Significance
Nicks are a very common form of DNA damage, but their threat to genomic integrity has been neglected because it is assumed that all nicks are repaired by simple religation. Here we challenge that assumption. We identify a robust pathway for homology-directed repair (HDR) that is active at DNA nicks. This alternative HDR pathway is stimulated upon down-regulation of key factors in canonical HDR at double-strand breaks. Alternative HDR at targeted nicks has immediate practical applications to genome engineering. Alternative HDR can promote repair of a nicked target by a nicked donor and may thereby contribute to loss of heterozygosity, a common form of genomic instability in tumors.
Keywords: loss of heterozygosity, targeted gene correction, recombination, cancer, gene conversion
Abstract
DNA nicks are the most common form of DNA damage, and if unrepaired can give rise to genomic instability. In human cells, nicks are efficiently repaired via the single-strand break repair pathway, but relatively little is known about the fate of nicks not processed by that pathway. Here we show that homology-directed repair (HDR) at nicks occurs via a mechanism distinct from HDR at double-strand breaks (DSBs). HDR at nicks, but not DSBs, is associated with transcription and is eightfold more efficient at a nick on the transcribed strand than at a nick on the nontranscribed strand. HDR at nicks can proceed by a pathway dependent upon canonical HDR factors RAD51 and BRCA2; or by an efficient alternative pathway that uses either ssDNA or nicked dsDNA donors and that is strongly inhibited by RAD51 and BRCA2. Nicks generated by either I-AniI or the CRISPR/Cas9D10A nickase are repaired by the alternative HDR pathway with little accompanying mutagenic end-joining, so this pathway may be usefully applied to genome engineering. These results suggest that alternative HDR at nicks may be stimulated in physiological contexts in which canonical RAD51/BRCA2-dependent HDR is compromised or down-regulated, which occurs frequently in tumors.
DNA nicks (single-strand breaks) are the most common form of DNA damage. Every day tens of thousands of DNA nicks occur and are repaired in each cell (1). Nicks can be caused by oxidative stress or ionizing radiation, which generates 30 nicks for every double-strand break (DSB). Reactive oxygen species (ROS), such as superoxide, hydrogen peroxide, and hydroxyl radicals, can damage a deoxyribose moiety to nick DNA directly, or modify DNA precursors (e.g., by converting guanine to 8-oxo-guanine) and thereby overload downstream repair to create a burden of nicked DNA (1–4). Nicks are also intermediates in essential DNA metabolism and repair pathways, including base excision repair, nucleotide excision repair, mismatch repair, ribonucleoside monophosphate removal, and regulation of superhelicity by topoisomerases.
Nicks are efficiently repaired by the single-strand break repair (SSBR) pathway, which assembles a repair complex at a nick in which X-ray repair cross-complementing protein 1 (XRCC1) is a critical but noncatalytic member (5–8). XRCC1 interacts with factors that clean up modified DNA ends to create a gap that is filled by polymerase POL β, or the replicative polymerases POL δ and ε. LIG3 or other ligases then reseal the DNA backbone (5–7).
Nicks can also initiate homology-directed repair (HDR) (9–12). This has drawn considerable interest as a strategy for gene therapy by targeted gene correction, because nicks cause less mutagenic end-joining (mutEJ) than do DSBs (13, 14). However, the mechanism of HDR at nicks has not been defined, either in mammalian cells or in model organisms such as Saccharomyces cerevisiae. In particular, it is not known whether HDR at nicks proceeds via the canonical HDR pathway that has been characterized in detail at DNA DSBs, in which free single-stranded 3′ ends are exposed, allowing BRCA2 to load RAD51, thus promoting strand invasion (15).
Here we compare frequencies of HDR at DNA nicks and DSBs in human cells, using both dsDNA and ssDNA donors for repair. We show that HDR at nicks, but not DSBs, is associated with transcription and occurs more efficiently at a nick on the transcribed strand than at a nick on the nontranscribed strand. We further show that two distinct pathways can promote HDR at nicks. One pathway resembles canonical HDR at DSBs and requires RAD51 and BRCA2 and primarily uses dsDNA donors. The other pathway, which we refer to as alternative HDR, is a unique pathway quite distinct from canonical HDR. Alternative HDR is inhibited by RAD51 and BRCA2 and is stimulated by transient knockdown of these factors or by expression of the RAD51 dominant negative mutant, RAD51K133R. Alternative HDR preferentially uses ssDNA or nicked dsDNA donors rather than intact dsDNAs. This newly identified alternative HDR pathway can efficiently process nicks targeted by the CRISPR/Cas9D10A nickase, with little accompanying mutEJ, so it is of potential practical utility for genome engineering by targeted gene correction. In physiological contexts, alternative HDR at nicks may be stimulated under conditions in which canonical HDR is compromised by mutation of key factors, or down-regulated in response to environmental conditions or drugs. Our evidence that a nicked dsDNA donor can promote alternative HDR at a nicked dsDNA target also raises the possibility that alternative HDR at nicks may contribute to loss of heterozygosity, a form of genomic instability frequently observed in tumors.
Results
HDR with a Duplex Donor Is More Efficient at a Nick on the Transcribed Strand.
