of NCDs that can be detected at the POC (5–7).
Rather than searching for endogenous biomarkers, a promising strategy is to engineer exogenous agents that can specifically probe for the presence of diseased tissue. Radiolabeled glucose is an example of a common exogenous agent used with PET to reveal the location of metabolically active tumors. Inspired by engineered approaches, our group recently outlined a framework whereby protease-sensitive nanoparticles (NPs) called “synthetic biomarkers” are administered to detect NCDs including liver fibrosis, cancer, and thrombosis noninvasively (8, 9). These peptide-coated NPs probe diseased sites and, in response to cleavage by local dysregulated proteases, release mass-encoded reporters that then filter into the urine for analysis by mass spectrometry. In practice, administering NPs and collecting urine samples are both well-suited for the POC, but the need for a mass spectrometer to analyze the urinary cleavage fragments limits the utility of mass-encoded synthetic biomarkers for global health applications.
To address these difficulties, we reformulated our synthetic biomarkers to release ligand-encoded reporters designed for detection by a companion POC diagnostic comprised of paper test strips, a well-established technology used to screen and monitor diseases with readily available biomarkers (10, 11). The benefits of paper testing include low cost, rapid diagnosis, and no need for complex equipment or technical expertise (11, 12). Paper tests operate by wicking a biological specimen (commonly urine, saliva, or blood) containing a target analyte to regions where subsequent chemical or antibody-mediated analyses are detected by direct observation (13) or inexpensive and accessible quantitative imaging (14–16). Recently, paper diagnostics have been developed for quantitative assays like monitoring transaminases released by liver damage (13) or detecting infectious diseases like HIV-1, malaria, or trichinellosis (17). Unfortunately, the scarcity of naturally occurring biomarkers has limited use of paper diagnostics for NCDs. Consequently, combining paper diagnostics with synthetic biomarkers that sensitively and specifically indicate disease from the urine may provide a simple and low-cost method to diagnose NCDs in resource-limited settings.
To pursue the goal of developing affordable POC tests for NCDs, we engineer synthetic biomarkers to detect thrombosis and colorectal cancer (CRC) from the urine by custom lateral flow assay (LFA), a variant of paper tests. Thrombosis, the formation of obstructive blood clots, occurs in many cardiovascular-associated disorders (e.g., stroke and heart attack) and is characterized by the activation of the plasma protease thrombin that mediates fibrin clot formation. In CRC and most solid cancers, tumors produce matrix metalloproteinases (MMPs) to facilitate growth, angiogenesis, and metastatic spread (18). To detect these diseases, we develop thrombin- and MMP-sensitive NPs by conjugating substrate–reporter tandem peptides to the surface of NPs (Fig. 1A). When administered, these NPs probe diseased tissues (blood clots or tumors) where local up-regulated proteases (thrombin or MMPs, respectively) cleave their surface coat of peptides, releasing reporters that are concentrated into the urine. The urinary reporters are functionalized with structurally distinct ligands for capture onto paper test strips adsorbed with ligand-binding antibodies (Fig. 1B). Beyond thrombosis and CRC, this approach may be amenable to many noncommunicable and infectious diseases in which aberrant protease activities are implicated.
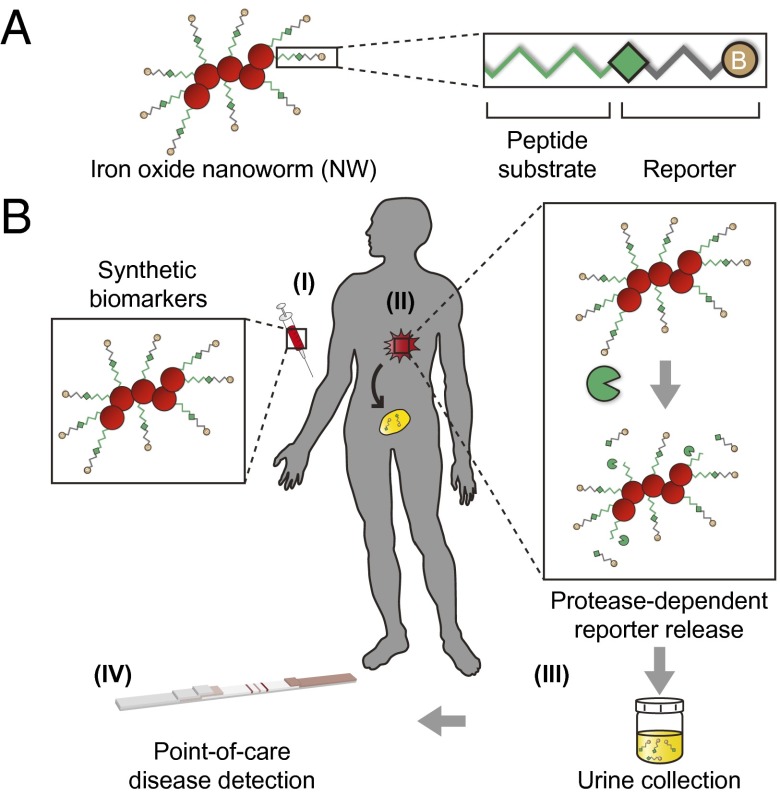
Fig. 1.
