Significance
Stem cells generate tissues, and they can be hijacked in cancer. A fundamental mechanism of stem cell regulation is signaling from their immediate microenvironment, or niche. Although many niches and signaling pathways have been identified, much less is known about how stem cells respond. Here, we report the discovery of two key niche signaling effector genes. These genes are activated in stem cells by Notch signaling, a conserved and clinically significant pathway, and together are essential for the stem cell state. Surprisingly, neither was previously implicated in stem cell regulation and their sequences yield few clues to their functions. We suggest that these newly discovered genes may be pioneers for a new class of stem cell regulators.
Abstract
A stem cell’s immediate microenvironment creates an essential “niche” to maintain stem cell self-renewal. Many niches and their intercellular signaling pathways are known, but for the most part, the key downstream targets of niche signaling remain elusive. Here, we report the discovery of two GLP-1/Notch target genes, lst-1 (lateral signaling target) and sygl-1 (synthetic Glp), that function redundantly to maintain germ-line stem cells (GSCs) in the nematode Caenorhabditis elegans. Whereas lst-1 and sygl-1 single mutants appear normal, lst-1 sygl-1 double mutants are phenotypically indistinguishable from glp-1/Notch mutants. Multiple lines of evidence demonstrate that GLP-1/Notch signaling activates lst-1 and sygl-1 expression in GSCs within the niche. Therefore, these two genes fully account for the role of GLP-1/Notch signaling in GSC maintenance. Importantly, lst-1 and sygl-1 are not required for GLP-1/Notch signaling per se. We conclude that lst-1 and sygl-1 forge a critical link between Notch signaling and GSC maintenance.
Stem cell self-renewal requires signaling from a specialized microenvironment, or stem cell niche (1, 2). Well-defined stem cell niches include the distal tip cell (DTC) for Caenorhabditis elegans germ-line stem cells (GSCs) (3), the cap and hub cells for Drosophila GSCs (e.g., refs. 4 and 5) and Paneth cells for murine intestinal stem cells (6). These niches use a variety of intercellular signaling pathways (e.g., Notch, BMP, JAK/Stat, Wnt), all of which have been implicated broadly in stem cell regulation throughout the animal kingdom (1). The identification of direct molecular links between niche signaling and downstream targets driving stem cell self-renewal is crucial for understanding niche function. However, few such links have been established.
Here, we focus on how Notch signaling controls stem cell self-renewal. Notch signaling typically occurs between adjacent cells, one signaling and the other receiving, and activates target genes using a transcription factor complex that includes the DNA binding protein CSL [human CBF1/RBPJκ, Drosophila Su(H), and C. elegans LAG-1] (7). Although Notch signaling regulates stem cells in vertebrates, including those in the mammalian muscle, brain, and intestine (reviewed in ref. 8), its use in the nematode C. elegans to maintain GSCs provides the best defined and most tractable paradigm (Fig. 1A). In this case, a single mesenchymal cell, the DTC, forms the niche for GSCs. The DTC uses GLP-1/Notch signaling to maintain GSCs, and when GSCs leave their DTC niche they are triggered to enter the meiotic cell cycle and begin differentiation (3, 9). Regardless of sex or developmental stage, laser ablation of the DTC or genetic ablation of GLP-1/Notch signaling causes all GSCs to cease self-renewal and differentiate, the so-called Glp (germ-line proliferation defective) phenotype (3, 9, 10). No other signaling pathway has the same profound effect on C. elegans GSC maintenance. Therefore, C. elegans GLP-1/Notch provides an unequalled entrée into understanding Notch regulation of stem cells.
Fig. 1.
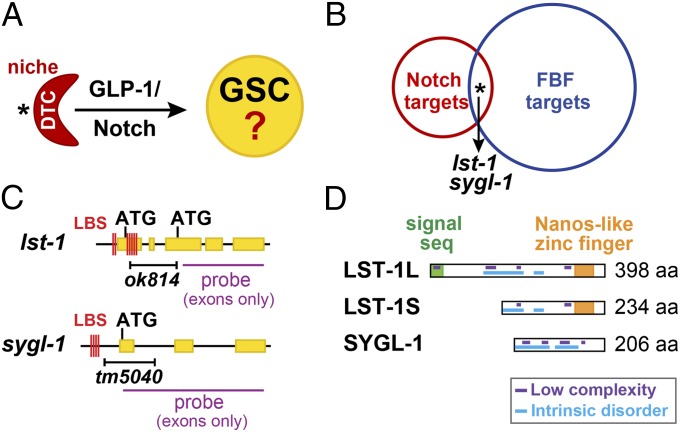
Identification of lst-1 and sygl-1 as candidate GSC regulators. (A) DTC (red) uses GLP-1/Notch signaling to maintain GSCs within the niche. Asterisk marks distal end. (B) Fifteen genes (asterisk) are shared between lists of putative Notch and FBF targets. Double RNAi of two common genes, lst-1 and sygl-1, caused a Glp phenotype. Also see Tables S1 and S2. (C) lst-1 and sygl-1 genes. Yellow, exons; red lines, LAG-1 binding sites (LBS); black bar, deletions; purple line, probe for in situ hybridizations (exons only). (D) LST-1 and SYGL-1 proteins and their predicted motifs.