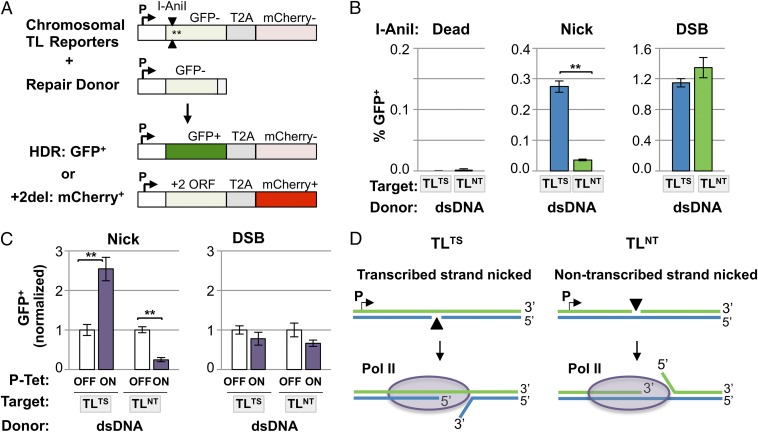
To study nick-initiated and DSB-initiated HDR, we used two versions of the I-AniI homing endonuclease. One generates DSBs, and the other, a “nickase” derivative, is disabled at one of its two active sites so it cleaves only one DNA strand to generate a nick rather than a DSB (12). To compare frequencies of HDR at nicks and DSBs, we used a Traffic Light (TL) reporter carrying an I-AniI site as the target for HDR (16). In cells bearing these reporters stably integrated in the genome, HDR by a homologous donor that replaces the I-AniI site and proximal stop codon will yield GFP+ cells, whereas mutEJ events that cause a +2 frameshift yield mCherry+ cells (Fig. 1A). To enable comparison of the outcomes of nicking either DNA strand, the I-AniI site was inserted in both forward and reverse orientations, to create the TLTS and TLNT reporters, which support nicking on the transcribed or nontranscribed strand, respectively (Fig. S1).
Fig. 1.
HDR occurs preferentially at a nick on the transcribed strand and is stimulated by transcription. (A) The chromosomal I-AniI TL reporter (16) consists of a defective GFP gene containing an I-AniI site and two stop codons (asterisks) near the 5′ end, joined by a T2A translational linker to the mCherry coding sequence in the +2 ORF. The I-AniI site is oriented to nick the transcribed strand in the TLTS reporter and the nontranscribed strand in the TLNT reporter, as indicated by the two arrowheads. GFP+ (green) cells will result from HDR of the chromosomal reporter using an exogenous donor, whereas mCherry+ (red) cells will result from cleavage at the I-AniI site followed by mutEJ that puts mCherry in the +2 reading frame. Details in Fig. S1. (B) HDR (GFP+) and mutEJ (mCherry+) frequencies calculated from independent transient transfections of 293T-TLTS or 293T-TLNT cells with indicated I-AniI derivatives (Dead, n = 2; Nick, n = 4; DSB, n = 4), one example of which is shown in Fig. S2. Mean and SEM are shown. Differences in HDR between the TLTS and TLNT cell populations are significant at nicks (**P < 0.005) but not at DSBs (P = 0.23). (C) HDR (GFP+) frequencies, based on pooled results from a total of 11 independent transfections of three different clonal 293-P-Tet-TLTS or 293-P-Tet-TLNT cell lines carried out in the absence (OFF) or presence (ON) of 1 μg/mL doxycycline and normalized to frequencies for cells cultured without inducer. Cell lines were analyzed individually, and frequencies for HDR at nicks or DSBs in each were normalized to frequencies for cells cultured without doxycycline (Dataset S1). (D) The model diagrams how transcription may stimulate repair at a nick on the transcribed strand (TLTS reporter) by unwinding DNA to expose the recombinogenic 3′ end, but inhibit repair of a nick on the nontranscribed strand (TLNT reporter) by occluding the 3′ end and exposing the less recombinogenic 5′ end.
Transcription-coupled nucleotide excision repair preferentially detects and repairs damage on the transcribed DNA strand (17), and a nick on the transcribed strand can arrest transcriptional elongation in human cell extracts (18). To determine whether a similar strand bias characterizes nick-initiated HDR, populations of 293T cells bearing either the TLTS or TLNT reporter were established using retroviral vectors, which integrate at heterogeneous integration sites, to minimize effects of chromosomal position. 293T cells are a commonly used cellular model for repair. These highly transformed cells derive from human embryonic kidney tissue, express SV40 large T antigen, and are p53 deficient and exhibit a complex karyotype. The 293T TLTS and TLNT populations were transiently cotransfected with two constructs. One transfected construct, plasmid pCS14GFP, provided the dsDNA repair donor, which is homologous with the TLTS and TLNT reporters over a region extending 2.47 kb upstream and 0.56 kb downstream of the I-AniI site (Fig. S1). The other transfected construct coexpressed wild-type I-AniI (DSB), or its nickase (Nick) or catalytically inactive (Dead) derivatives, along with the blue fluorescent protein mTagBFP (BFP) to enable transfectants to be identified by FACS as BFP+ cells. GFP+ cells among I-AniI–expressing (BFP+) cells were quantified at 3 d after transfection. Control experiments showed that no GFP+ cells were generated after expression of catalytically inactive I-AniI in the presence of donor (“Dead”; Fig. 1B, Left, and Fig. S2A). Additional controls verified that generation of GFP+ cells depends upon HDR, by showing that no GFP+ cells were generated upon transfection of constructs that expressed active I-AniI nickase or wild-type enzyme in the absence of a DNA repair donor (Fig. S2B and Dataset S1).
Comparison of HDR frequencies in 293T TLTS or 293 TLNT reporter populations after cotransfection with the I-AniI nickase expression construct and the dsDNA repair donor showed that a nick on the transcribed strand initiated HDR with nearly eightfold greater frequency than a nick on the nontranscribed strand (Fig. 1B, Center). Frequencies of mutEJ at nicks were comparable in TLTS and TLNT populations (Fig. S3A, Left), so the elevated frequency of HDR at a transcribed strand nick is unlikely to reflect differences in efficiency of nicking the two strands. A nick initiated many fewer mutEJ events (mCherry+ cells) than did a DSB, as anticipated (13, 14). DSBs initiated both HDR (Fig. 1B, Right) and mutEJ (Fig. S3A, Right) at comparable frequencies in TLTS and TLNT populations. These results suggest that I-AniI recognizes its site in the TLTS and TLNT reporters with comparable efficiency but that HDR at a nick proceeds with a strand bias that promotes more efficient HDR at a nick on the transcribed strand.
Transcription Stimulates HDR at a Nick on the Transcribed Strand and Inhibits HDR at a Nick on the Nontranscribed Strand.