Protease-sensitive NPs for POC urinary monitoring of disease. (A) Synthetic biomarkers were synthesized by conjugating substrate–reporter tandem peptides to carrier iron oxide NWs. Proteolytic cleavage of the linking peptide substrate liberates ligand-encoded reporters that filter into urine. (B) (I) A patient suspected of harboring a disease receives a disease-tuned diagnostic nanoworm (NW) mixture. (II) NWs infiltrate the disease site and release reporters upon proteolytic cleavage of peptide substrates. Although intact NWs are too large to pass the glomerular basement membrane, liberated reporters passively filter through the kidney. (III) The patient collects a urine sample. (IV) Application of unprocessed urine to a low-cost POC paper lateral flow assay (LFA) enables diagnosis.
Results
Protease-Sensitive NPs for Urinary Monitoring of Disease.
To develop synthetic biomarkers for thrombosis and cancer, we first sought to design NPs for sensing the activity of the proteases thrombin and matrix metalloproteinase 9 (MMP9). We functionalized poly(ethylene glycol)-coated iron oxide nanoworms (NWs)—a long-circulating NP formulation previously characterized by collaborators and our laboratory (19, 20)—with fluorescein-labeled derivatives of thrombin- and MMP9-cleavable substrates [PLGLRSW and PLGVRGK, respectively (8)] at a surface valency of 20–30 peptides per NW to induce intermolecular quenching (Fig. 1A). To test the efficiency of peptidolysis, we incubated thrombin-sensitive NWs with thrombin and observed a rapid increase in sample fluorescence as cleaved peptide fragments released into solution fluoresced freely. By contrast, no increase in fluorescence was observed in the presence of Argatroban, a direct thrombin inhibitor, or when the substrate was synthesized with protease-resistant d-stereoisomers (Fig. 2A), indicating that thrombin activity was required to activate the NWs. We observed similar increases in sample fluorescence when MMP-sensitive NWs were incubated with MMP9 and no activity when the broad-spectrum MMP inhibitor Marimastat or d-isomer substrates were used (Fig. 2B). Together, these findings showed that peptides on the surface of NWs can be efficiently cleaved by thrombin or MMP9.
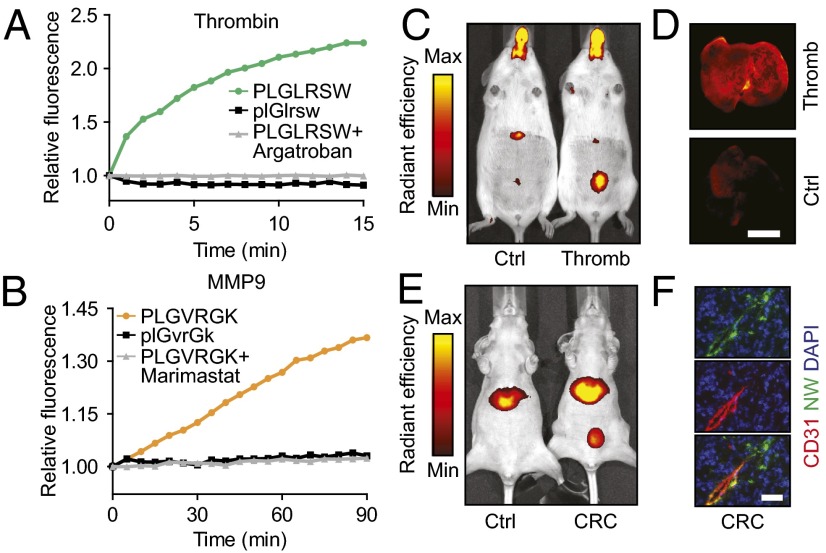
Fig. 2.
Nanoparticle protease sensitivity and animal models. (A and B) Fluorescein-functionalized thrombin- or MMP-sensitive (A and B, respectively) NWs were incubated with target enzyme. Proteolytic release of quenched reporters increased fluorescence. Addition of thrombin inhibitor Argatroban (A) or MMP inhibitor Marimastat (B) abrogated fluorescent increase, as did use of d-isomer substrates. (C and D) Induction of thrombosis increased bladder (C) and lung (D) localization of near-infrared fluorophore-labeled thrombin-sensitive NWs over controls. (Scale bar: 5 mm.) (E) Intravenous injection of near-infrared fluorophore-labeled MMP-sensitive NWs resulted in increased bladder localization of reporters in tumor-bearing mice compared with controls. (F) NW mixture administration showed colocalization of blood vessels (CD31; red) and NWs/reporters (fluorescein; green) and significant NW/reporter tumor infiltration. Nuclei were DAPI counterstained (blue). (Scale bar: 50 µm.)
We previously showed that synthetic biomarkers composed of long-circulating NWs accumulate in diseased tissue by diffusing across fenestrated vessels such as in liver fibrosis and cancer (8), or when the sites of disease are intravascular as in thrombosis, sense protease activity while in systemic circulation (9). These studies also showed that free peptides administered i.v. are rapidly cleared by renal filtration, but their conjugation to NWs makes urinary clearance conditional upon cleavage by disease-specific proteases. To confirm that our synthetic biomarkers tailored for thrombin and MMP9 exhibit similar pharmacokinetics, we synthesized NWs with substrates labeled with carrier peptide-linked near-infrared fluorophores to monitor peptide traffic and cleavage by in vivo fluorescence imaging. To promote renal clearance and to enable in vivo fluorescent visualization of the peptide-fluorophore reporter released by substrate proteolysis, we conjugated the near-infrared fluorophore VT750 (N-terminal) to the peptide glutamate-fibrinopeptide B (GluFib) (sequence eGvndneeGffsar), which we synthesized with d-amino acids (lowercase) to confer stability against protease activity (8, 9, 21).