The key direct targets of GLP-1/Notch signaling that promote GSC self-renewal remain an open question. A partial answer is that GLP-1/Notch signaling activates fbf-2 transcription (11). FBF-2, a PUF family mRNA binding protein, is a broad-spectrum inhibitor of differentiation and a regulator of GSC self-renewal together with its nearly identical paralog FBF-1 (see ref. 12 for review). FBF-1 and FBF-2 function redundantly to maintain GSCs in late larvae and adults (13). GLP-1/Notch signaling also activates lip-1, which encodes a dual specificity phosphatase and ERK/MAPK inhibitor (14). LIP-1 promotes robust germ-line proliferation but has not been implicated in GSC self-renewal per se (15). Therefore, two GLP-1/Notch target genes are known, but neither accounts for GLP-1/Notch regulation of GSC self-renewal.
Additional GLP-1/Notch targets must exist to drive GSC self-renewal, but such genes have been elusive. Thirty years of genetics have not found them, either by screening for mutants with a Glp phenotype or by isolating suppressors and enhancers of genes in the GSC control pathway. Here, we report that lst-1 and sygl-1 function redundantly as pivotal GSC regulators with a double-mutant GSC phenotype indistinguishable from that of glp-1/Notch mutants. We also provide evidence that both genes are GLP-1/Notch targets but not components of GLP-1/Notch signaling per se. The lst-1 and sygl-1 genes establish a previously unidentified and important link between niche signaling and stem cell maintenance.
Results
GLP-1/Notch Targets Essential for Larval GSC Self-Renewal Not Yet Known.
Wild-type adults possess a total of ∼2,000 germ cells, with ∼1,000 in each of two gonadal arms (Fig. 2A and Fig. S1 A and E) (e.g., ref. 16), whereas glp-1 null mutants make a total of only ∼four to eight germ cells that all differentiate precociously (Fig. S1 B and E) (9). Previous studies identified fbf-2 and lip-1 as germ-line targets of GLP-1/Notch signaling, but neither on its own was essential for GSC self-renewal (see Introduction). One possibility might have been that fbf-2 and lip-1 function redundantly to maintain GSCs. To test this idea, we examined fbf-2; lip-1 double mutants, but they were self-fertile with essentially normal germ lines (Fig. S1E). Another possibility might have been that lip-1 functions redundantly with the two nearly identical fbf-1 and fbf-2 genes, but fbf-1 fbf-2; lip-1 triple mutant germ lines were similar to fbf-1 fbf-2 double-mutant germ lines (Fig. S1 C–E). Therefore, we reasoned that one or more additional GLP-1/Notch target genes must await discovery.
Fig. 2.
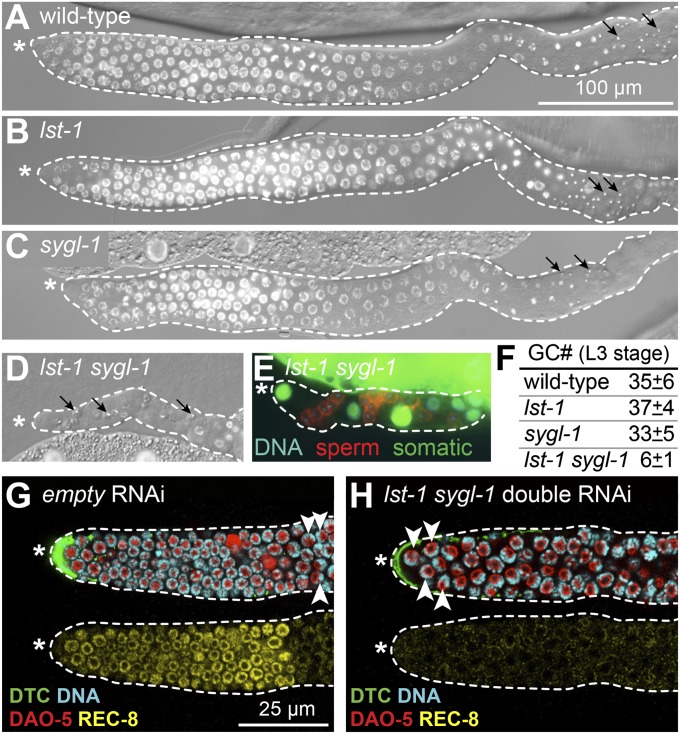
lst-1 and sygl-1 function redundantly to promote GSC self-renewal in larvae and adults. (A–D) DIC- and DAPI-stained images of gonads dissected from L4 hermaphrodites. Asterisk marks distal end; dotted line outlines germ line plus somatic gonadal cells; arrows mark mature sperm. (A–C) Wild type (100% non-Glp; n > 100), lst-1(ok814) (100% non-Glp; n = 146), and sygl-1(tm5040) (100% non-Glp; n = 159) all produce normal germ lines. (D) lst-1 sygl-1 double mutants produce Glp germ lines with only a few differentiated sperm (100% Glp; n = 76). (E) Gonad from lst-1 sygl-1 L4 with a somatic GFP marker (green), a sperm marker (red) and DNA staining (blue). All nonsperm cells expressed somatic GFP. Each gonadal arm contained 14 ± 3 sperm (n = 9) on average (from three to four premeiotic germ cells). (F) Total