To determine whether active transcription was required for the strand bias in HDR at nicks, we created derivatives of the TLTS and TLNT reporters in which a tetracycline-inducible promoter (P-Tet) was substituted for the constitutive SFFV promoter upstream of GFP. These P-Tet TLTS and P-Tet TLNT reporters were stably integrated at the unique FRT site in Flp-In T-REx-293 cells. Cells were cultured with (ON) or without (OFF) 1 μg/mL doxycycline, transfected with the pCS14GFP plasmid dsDNA donor and an I-AniI expression construct, and after 8–9 d of culture, doxycycline was added to the OFF cells to permit detection of HDR (GFP+) and mutEJ (mCherry+) events that had occurred in the absence of transcription. Active transcription increased the frequency of HDR 2.5-fold at a nick on the transcribed strand but reduced the frequency of HDR fourfold at a nick on the nontranscribed strand (Fig. 1C). These opposing effects together account for the eightfold greater frequency of HDR at a nick on the transcribed strand in an actively transcribed gene (Fig. 1B, Center). Frequencies of mutEJ events at nicks were not affected by transcription (Fig. S3B, Left), suggesting that the effect of transcription on HDR is not due to stimulation or inhibition of nicking on the transcribed and nontranscribed strands. Transcription did not affect the frequency of HDR at a DSB generated by wild-type I-AniI (Fig. 1C, Right), consistent with other reports (19–21), but did cause a twofold reduction in the frequency of mutEJ at DSBs in both P-Tet TLTS and P-Tet TLNT cells (Fig. S3B, Right), suggesting that factors associated with the transcription apparatus may protect a DSB from nonhomologous end-joining (NHEJ).
The results above suggest that the transcription-associated strand bias of HDR at nicks does not reflect the level of nicking but subsequent events in nick repair. One simple possibility is that DNA unwinding ahead of the advancing RNA polymerase may expose the recombinogenic 3′ end of a nick on the transcribed strand and thereby stimulate HDR; and conversely, that occlusion of the 3′ end and exposure of the less recombinogenic 5′ end of a nick on the nontranscribed strand may impair HDR (Fig. 1D).
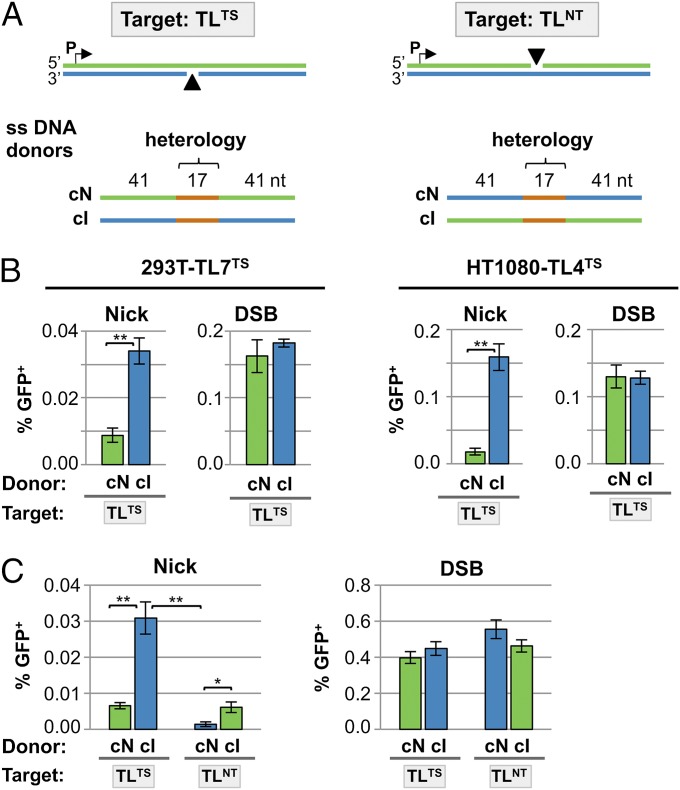
HDR at Nicks with ssDNA Donors Displays Strand Bias.
ssDNA molecules can serve as donors for HDR at DSBs (22–24). We asked whether they can also serve as donors for HDR at nicks and whether strand bias was evident in use of ssDNA donors for repair. In these experiments the donors for HDR were 99-nt ssDNA oligonucleotides in which a central 17-nt heterologous region corrects the mutations in the defective target gene (Fig. 2A; here and elsewhere the transcribed strand is shown in blue and the nontranscribed strand in green, independent of the orientation of the nick). HDR frequencies were compared in parallel in cells transfected with ssDNA donors complementary to either the intact (cI) or nicked (cN) DNA strand. In 293T-TL7TS cells, a clonal derivative of 293T carrying the TLTS reporter, the cI donor supported HDR at nicks threefold more efficiently than the cN donor (Fig. 2B, Left). There was no significant difference in the frequencies of HDR at a DSB by these donors. However, even with the preferred cI donor, HDR at a transcribed strand nick was more than eightfold less efficient than with the plasmid dsDNA donor (0.034% vs. 0.28%; Fig. 1B, Center).
Fig. 2.
Donor use and donor strand bias in HDR at nicks. (A) TLTS and TLNT reporters. Blue, transcribed strand; green, nontranscribed strand. ssDNA oligonucleotide donors complementary to the nicked (cN) or intact (cI) strand are shown below. Donors were 99 nt in length, centered at the nick, and complementary to the indicated strand except for a 17-nt region of heterology, shown in orange. This region of heterology contains the GFP coding sequence that replaces the stop codons and I-AniI site in the reporter to enable GFP expression (Fig. S1). (B) HDR (GFP+) frequencies at nicks or DSBs, using ssDNA donors shown in A, in 293T-TL7TS and HT1080 TL4TS clonal lines. Mean and SEM calculated from at least six transfections; **P < 0.005. (C) HDR (GFP+) frequencies at nicks (Left) or DSBs (Right), using ssDNA donors shown in A, in 293T-TLNT and 293T-TLTS cell populations. Mean and SEM calculated from at least five transfections; *P < 0.05, **P < 0.005.