We chose a murine model of thrombosis in which the onset of clotting is controlled by the i.v. administration of collagen and epinephrine to activate platelets and thrombin, forming blood clots that embolize to the lungs (22). Consistent with our previous findings, coadministration of NWs to mice challenged with collagen and epinephrine resulted in a pronounced increase in their urinary and lung fluorescence relative to healthy controls (Fig. 2 C and D), indicating in vivo cleavage and renal clearance of peptides. To apply to CRC, we infused MMP9-sensitive NWs into nude mice bearing s.c. human colorectal tumors (LS174T), formed by a cell line that secretes MMP9 (23), and observed similar increases in fluorescence localized to the bladder (Fig. 2E). Immunofluorescent staining of tumor sections confirmed NW (green) extravasation from the vasculature (red) into the tumor interstitium (Fig. 2F and Fig. S1). Collectively, these results verified the ability of our synthetic biomarkers to probe disease sites and release cleaved peptide fragments into the host urine.
Detecting Ligand-Encoded Reporters by Sandwich Complexes.
We next sought to design a panel of ligand-encoded reporters that can be detected by protein-based sandwich complexes (Fig. 3A). The formation of a sandwich complex requires a target antigen to express two distinct epitopes that bind separately to a capture and detection agent; thus to construct the synthetic heterobifunctional reporter R1, we conjugated the ligands fluorescein (FAM; capture) and biotin (detection) to the opposing termini of the same d-stereoisomer GluFib as previously (Fig. 3A and Fig. S2F). Critically, GluFib functions as a molecular spacer, allowing FAM and biotin to bind freely to their cognate proteins α-FAM antibody (α-R1) and streptavidin, respectively, and, as before, promotes clearance of the reporter as GluFib is biologically inert and efficiently filtered by the kidneys (8, 9, 21). We first determined whether urine samples spiked with R1 could be detected by sandwich ELISA, a standard assay used in clinical laboratories. Serial dilutions of R1 were applied to a 96-well plate coated with α-R1 antibodies to immobilize R1 before NeutrAvidin-horseradish peroxidase (NA-HRP) was added to catalyze the development of the chromogenic substrate 3,3′,5,5′-tetramethylbenzidine (TMB), revealing a linear dose dependence from ∼6 pM down to the limit of detection (LOD) of ∼0.1 pM (Fig. 3B and Fig. S2E). This LOD compared favorably with the sensitivity at which naturally occurring biomarkers such as prostate-specific antigen (24) can be detected by ELISA, establishing our ability to design synthetic sandwich assays by harnessing ligand–protein interactions.
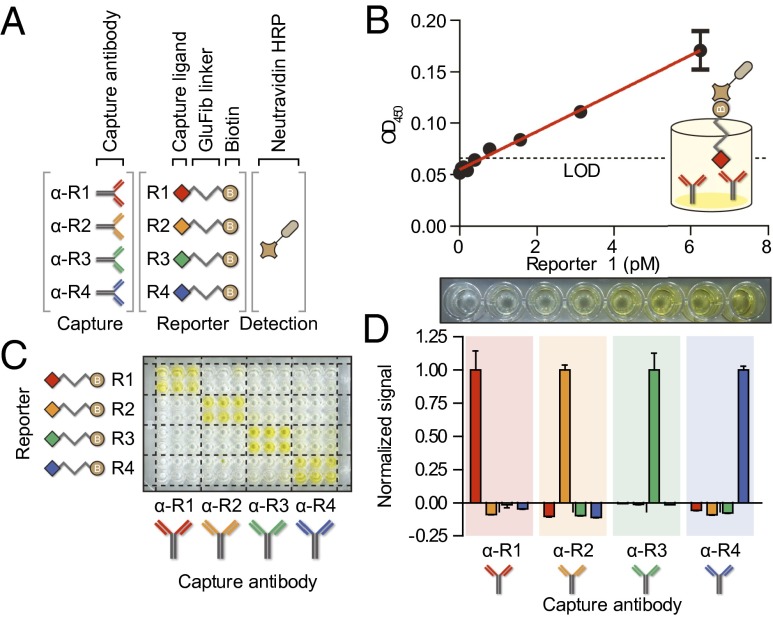
Fig. 3.
Detecting ligand-encoded reporters by sandwich complexes. (A) Reporters R1–R4 consist of a nondegradable spacer, GluFib, conjugated to a specific capture ligand on one terminus and a common detection ligand, biotin, on the other. Each reporter is captured by a paired antibody (α-R1–α-R4), and all are detected by NA-HRP. (B) Serial dilutions of R1 spiked in urine were captured on an α-R1-coated plate. Oxidation of chromogenic substrate by HRP (Lower) resulted in a linear detection range of ∼0.1–6 pM (Upper, linear fit R2 = 0.95). (C) A 96-well plate coated with four different capture antibodies visually demonstrated the ability of each capture antibody to specifically bind only its cognate reporter. (D) Quantification of marker specificity demonstrated that noise generated by other probes was below the LOD of the correct signal. Error bars are SEM.