We extended this analysis to a different cell line to ensure that results did not reflect unusual properties of 293T cells. HT1080 cells derive from a human fibrosarcoma and, in contrast to 293T cells, are p53-positive and exhibit a stable diploid karyotype. In HT1080-TL4TS cells, a clonal derivative of HT1080 carrying the TLTS reporter, the cI donor supported HDR at nicks with eightfold greater efficiency than the cN donor (0.16% and 0.02%, respectively; Fig. 2B, Right), the same donor strand bias as observed in 293T-TL7TS cells. The absolute frequency of HDR at a nick by ssDNA donors was several-fold higher in HT1080 cells than in 293T cells (Fig. 2B). The difference in absolute HDR frequency between the 293T and HT1080 reporter lines is not surprising considering that the two cell lines originated in different tissues and are likely to have disregulated different repair pathways upon transformation. The frequency of HDR at DSBs displayed no donor strand bias and was comparable in 293T-TL7TS and HT1080-TL4TS cells (Fig. 2B). We conclude that, in both 293T and HT1080 cells, at nicks but not DSBs the ssDNA donor complementary to the intact strand supported HDR several-fold better than the donor complementary to the nicked strand (Fig. 2B).
We tested the effect of transcriptional orientation on ssDNA donor strand preference by comparing HDR by the cI and cN ssDNA donors in the 293T TLTS and 293T TLNT populations, in which reporters are integrated at heterogeneous chromosomal positions to minimize possible effects of replication direction. The cI ssDNA donor, complementary to the intact strand, supported HDR more efficiently than the cN donor regardless of whether the initiating nick was on the transcribed or nontranscribed strand (Fig. 2C, Left). No donor strand bias was evident in HDR at DSBs in these populations (Fig. 2C, Right).
We conclude that there is clear donor strand bias in repair of nicks, which is independent of transcriptional orientation. Bias is determined by which DNA strand is nicked, and the HDR pathway preferentially uses an ssDNA donor that can anneal to the intact strand.
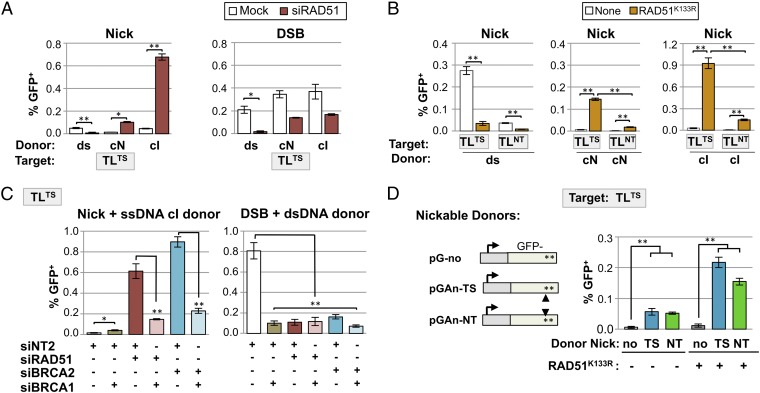
HDR at Nicks with ssDNA Donors Can Proceed via an Alternative Pathway Normally Suppressed by RAD51 and BRCA2.
RAD51 promotes strand exchange and is a critical component of the canonical HDR pathway (15). To determine whether HDR at nicks uses this canonical pathway, we examined the effect of RAD51 knockdown by siRNA treatment of the clonal 293T-TL7TS line. Strikingly, siRAD51 greatly increased the frequency of HDR at nicks by ssDNA donors complementary to either strand, but reduced the frequency of HDR by a dsDNA donor (Fig. 3A, Left). siRAD51 reduced the frequency of HDR at DSBs, as expected, but had a much greater effect on HDR by a dsDNA than a ssDNA donor (12-fold vs. twofold; Fig. 3A, Right). Similar results were observed upon transient expression of RAD51K133R, a dominant negative mutant that does not hydrolyze ATP (25), in assays of TLTS and TLNT populations (Fig. 3B) and in both the 293T-TL7TS and the HT1080-TL4TS clonal lines (Fig. S4 A and B). Thus, reduction of RAD51 activity, either by knockdown or expression of a dominant negative mutant, stimulated HDR with ssDNA donors at nicks but not at DSBs, and stimulation was not specific to a single cell type. This demonstrates that HDR at nicks using ssDNA donors uses a pathway distinct from the canonical RAD51-dependent pathway that supports HDR at DSBs.
Fig. 3.
Down-regulation of canonical HDR stimulates HDR at nicks by ssDNA or nicked dsDNA donors. (A) HDR (GFP+) frequencies at nicks (Left) or DSBs (Right) in the 293T-TL7TS clonal line, either untreated or treated with siRAD51, using the pCS14GFP dsDNA donor (n = 4) or cN (n = 2) or cI (n = 4) ssDNA donors, as indicated. Mean and SEM are presented; *P < 0.05, **P < 0.005. (B) HDR (GFP+) frequencies at nicks in 293T-TLNT and 293T-TLTS control cell populations or populations transiently expressing RAD51K133R, using indicated donors (n = 4–6). (C) HDR (GFP+) frequencies at nicks using the cI ssDNA donor or at DSBs using the dsDNA donor, in the 293T-TL7TS clonal line treated with the indicated siRNA (n = 6–12; siNT2 is a nontargeting control siRNA from Life Technologies). (D) (Left) Diagram of dsDNA donors, with no I-AniI site (pG-no) or carrying an I-AniI site oriented for intracellular nicking of the transcribed (pGAn-TS) or nontranscribed (pGAn-NT) strand. The I-AniI site in pGAn-TS and pGAn-NT is ∼500 bp downstream of the I-AniI site in the reporter GFP gene targeted for repair. The donors contain 100 bp of upstream and 500 bp of downstream homology with the chromosomal reporter, and the promoters are not homologous to the TL promoter (Fig. S1). (Right) HDR (GFP+) frequencies at nicks in the 293T-TL7TS clonal line, either control cells or cells transiently expressing the RAD51K133R dominant negative mutant, as indicated, with intact or nicked dsDNA donors (n = 5); **P < 0.005.
Single-strand annealing (SSA) in human cells can repair DSBs by joining flanking repeated sequences in cis, leading to deletion (23, 26–29). SSA is inhibited by RAD51, like HDR at nicks by ssDNA donors. SSA is also inhibited by BRCA2 but requires BRCA1. As a first step toward defining additional similarities and differences between alternative HDR and SSA, we assayed the effects of siBRCA2 and siBRCA1 on HDR at nicks. HDR at nicks using an ssDNA donor was stimulated 60-fold by siBRCA2 in 293T-TL7TS cells (Fig. 3C, Left). siBRCA1 alone caused some stimulation (2.5-fold), which may reflect a contribution of BRCA1 to the activity of BRCA2/RAD51. siBRCA1 had the opposite effect on the alternative HDR pathway: siBRCA1 reduced by fourfold the stimulation of HDR at nicks observed in response to treatment with either siRAD51 or siBRCA2 (Fig. 3C, Left). At DSBs, using a plasmid donor, siBRCA1, siBRCA2, or siRAD51 inhibited HDR, as expected (Fig. 3C, Right). We conclude that HDR at nicks can proceed by an alternative pathway that is normally inhibited by the canonical RAD51/BRCA2-dependent HDR pathway but requires BRCA1.
Nicked dsDNA Donors Promote Efficient Alternative HDR at a Nicked Target.
Concerted nicking of both donor and target DNA has been reported to stimulate HDR (30). This raises the possibility that the prevalence of nicks in the genome may stimulate HDR not only between sister chromatids, which would not alter genomic sequence, but also between homologous chromosomes, which could result in loss of heterozygosity, a common form of genomic instability in tumor cells. We therefore compared HDR of a nicked target using either an intact or nicked dsDNA plasmid donor in the 293T-TL7TS cell line. The donors were dsDNA plasmids carrying a GFP gene that had been inactivated by insertion of two stop codons, bearing no I-AniI site (pG-no) or an I-AniI site at the 3′ end of GFP on either the transcribed (pGAn-TS) or nontranscribed (pGAn-NT) DNA strand, to enable intracellular nicking in cells expressing I-AniI nickase (Fig. 3D, Left and Fig. S1). The nicked dsDNA donors were more active than the intact dsDNA donor, regardless of which donor strand was nicked, both in cells carrying out canonical HDR and in cells in which canonical HDR was suppressed by transient expression of RAD51K133R (Fig. 3D, Right). Thus, genomic dsDNA that has been nicked in the course of replication, transcription, recombination, or repair may serve as an intracellular donor or target for canonical or alternative HDR at a nick in a homologous sequence.
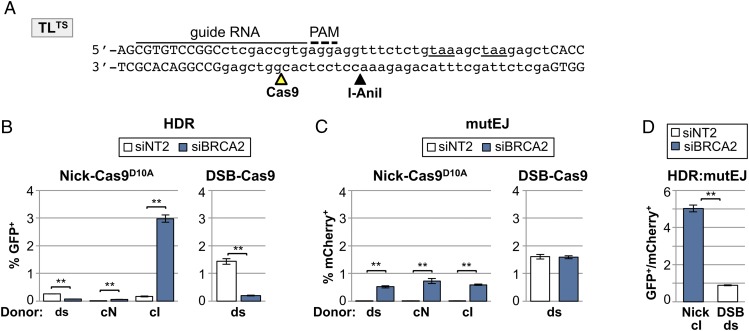
Efficient Alternative HDR at Nicks Generated by CRISPR/Cas9D10A.
The very high efficiency of alternative HDR at nicks suggested that this pathway might be useful in targeted gene correction. The CRISPR/Cas9 system is ideal for this application because target specificity is easily modified and, as with I-AniI, targeting by the nickase derivative CRISPR/Cas9D10A is accompanied by less local deletion than targeting by the CRISPR/Cas9 cleavase (31). To determine whether nicks generated by CRISPR/Cas9D10A could be repaired by alternative HDR, Cas9D10A or Cas9WT was coexpressed with a CRISPR guide RNA designed to target the enzyme to a site 9 bp upstream of the I-AniI recognition sequence in the TLTS reporter in 293T-TL7TS cells (Fig. 4A). HDR frequencies were compared in cells treated with a nontargeting control siRNA (NT2) or siRNAs directed toward the key canonical HDR factor, BRCA2 (Fig. 4B). These experiments showed that siBRCA2 inhibited HDR with a dsDNA donor at either a nick or DSB but stimulated HDR at a nick using an ssDNA donor. In addition, an ssDNA donor complementary to the intact strand (cI) supported HDR at levels 30-fold higher than the donor complementary to the nicked strand (cN). Thus, the effects of siBRCA2 treatment and the strand bias of the ssDNA repair donor were the same at nicks targeted by CRISPR/Cas9D10A and the I-AniI nickase.
Fig. 4.
Nicks generated by CRISPR/Cas9D10A initiate alternative HDR and are associated with less mutEJ than are DSBs. (A) Sequence of the portion of the TLTS reporter containing CRISPR/Cas9 and I-AniI target sites (open and filled arrowheads). The insertion (lowercase) bearing the I-AniI site and stop codons (underlined) and a portion of the GFP coding sequence (uppercase) are shown. The CRISPR guide RNA and protospacer adjacent motif (PAM) sequence are indicated. (B) HDR (GFP+) frequencies at nicks (Left) or DSBs (Right) in the 293T-TL7TS clonal line after transient transfection of a Cas9D10A (Nick) or Cas9 (DSB) expression plasmid, a guide RNA expression plasmid, and either a dsDNA plasmid donor pCS14GFP or cI or cN ssDNA donors, as indicated. Mean and SEM calculated from three transfections; **P < 0.005. (C) MutEJ (mCherry+) frequencies at nicks or DSBs in the 293T-TL7TS clonal line; same cells as in B. (D) The ratio of HDR to mutEJ (GFP+:mCherry+ cells) compiled from transfections of 293T-TL7TS cells in B, analyzing HDR at a nick in the transcribed strand using the cI ssDNA donor in siBRCA2-treated cells; and HDR at DSBs using a dsDNA donor in untreated cells.