To generalize our approach and construct a multiplexed library of reporters, we synthesized additional heterobifunctional derivatives of GluFib by pairing the capture ligands dinitrophenyl (DNP), tetramethylrhodamine (TMR), and Alexa Fluor 488 (AF488) with biotin to create reporters R2–R4, respectively (Fig. 3A and Fig. S2F). Similar to fluorescein, these ligands were selected because they are stable, well tolerated by living organisms, and do not elicit immune responses unless coupled to a potent immunogen (25). We developed independent sandwich ELISAs by using the antibodies α-DNP (α-R2), α-TMR (α-R3), or α-AF488 (α-R4) to capture R2, R3, or R4, respectively, and identified the LOD and working linear ranges for each, which were also within the typical values expected for sandwich ELISAs (Fig. S2 A–E). Assaying for specificity revealed an increase in sample intensity in wells that contained matched antibody–ligand pairs (Fig. 3C), whereas cross-reactivity in nonmatched wells was below the LOD of the cognate detection signal (Fig. 3D). These results showed that ligand-encoded reporters can be sensitively and specifically detected in urine by sandwich ELISA.
Paper Assay Development and Detection of Protease Activity.
First developed more than two decades ago to detect human chorionic gonadotropin as a home pregnancy test, paper-based LFAs have since been expanded for use in diverse settings to detect pathogens, drugs, hormones, and metabolites (17). LFAs detect antigens by a sandwich complex in which capture antibodies are adsorbed onto a highly porous test strip, such as a nitrocellulose membrane, which serves to wick fluids and transport analytes from the sample pad to the capture regions (Fig. 4A). The immobilized analytes are then visualized by a detection agent coupled to NPs (typically gold or latex nanospheres) that create a colored line detectable by eye without enzymatic amplification.
Fig. 4.
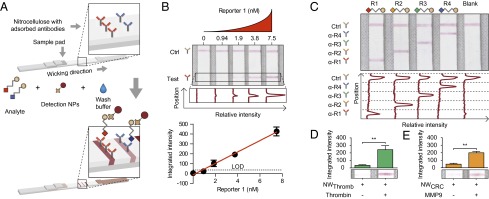
Paper assay development and detection of protease activity. (A) Capture antibodies were adsorbed in spatially multiplexed lines on nitrocellulose membrane. The LFAs were developed by application of analyte, wash buffer, and streptavidin-gold detection NPs that wick past the capture antibodies and develop lines. LFAs may be scanned and reporter concentration is proportional to band intensity. (B) Paper LFAs demonstrated a linear increase in band intensity with reporters diluted in urine by eye (Top) and automated image analysis (Middle), resulting in a linear detection region for R1 of ∼1–7 nM (Bottom, linear fit R2 = 0.99). (C) Spatially multiplexed detection antibodies demonstrated specific detection of each of the four reporters and a test control line. (D) LFA detection of R3 released by incubation of thrombin-sensitive NWs with thrombin demonstrated darker bands visually (Lower) and by image analysis (Upper, P = 0.0022) in the presence of enzyme than without enzyme. (E) Increased R2 was detected by LFA upon incubation of MMP-sensitive NWs with MMP9 than without enzyme visually (Lower) and by image analysis (Upper, P = 0.0022). Error bars are SEM.
Here, we sought to determine whether ligand-encoded reporters could be detected on paper. Using a low-volume robotic liquid handler, we deposited α-R1 and α-streptavidin antibodies to create test and control lines, respectively, on nitrocellulose paper strips. Unprocessed mouse urine samples spiked with R1 were then applied to the sample pads followed by a solution containing gold NP-conjugated streptavidin. Colored lines appeared where the test antibodies were printed, indicating R1 capture from urine and detection as a sandwich complex (Fig. 4B). Quantitative scans of LFAs used to analyze serial dilutions of R1 revealed a LOD of ∼1 nM and a working linear range of ∼1–7 nM (Fig. 4B and Fig. S3E). Similar performance metrics were observed for separate LFAs customized for the remaining reporters (Fig. S3 A–E).
To enable multiplex reporter detection, we printed capture antibodies into four parallel test lines relative to a control line and analyzed urine samples that contained one of the four reporters. Similar to the ELISA results, only the test lines printed with the cognate capture antibody developed a positive signal (Fig. 4C), highlighting the LFA’s specificity and the capacity to detect distinct reporters with single spatially encoded paper strips.
To detect protease activity by LFAs, thrombin-sensitive substrates were conjugated in tandem with R3 onto NWs. Following in vitro substrate cleavage by thrombin, we collected the peptide fragments by size-exclusion filtration. Cleaved R3 was readily detected from the filtrate by LFA, developing into significantly darker test lines compared with control samples not exposed to thrombin (Fig. 4D, P = 0.0022). Similar results were obtained when filtrate collected after incubation of R2-encoded MMP-sensitive NWs with MMP9 was analyzed by LFA (Fig. 4E, P = 0.0022). Together, these results demonstrated that the activity of distinct proteases can be detected by paper-based LFAs.
Disease Detection on Paper with Synthetic Urinary Biomarkers.