The frequency of mutEJ (mCherry+ cells) at nicks generated by CRISPR/Cas9D10A was elevated by siBRCA2 treatment, but nonetheless was significantly lower than the frequency of mutEJ at a DSB generated by CRISPR/Cas9WT (Fig. 4C). Moreover, the ratio of HDR:mutEJ at nicks by the alternative HDR pathway, assayed in siBRCA2-treated cells, was fivefold higher than at DSBs by canonical HDR (Fig. 4D). Similar results were obtained in parallel assays of mutEJ initiated by I-AniI nickase in cells in which the alternative HDR pathway was stimulated by transient expression of dominant negative RAD51K133R (Fig. S3C). Nicks on the nontranscribed strand yielded fewer HDR events than nicks on the transcribed strand but comparable levels of mutEJ, resulting in a higher HDR:mutEJ ratio for nicks on the transcribed than nontranscribed strand (compare Fig. 1B and Fig. S3A). Thus, alternative HDR targeted by CRISPR/Cas9D10A to a nick on the transcribed strand is twice as efficient as canonical HDR targeted to a DSB by CRISPR/Cas9 and accompanied by significantly less local mutagenesis.
We conclude that the key features of HDR at a nick via the alternative pathway are characteristic of the pathway and independent of the enzyme that targets the nick. It should therefore be straightforward to achieve very efficient gene targeting accompanied by low mutEJ in cells treated transiently with siBRCA2 to stimulate alternative HDR.
Unwinding or Resection of the Target May Promote HDR at a Nick.
Donor DNA strands complementary to either the nicked or intact target strand are competent to engage the alternative HDR pathway (Figs. 2 and 3), suggesting an HDR mechanism in which unwinding or resection occurs at the nick to make both strands of the chromosomal target accessible for donor annealing. To determine whether unwinding or resection may occur preferentially at one side of the nick or the other, we compared HDR at a nick on the transcribed (Fig. 5A) or nontranscribed (Fig. 5B) strand by 75-nt ssDNA donors that were centered on the nick or extended either 3′ or 5′ of it. All of the donors tested could support HDR (Fig. 5 and Dataset S1), but there were significant position-dependent differences in donor efficiency. In all cases the donor centered on the nick was most efficient, and the donor with extended homology 3′ of the nick was least efficient. Clear statistical significance was evident between HDR frequencies for cI (but not cN) donors extending 5′ and 3′ of the nick (P = 1.19 × 10−5 at a nick on the transcribed strand nick; P = 6.02 × 10−3 at a nick on the nontranscribed strand). These results suggest that in the alternative HDR pathway, a helicase or nuclease unwinds or resects the nick to expose a gap and that the predominant initial step is exposure of a gap on the 5′ side of the nick.
Fig. 5.
ssDNA donor homology and HDR at nicks. (A) 293T-TLTS and (B) 293T-TLNT cell populations, transiently expressing the I-AniI nickase and RAD51K133R, were provided with a 75-nt ssDNA donor centered at the nick or extending either 3′ or 5′ of the nick, as diagrammed, and HDR assayed. Target: blue, transcribed strand; green, nontranscribed strand. ssDNA donors were complementary to the nicked (cN) or intact (cI) strands and carried a 17-nt region of heterology (orange) and homologous flanking sequences of indicated lengths. For each target/donor combination, the frequency of HDR (% HDR) is shown, and HDR relative to the donor centered on the nick is graphed. Data represent mean and SEM of at least seven transfections.
Discussion
We have shown that nicks that bypass the SSBR pathway may undergo HDR via two distinct pathways. One pathway requires RAD51 and BRCA2 and uses a dsDNA donor, like canonical DSB repair. The second, a unique alternative pathway, uses ssDNA or nicked dsDNA donors and is normally suppressed by RAD51 and BRCA2. Both canonical and alternative HDR were especially efficient at a nick in the transcribed strand of an actively transcribed gene. HDR at nicks by the alternative pathway was independent of the source of the nick and could be initiated by either the I-AniI or CRISPR/Cas9D10A nickase.
Use of the CRISPR/Cas9D10A nickase to initiate alternative HDR at nicks (Fig. 4) represents a useful strategy for genome engineering. CRISPR/Cas9D10A is a versatile targeting enzyme. The use of short ssDNAs as HDR donors further streamlines this application. HDR by this pathway is not only efficient but also accompanied by relatively little associated mutEJ, a key advantage for targeted gene correction.
Our results establish that HDR at nicks is distinct from HDR at DSBs in three ways. HDR at nicks—but not DSBs—is (i) transcription-associated; (ii) preferentially uses a ssDNA donor complementary to the intact strand of the target; and (iii) can proceed by an alternative HDR pathway that is stimulated by down-regulation of RAD51 or BRCA2 expression or activity. Previous experiments had provided compelling evidence that a nick can initiate HDR (9–13) but left open the possibility that it might be necessary for a nick to be converted to a DSB for subsequent processing by the DSB repair pathway. The differences we have documented between HDR at nicks and DSBs make it very unlikely that a replicative DSB is an obligatory intermediate in HDR initiated by a nick.
The results reported here lead us to propose a working model for HDR at a nick (Fig. 6). In HDR using a dsDNA donor (Fig. 6, Left), BRCA2 loads RAD51 on the free 3′ end to promote homology-dependent strand invasion, as in canonical DSB repair. The requirement for a free 3′ end is consistent with the transcription-dependent strand bias of HDR using a dsDNA donor, because the unwinding that accompanies transcription will stimulate HDR at a nick on the transcribed strand by promoting release of a 3′ end and inhibit HDR at a nick on the nontranscribed strand by occluding the 3′ end (Figs. 1 C and D). After strand invasion, the 3′ end of the target is extended by repair synthesis, again as in canonical DSB repair (Fig. 6). The donor strand is then released, the target strand reanneals, and flaps are removed and DNA ligated. This resembles gene conversion by synthesis-dependent strand annealing (15), but recombination might also involve crossover via a single Holliday junction intermediate (32, 33).