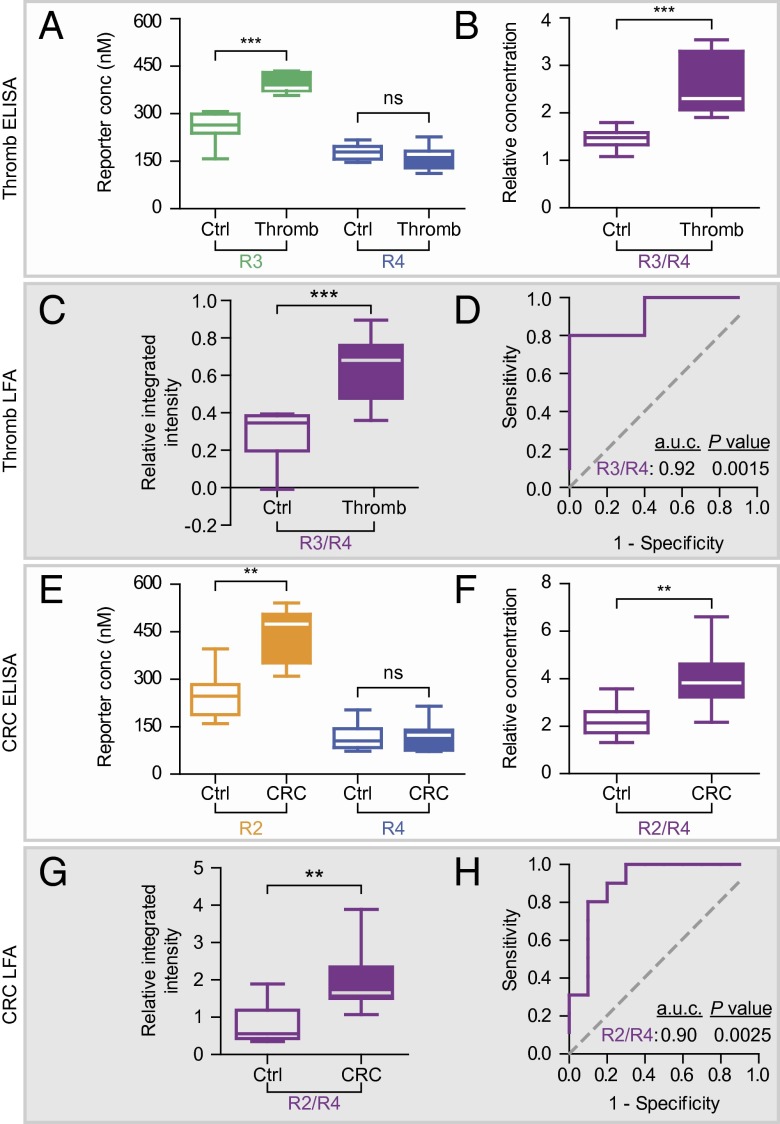
Urine concentration is dependent on many host and environmental factors (e.g., diet, activity level, circadian rhythm, medical history); therefore, we sought to develop a normalization strategy for our test. We hypothesized that coadministered free reporters would pass into the urine independent of disease state and could be used to normalize the level of reporters released by protease activity. To investigate this approach, we infused a mixture of free R4 and thrombin-sensitive NWs (labeled with R3) into healthy or thrombotic cohorts of mice and collected all urine for 30 min postinjection. As anticipated, urinary concentrations of R4 were statistically equivalent between the two groups by ELISA, indicating unbiased clearance of the free reporter (Fig. 5A, Right, P = 0.25). By contrast, urinary levels of R3, the reporter of thrombin activity, significantly increased in mice harboring thrombi when quantified independently (Fig. 5A, Left, P < 10−4) or when normalized against R4 (Fig. 5B, P < 10−4). Using a paper strip printed with multiple capture antibodies, we analyzed the urinary levels of R3 and R4 simultaneously (Fig. S4 C and D) and similarly observed a statistically significant increase in the ratio of R3/R4 in diseased urine samples compared with healthy controls (Fig. 5C, P = 0.0015). To determine the diagnostic accuracy of the assay, we analyzed the rate of true positives (sensitivity) and false positives (one-specificity) by receiver-operating characteristic (ROC) curves and found that the multiplexed paper test discriminated urine from thrombotic versus control mice accurately, with an area under the curve (a.u.c.) of 0.92 (Fig. 5D, P = 0.0015).
Fig. 5.
Paper-based disease detection using synthetic urinary biomarkers. (A) Urine was collected from mice (n = 10) coinjected with R3-encoded thrombin-sensitive NWs, free R4, and either PBS or collagen/epinephrine (to induce thrombosis). By ELISA, urinary clearance of free reporter R4 was not different between control and induced animals (Right, P = 0.25), but liberated R3 was significantly increased in animals that underwent thrombosis (Left, P < 0.0001). (B) Normalization of proteolytically liberated reporter to free reporter (R3/R4) was significantly increased in diseased animals (P < 0.0001). (C) LFAs also detected significantly increased R3/R4 in urine from diseased mice from the same cohort (P < 0.0001). (D) Receiver-operating characteristic (ROC) classification by LFA detection of R3/R4 discriminated healthy from diseased mice with an a.u.c. of 0.92 (P = 0.0015 from random classifier). (E) Control or tumor-bearing mice (n = 10) were administered free R4 and R2-encoded MMP-sensitive NWs. By ELISA, R4 clearance was not affected by disease state (Right, P = 0.92) and R2 was significantly higher in CRC-positive mice (Left, P = 0.0039). (F) The liberated to free reporter ratio (R2/R4) was increased in tumor-bearing animals compared with controls (P = 0.0098). (G) LFA analysis of the same urine demonstrated increased R2/R4 ratio in diseased mice (P = 0.0020). (H) LFA quantification of R2/R4 discriminated urine from CRC-positive mice with an a.u.c. of 0.90 (P = 0.0025 from random classifier). Box plots show extremes, quartiles, and median.