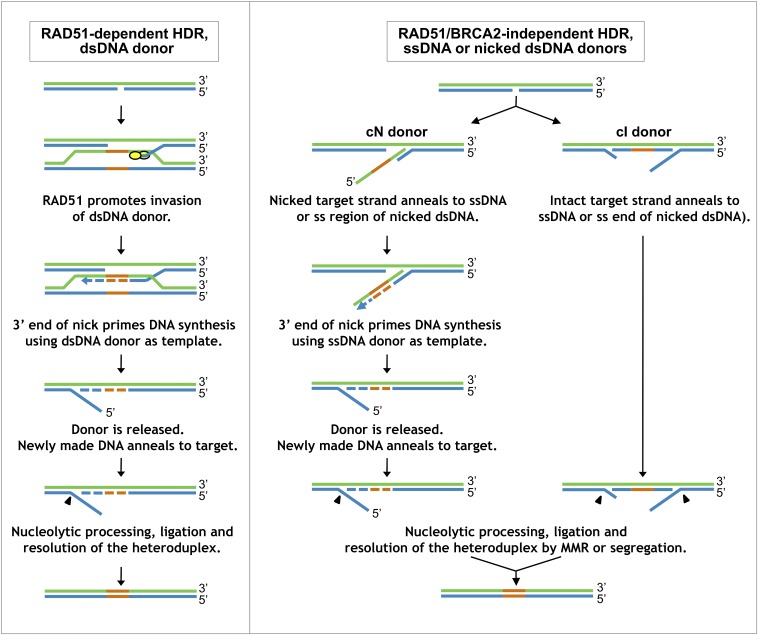
Fig. 6.
Working model for pathways of HDR at nicks. (Left) RAD51-dependent HDR using a dsDNA donor. A gap is exposed at the nicked target, and BRCA2 loads RAD51 on the free 3′ end, enabling invasion of a homologous dsDNA donor, as in canonical DSB repair. (Right) RAD51/BRCA2-independent HDR. A gap is exposed at the nicked target, and the donor anneals to either the nicked (Left) or intact (Right) strand of the duplex, independent of RAD51/BRCA2. Heterology (orange) and repair synthesis (dashed line) are shown. Arrowheads represent nucleolytic removal of DNA, either by excision or flap cleavage. Refer to Discussion for more detailed description; Fig. S5 for more complete diagrams of mismatch repair and ligation steps; and Figs. S6 and S7 for diagrams of how nicked dsDNA donors may participate in this pathway. MMR, mismatch repair.
HDR using an ssDNA donor (Fig. 6, Right) occurs via an alternative pathway that is not only independent of RAD51/BRCA2 but strongly inhibited by these factors (Figs. 3 and 4). First, DNA unwinding and/or excision at the nick exposes a single-stranded region in the repair target, and then the ssDNA donor anneals to the target. RAD51 and BRCA2 inhibit annealing and drive recombination via the pathway that uses dsDNA donors (Fig. 6, Left), which seems to compete with the alternative HDR pathway to carry out repair.
The alternative HDR pathway can use donors complementary to either strand, albeit with differing efficiencies (Figs. 2 and 3), and subsequent events depend upon the strand used for repair. The less efficient pathway of alternative HDR uses a donor complementary to the nicked strand (cN), which can anneal to the free 3′ end of the target and then serve as the template for repair synthesis primed by that 3′ end. Donor release then enables reannealing of the DNA duplex, followed by flap removal and ligation to complete HDR. Note the similarities with HDR at a nick using a dsDNA donor (Fig. 6, Left), especially the requirement for unwinding to expose a free 3′ end in both pathways. The more efficient pathway of alternative HDR uses a donor complementary to the intact strand (cI), which can anneal to the gap generated at the nick to form a heteroduplex with the intact strand of the target DNA. The preference for an ssDNA donor with extended homology to one side of the nick (Fig. 5) suggests that gap exposure occurs predominantly 5′ of the nick. The donor may then be ligated into the target (possibly requiring processing of donor or target ends), and heterology eliminated by mismatch repair or upon segregation (Fig. 6, Left). Alternatively, the donor may direct mismatch repair and be released in the course of repair synthesis (Fig. S5).
The alternative HDR pathway is inhibited by RAD51 or BRCA2 but requires BRCA1 (Fig. 3D). These features are shared with the SSA pathway of HDR of DSBs, which repairs DSBs using as donors repetitive sequences in cis. It will be interesting to learn in future experiments whether alternative HDR shares other features with the SSA pathway, such as dependence upon RAD52. BRCA1 seems to play contrasting roles in HDR at nicks, depending on whether cells have or have not been treated with siRAD51 or siBRCA2 to stimulate alternative HDR. Treatment with siBRCA1 alone caused modest stimulation in HDR by an ssDNA donor, in cells not treated with either siRAD51 or siBRCA2. This may reflect the well-documented role of BRCA1 in promoting BRCA2 function. In contrast, siBRCA1 inhibited HDR at nicks in cells treated with either siRAD51 or siBRCA2. This suggests that one physiological function of BRCA1 may be to promote alternative HDR. We do not yet know how this occurs, but BRCA1 is multifunctional, and it is plausible that BRCA1 recruits or regulates other factors that are critical to alternative HDR.
The pathways that support use of an ssDNA donor may also be applicable to repair by a single-stranded region of a nicked dsDNA donor (Figs. S6 and S7). In that context, this mechanism may be relevant to regulated diversification of Ig V regions by gene conversion (34), where cytidine deamination by activation-induced deaminase is processed to generate a nick in the target, and repair is templated by upstream pseudogene donors. More generally, this mechanism may contribute to loss of heterozygosity (LOH). LOH without accompanying change in gene copy number occurs frequently in cancer cells and is an important source of mutations that drive tumorigenesis (35–37). LOH can occur if HDR uses an allelic region of the homologous chromosome as donor. If HDR between nicked homologs promotes LOH, then DNA nicks may constitute a more serious threat to genomic integrity than previously appreciated.