To establish the ability to detect solid cancers, we adopted the normalization strategy developed for thrombosis by infusing a solution containing free R4 and R2-encoded MMP-sensitive NPs into nude mice bearing s.c. LS174T colorectal tumors and collecting all urine up to 1 h postinjection. As before, diseased mice cleared R4 with an efficiency statistically equivalent to healthy animals (Fig. 5E, Right, P = 0.92), whereas the urinary concentrations of R2, the reporter of in vivo MMP activity, or its normalized intensity (R2/R4) were both significantly elevated in tumor-bearing mouse urine by ELISA (Fig. 5E, Left, P = 0.0039; Fig. 5F, P = 0.0098). Analysis of the same urine samples by LFA demonstrated a significant increase in the ratio of R2/R4 in urine collected from tumor-bearing but not from control mice (Fig. 5G, P = 0.002). By ROC analysis, this urine test was highly accurate and discriminated CRC with an a.u.c. of 0.90 (Fig. 5H, P = 0.0025). Collectively, these results showed that LFAs can both detect synthetic biomarkers directly from the urine and discriminate NCDs with significant predictive power.
Discussion
In resource-limited environments, POC tests should be simple to operate, built from inexpensive components, and able to detect disease directly from biological fluids. Here, we outlined a strategy whereby NCDs are detected by a single infusion of synthetic biomarkers that release reporters into the urine in the presence of disease. Collected urine samples are then applied to custom LFAs that quantify reporter levels directly on paper without additional sample preparation or expensive instrumentation.
Building on our previous work on mass-encoded NPs, we developed NPs that sense protease activity by releasing rationally designed ligand-encoded reporters after substrate cleavage. We showed that these heterobifunctional reporters mediate the formation of sandwich complexes detectable by standard ELISA and LFA to allow POC testing as an alternative to expensive diagnostics platforms like CT scanners or mass spectrometers, which can cost over 100-fold more than a standard microplate reader (26). Because our ligand-encoded reporters are engineered to work with standard sandwich immunoassays, we anticipate their compatibility with a broad variety of technologies including single-molecule assays (27), nanowire-based techniques (28), and plasmonic ELISA (29). Furthermore, we expect our formulation to be well tolerated by patients, as our NPs are synthesized from Food and Drug Administration-approved materials including polypeptides (e.g., bivalirudin, enfuvirtide), ligands (e.g., fluorescein), and iron oxide NPs which are already in use in humans at gram-scale doses to treat iron deficiency anemia (30–32) (e.g., Feridex, Lumirem). Additionally, these iron oxide nanoparticles have been characterized extensively, exhibiting long circulation half-life (19, 20) and an inability to cross the glomerular basement membrane due to a hydrodynamic radius of ∼60 nm (8, 33). The development of additional NP formulations or methods of administration such as transdermal (34) or inhalation (35) delivery may also provide an alternative to i.v. infusions.
The ability to engineer synthetic biomarkers to produce a detection signal from urine has several advantages over POC tests that detect biomarkers from blood. Endogenous biomarkers are often limited by fundamental biological constraints, making detection challenging, especially at the earliest stages of disease when treatments are more likely curative (36, 37). By contrast, small inert analytes in plasma such as our ligand-encoded reporters are concentrated into the urine: in our animal models, reporters were enriched to levels that required the urine samples to be diluted (fourfold to fivefold for LFA; 102- to 104-fold for ELISA) to prevent signal saturation when NPs were administered at a dose typical of nanomedicines (∼1 mg/kg) (30, 31). In addition, detecting blood biomarkers depends on a blood draw that requires technical expertise unnecessary for urine collection.
In settings beyond the clinical laboratory that may lack critical infrastructure for refrigeration or sample preparation, we developed paper-based LFAs for urinalysis. Rapid diagnostics based on LFAs, like those for HIV, are revolutionizing health care in resource-limited settings by reducing the time for diagnosis from weeks to minutes, enabling clinical decisions to be made in real time at the POC, and significantly reducing costs while maintaining assay quality comparable to hospital tests (38). To date, the majority of LFAs and other POC devices have been developed for infectious disease pathogens, such as HIV and malaria (39). To detect NCDs from the urine, we designed multiplexed LFAs that allowed the level of cleaved reporter to be normalized against an internal standard to account for the hydration state of the host. Use of image analysis for assay quantification rather than visual discrimination of disease status enables improved analysis, repeatability, transmission, and secondary validation of assay results at little increased cost: many have demonstrated effective LFA quantification using low-cost and ubiquitous cell phone cameras (14, 40). Using our paper LFAs, we detected thrombosis and CRC with diagnostic accuracies (a.u.c. of 0.92 and 0.90, respectively) that compared favorably with clinically used blood biomarkers such as prostate-specific antigen for prostate cancer (a.u.c. of 0.68) (41). To further improve disease specificity, we could take advantage of the quantitative and multiplexed nature of our assay and incorporate additional disease-specific protease substrates, as demonstrated previously in our group (8).
Here, we outlined a strategy for designing integrated POC diagnostics—from injectable synthetic biomarkers to companion paper tests for urinalysis—that can be readily implemented in resource-limited settings. With over 500 proteases encoded by the human genome (and many more by pathogens), this approach could be further tailored for additional NCDs (e.g., fibrosis, atherosclerosis, and inflammation) as well as infectious diseases (e.g., malaria, viral hepatitis) to meet the pressing challenge of providing low-cost diagnostics for global health.
Methods
Expanded methods may be found in SI Methods.
Synthesis of Synthetic Biomarkers.
Heterobifunctional ligand-encoded reporters R1–4 were synthesized by derivatizing GluFib with a capture ligand (fluorescein, DNP, TMR, or AF488) on the C terminus and a detection ligand (biotin) on the opposing terminus (Fig. S2F). NWs (∼60 nm) were synthesized from the reaction of iron(III) chloride hexahydrate and iron(II) chloride tetrahydrate with dextran (15–25 kDa) as previously described (19, 20). Aminated NWs were derivatized with N-succinimidyl iodoacetate and reacted with sulfhydryl-terminated protease-sensitive reporters. Reporter valency (∼20–30) was quantified by absorbance or ELISA.
In Vitro Protease Activity Assays.
Fluorescent reporter-bound thrombin- or MMP-sensitive NWs (substrates PLGLRSW or PLGVRGK, respectively) were introduced to recombinant thrombin or MMP9 (respectively). Release of homoquenched fluorophores upon proteolysis was read as increased fluorescence by plate reader at 37 °C. Inhibitors Argatroban or Marimastat were incubated with the protease–NW mixture at 100 µM. To quantify reporter release by LFA, reporter-functionalized NWs were incubated with cognate proteases, passed through a 30-kDa Mr cutoff filter, quantified by LFA, and analyzed by Mann–Whitney test.
In Vivo Imaging.
All animal studies were approved by Massachusetts Institute of Technology’s committee on animal care (protocol 0411-036-14). Thrombin- or MMP-sensitive NWs were functionalized with infrared fluorescent reporter VT750. Bladder and/or lung localization of proteolytically released fluorescent reporter was imaged in control and diseased mice. Thrombosis was induced by coinjection of collagen and epinephrine with synthetic biomarkers in female Swiss Webster mice; colorectal flank tumors were induced by s.c. injection of human cell line LS147T in female NCr nude mice.
ELISA Characterization.
Ninety-six–well plates were adsorbed with capture antibodies and blocked with 1% (wt/vol) bovine serum albumin (BSA) in 1× PBS. Reporter standards were applied and detected by addition of NeutrAvidin-horseradish peroxidase. Oxidation of chromogenic substrate TMB for 1–5 min allowed quantification of reporter concentration. All incubations were 1 h and plates were washed with 1× PBS with 0.5% (wt/vol) Tween 20 between steps. Urine interference was assayed by spiking R1 in 1:100 control mouse urine. Assay specificity was measured by quantifying capture specificity of each antibody to all reporters and normalizing signal to a cognate reporter ladder.
Paper LFA Characterization.
Capture (same as for ELISA) or control (α-streptavidin) antibodies were printed in 2-mm–spaced lines with 50-nL droplets at 0.5-mm pitch onto cellulose ester membrane. Membranes were laminated to a plastic backing with glass fiber conjugate and absorbent pads. The resultant construct was cut into 4-mm strips and stored at 4 °C. Reporters diluted 1:1 in urine were applied to the conjugate pad and flushed with wash buffer [1× PBS with 1% (wt/vol) Tween 80]. Reporters were detected using 40-nm streptavidin-gold nanoparticles. Dried strips were scanned and processed by a custom script that integrated and quantified band intensity.
Collection and Analysis of Urinary Peptides.
Urine was collected from mice i.v. infused with synthetic biomarker mixtures (free R4 plus either R3-functionalized thrombin-sensitive NWs to detect thrombosis or R2-functionalized MMP-sensitive NWs to detect CRC) for 30 or 60 min postinjection (to detect thrombosis or CRC, respectively). Urine collection times were optimized from previous studies using these disease models (8, 9) and are dependent on site of disease and rate of enzymatic substrate cleavage. Reporter concentration in unprocessed urine was assayed by above protocols from urine diluted 1:102 to 104 for ELISA or 1:4–5 for LFA. Data were analyzed using ROC curves (both) and Wilcoxon signed rank test (CRC) or Mann–Whitney test (thrombosis).
Supplementary Material
Acknowledgments
We thank Prof. B. Engelward for use of the robotic liquid handler, Dr. H. Fleming for helpful guidance, and J. Gómez-Márquez for LFA development insights. A.D.W. thanks the National Science Foundation Graduate Research Fellowship Program for support. D.K.W. acknowledges support by the Mazumdar–Shaw International Oncology Fellowship. G.A.K. acknowledges support from National Research Service Award F32CA159496-02 and holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. K.Y.L. acknowledges support from Center of Cancer Nanotechnology Excellence Grant 5 U54 CA151884-03. We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically Richard Cook and the Biopolymers and Proteomics Core. This work was supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. S.N.B. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314651111/-/DCSupplemental.