Alternative HDR at nicks is suppressed by canonical HDR and may therefore be active in contexts in which canonical HDR is inactive. Examples include breast and ovarian cancers bearing BRCA2 mutations, and regions of solid tumors in which local hypoxic conditions down-regulate canonical HDR (38–41). Recently, highly significant correlations have been documented between increased frequencies of LOH and deficiencies in canonical HDR in primary breast and ovarian tumors and cell lines (42). This otherwise paradoxical observation may be explained if the alternative HDR pathway mediates LOH in these tumors, using the nicked homologous chromosome as donor.
Materials and Methods
Details regarding materials and methods are in SI Materials and Methods.
Cell Culture and Transfection.
Human cells lines were cultured as previously described (13). siRNA and DNA transfections were performed according to the manufacturer’s protocol (Lipofectamine RNAiMAX and LTX, repectively; Life Technologies). To determine the efficiency of siRNA knockdown (Fig. S8), mRNA levels of BRCA1, BRCA2, and RAD51 were quantified by RT-PCR, relative to a control that assayed transcripts from the lactate dehydrogenase A (LDHA) gene. The effect of siRNA treatment was corroborated by control experiments that showed that siBRCA1, siBRCA2, and siRAD51 treatments all inhibited HDR at a DSB, as predicted by the extensive literature on the canonical HDR pathway.
Flow Cytometry, HDR, and mutEJ Frequencies.
Cells were processed for flow cytometry as described previously (13). In experiments with I-AniI, which was coexpressed with mTagBFP (BFP), data are presented as GFP+ and mCherry+ frequencies among BFP+ cells, except in cases in which flow was carried out more than 8 d after transfection, by which time the BFP signal was largely extinguished. Raw data for each experiment, including frequencies of GFP+, mCherry+, and BFP+ cells among total live cells is in Dataset S1. HDR and mutEJ frequencies were displayed as mean and SEM.
Supplementary Material
Acknowledgments
Research reported here was supported by National Institutes of Health Grant RL1 GM084434 and by National Cancer Institute Grant P01 CA077852.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400236111/-/DCSupplemental.
References
- 1.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 2.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: Induction, repair and significance. Mutat Res. 2004;567(1):1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Loeb LA, Harris CC. Advances in chemical carcinogenesis: A historical review and prospective. Cancer Res. 2008;68(17):6863–6872. doi: 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336(6079):315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao Y, et al. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471(7337):240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katyal S, McKinnon PJ. Disconnecting XRCC1 and DNA ligase III. Cell Cycle. 2011;10(14):2269–2275. doi: 10.4161/cc.10.14.16495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simsek D, et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471(7337):245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanssen-Bauer A, et al. The region of XRCC1 which harbours the three most common nonsynonymous polymorphic variants, is essential for the scaffolding function of XRCC1. DNA Repair (Amst) 2012;11(4):357–366. doi: 10.1016/j.dnarep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strathern JN, Weinstock KG, Higgins DR, McGill CB. A novel recombinator in yeast based on gene II protein from bacteriophage f1. Genetics. 1991;127(1):61–73. doi: 10.1093/genetics/127.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arcangioli B. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 1998;17(15):4503–4510. doi: 10.1093/emboj/17.15.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117(2):171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 12.McConnell Smith A, et al. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc Natl Acad Sci USA. 2009;106(13):5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis L, Maizels N. DNA nicks promote efficient and safe targeted gene correction. PLoS ONE. 2011;6(9):e23981. doi: 10.1371/journal.pone.0023981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez CL, et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40(12):5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Certo MT, et al. Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods. 2011;8(8):671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 18.Kathe SD, Shen GP, Wallace SS. Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J Biol Chem. 2004;279(18):18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- 19.Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17(11):6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen C, Miller CA, Nickoloff JA. The mutagenic potential of a single DNA double-strand break in a mammalian chromosome is not influenced by transcription. DNA Repair (Amst) 2003;2(10):1147–1156. doi: 10.1016/s1568-7864(03)00139-3. [DOI] [PubMed] [Google Scholar]
- 21.Langston LD, Symington LS. Gene targeting in yeast is initiated by two independent strand invasions. Proc Natl Acad Sci USA. 2004;101(43):15392–15397. doi: 10.1073/pnas.0403748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci USA. 2003;100(25):14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24(21):9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8(9):753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark JM, et al. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277(23):20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 26.Tutt A, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20(17):4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol Cell Biol. 2002;22(18):6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstock DM, Richardson CA, Elliott B, Jasin M. Modeling oncogenic translocations: Distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst) 2006;5(9-10):1065–1074. doi: 10.1016/j.dnarep.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 29.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4(6):e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonçalves MA, van Nierop GP, Holkers M, de Vries AA. Concerted nicking of donor and chromosomal acceptor DNA promotes homology-directed gene targeting in human cells. Nucleic Acids Res. 2012;40(8):3443–3455. doi: 10.1093/nar/gkr1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cromie GA, et al. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127(6):1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith GR. How homologous recombination is initiated: Unexpected evidence for single-strand nicks from v(d)j site-specific recombination. Cell. 2004;117(2):146–148. doi: 10.1016/s0092-8674(04)00338-1. [DOI] [PubMed] [Google Scholar]
- 34.Maizels N. Immunoglobulin gene diversification. Annu Rev Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 35.Makishima H, Maciejewski JP. Pathogenesis and consequences of uniparental disomy in cancer. Clin Cancer Res. 2011;17(12):3913–3923. doi: 10.1158/1078-0432.CCR-10-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Score J, Cross NC. Acquired uniparental disomy in myeloproliferative neoplasms. Hematol Oncol Clin North Am. 2012;26(5):981–991. doi: 10.1016/j.hoc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Pedersen BS, De S. Loss of heterozygosity preferentially occurs in early replicating regions in cancer genomes. Nucleic Acids Res. 2013;41(16):7615–7624. doi: 10.1093/nar/gkt552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindra RS, et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24(19):8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng AX, et al. Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother Oncol. 2005;76(2):168–176. doi: 10.1016/j.radonc.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 40.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8(3):180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 41.Klein TJ, Glazer PM. The tumor microenvironment and DNA repair. Semin Radiat Oncol. 2010;20(4):282–287. doi: 10.1016/j.semradonc.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abkevich V, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107(10):1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.