References
- 1.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanavos P. The rising burden of cancer in the developing world. Ann Oncol. 2006;17(Suppl 8) doi: 10.1093/annonc/mdl983. i15, i23. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization . The Global Burden of Disease. Geneva: World Health Organization; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yach D, Hawkes C, Gould CL, Hofman KJ. The global burden of chronic diseases: Overcoming impediments to prevention and control. JAMA. 2004;291(21):2616–2622. doi: 10.1001/jama.291.21.2616. [DOI] [PubMed] [Google Scholar]
- 5.Gutman S, Kessler LG. The US Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer. 2006;6(7):565–571. doi: 10.1038/nrc1911. [DOI] [PubMed] [Google Scholar]
- 6.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 7.Brooks JD. Translational genomics: The challenge of developing cancer biomarkers. Genome Res. 2012;22(2):183–187. doi: 10.1101/gr.124347.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong GA, et al. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nat Biotechnol. 2013;31(1):63–70. doi: 10.1038/nbt.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin KY, Kwong GA, Warren AD, Wood DK, Bhatia SN. Nanoparticles that sense thrombin activity as synthetic urinary biomarkers of thrombosis. ACS Nano. 2013;7(10):9001–9009. doi: 10.1021/nn403550c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin CD, Linder V, Sia SK. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab Chip. 2007;7(1):41–57. doi: 10.1039/b611455e. [DOI] [PubMed] [Google Scholar]
- 11.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the developing world: Microfluidic paper-based analytical devices. Anal Chem. 2010;82(1):3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 12.Yager P, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442(7101):412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 13.Pollock NR, et al. A paper-based multiplexed transaminase test for low-cost, point-of-care liver function testing. Sci Transl Med. 2012;4(152):ra129. doi: 10.1126/scitranslmed.3003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mudanyali O, et al. Integrated rapid-diagnostic-test reader platform on a cellphone. Lab Chip. 2012;12(15):2678–2686. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H-S, Ko H, Kang M-J, Pyun J-C. Highly sensitive rapid test with chemiluminescent signal bands. BioChip J. 2010;4(2):155–160. [Google Scholar]
- 16.Ellerbee AK, et al. Quantifying colorimetric assays in paper-based microfluidic devices by measuring the transmission of light through paper. Anal Chem. 2009;81(20):8447–8452. doi: 10.1021/ac901307q. [DOI] [PubMed] [Google Scholar]
- 17.Posthuma-Trumpie GA, Korf J, van Amerongen A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal Bioanal Chem. 2009;393(2):569–582. doi: 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- 18.Zucker S, Vacirca J. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev. 2004;23(1-2):101–117. doi: 10.1023/a:1025867130437. [DOI] [PubMed] [Google Scholar]
- 19.Park J-H, et al. Magnetic iron oxide nanoworms for tumor targeting and imaging. Adv Mater. 2008;20(9):1630–1635. doi: 10.1002/adma.200800004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J-H, et al. Systematic surface engineering of magnetic nanoworms for in vivo tumor targeting. Small. 2009;5(6):694–700. doi: 10.1002/smll.200801789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris TA, et al. Urine and plasma levels of fibrinopeptide B in patients with deep vein thrombosis and pulmonary embolism. Thromb Res. 2003;110(2-3):159–165. doi: 10.1016/s0049-3848(03)00334-7. [DOI] [PubMed] [Google Scholar]
- 22.Smyth SS, Reis ED, Väänänen H, Zhang W, Coller BS. Variable protection of beta 3-integrin–deficient mice from thrombosis initiated by different mechanisms. Blood. 2001;98(4):1055–1062. doi: 10.1182/blood.v98.4.1055. [DOI] [PubMed] [Google Scholar]
- 23.Brand K, et al. Treatment of colorectal liver metastases by adenoviral transfer of tissue inhibitor of metalloproteinases-2 into the liver tissue. Cancer Res. 2000;60(20):5723–5730. [PubMed] [Google Scholar]
- 24.Giljohann DA, Mirkin CA. Drivers of biodiagnostic development. Nature. 2009;462(7272):461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermanson GT. Bioconjugate Techniques. 2nd Ed. New York: Academic; 2008. [Google Scholar]
- 26. Berenson A, Abelson R (June 29, 2008) The evidence gap: Weighing the costs of a CT scan’s look inside the heart. NY Times, Section A, p 1.
- 27.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28(6):595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern E, et al. A nanoelectronic enzyme-linked immunosorbent assay for detection of proteins in physiological solutions. Small. 2010;6(2):232–238. doi: 10.1002/smll.200901551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Rica R, Stevens MM. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat Nanotechnol. 2012;7(12):821–824. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- 30.Harisinghani MG, et al. Noninvasive detection of clinically occult lymph-node metastases in prostate cancer. N Engl J Med. 2003;348(25):2491–2499. doi: 10.1056/NEJMoa022749. [DOI] [PubMed] [Google Scholar]
- 31.Heesakkers RAM, et al. Prostate cancer: Detection of lymph node metastases outside the routine surgical area with ferumoxtran-10-enhanced MR imaging. Radiology. 2009;251(2):408–414. doi: 10.1148/radiol.2512071018. [DOI] [PubMed] [Google Scholar]
- 32.Danielson BG. Structure, chemistry, and pharmacokinetics of intravenous iron agents. J Am Soc Nephrol. 2004;15(Suppl 2):S93–S98. doi: 10.1097/01.ASN.0000143814.49713.C5. [DOI] [PubMed] [Google Scholar]
- 33.Choi HS, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25(10):1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitragotri S. Devices for overcoming biological barriers: The use of physical forces to disrupt the barriers. Adv Drug Deliv Rev. 2013;65(1):100–103. doi: 10.1016/j.addr.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Yang W, Peters JI, Williams RO., 3rd Inhaled nanoparticles—a current review. Int J Pharm. 2008;356(1-2):239–247. doi: 10.1016/j.ijpharm.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Hori SS, Gambhir SS. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci Transl Med. 2011;3(109):ra116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutz AM, Willmann JK, Cochran FV, Ray P, Gambhir SS. Cancer screening: A mathematical model relating secreted blood biomarker levels to tumor sizes. PLoS Med. 2008;5(8):e170. doi: 10.1371/journal.pmed.0050170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branson BM. Point-of-care rapid tests for HIV antibodies. Laboratoriums Medizin. 2003;27(7–8):288–295. [Google Scholar]
- 39.Chin CD, Linder V, Sia SK. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip. 2012;12(12):2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 40.Lee L, Nordman E, Johnson M, Oldham M. A low-cost, high-performance system for fluorescence lateral flow assays. Biosensors. 2013;3(4):360–373. doi: 10.3390/bios3040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson IM, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/ml or lower. JAMA. 2005;294(1):66–70. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.