premeiotic germ cells (GC#) in entire L3 gonad, scored with PGL-1 germ cell marker. For wild-type and each single mutant, n = 5; for lst-1 sygl-1 double mutant, n = 9. (G–H) Representative confocal images of wild-type late L4 larval germ lines treated with RNAi for 48 h; DTC expresses GFP (green). Same conventions as in A–D; mitotic zone scored by presence of mitotic marker REC-8 (yellow) and absence of crescent-shaped DAPI staining typical of early meiotic prophase nuclei (white arrowheads). Anti–DAO-5 (red, nucleolar marker) counter stain facilitates scoring of DAPI crescents. (G) Germ line treated with empty RNAi vector possesses a mitotic zone. (H) Germ line treated with lst-1 sygl-1 double RNAi lacks a mitotic zone and hence lacks GSCs.
Identification of lst-1 and sygl-1 as Functionally Redundant Stem Cell Regulators.
Our strategy to identify the key GLP-1/Notch targets essential for GSC maintenance was as follows. We hypothesized that (i) such genes are targets of both GLP-1/Notch signaling and the FBF RNA-binding protein, (ii) they are expressed in GSCs, and (iii) their functions in GSC self-renewal are masked by redundancy or pleiotropy. The idea that such genes might be targets of FBF regulation was entirely speculative and based simply on fbf-2 and lip-1 being FBF targets (11, 15); the idea that they might function redundantly or have pleiotropic effects was based on the failure to find them previously despite many genetic screens.
We compared lists of putative Notch and FBF-1 targets. The Notch target list of 163 genes was derived bioinformatically with all harboring clusters of at least four LAG-1 binding sites (LBSs) (17); the FBF-1 target list of 1,350 mRNAs was obtained experimentally using immunoprecipitation of FBF-1 with associated mRNAs followed by microarray analysis (18). Fifteen genes were common to both lists (Fig. 1B and Table S1). We depleted each of the 15 using RNAi, but none caused a Glp phenotype (Table S2). Among those 15 genes, T27F6.4 was expressed within the GSC niche, according to a database of mRNA in situ hybridizations to ∼10,000 C. elegans genes (Nematode Expression Pattern DataBase; http://nematode.lab.nig.ac.jp); the other 14 were either not in the database or did not stain detectably in the niche. With the idea that T27F6.4 might function redundantly with one of the other 14 genes, we performed double RNAi against T27F6.4 and other genes in the common pool. The double RNAi knockdown of T27F6.4 plus lst-1 caused a Glp phenotype but others did not (Tables S2 and S3); the double RNAi of T27F6.4 plus lst-1 gave similar results in either a wild-type strain or the rrf-1 mutant, which is sensitive to RNAi in only some tissues, including the germ line (19) (Table S3). We therefore named T27F6.4 sygl-1 (synthetic Glp).
Molecularly, the lst-1 gene is predicted to produce two mRNAs and polypeptides (Fig. 1 C and D, Upper), whereas sygl-1 is predicted to produce a single mRNA and polypeptide (Fig. 1 C and D, Lower). BLAST analysis of lst-1 and sygl-1 sequences revealed homologs in closely related nematodes, including Caenorhabditis briggsae and Caenorhabditis remanei, but not in more distantly related organisms. We queried LST-1 and SYGL-1 amino acid sequences using a variety of algorithms. A structure prediction program [Phyre2 (20)] predicted a single Nanos-like zinc finger in LST-1 (Fig. 1D), but no motifs or folded domains in SYGL-1; a signal sequence prediction program [SignalP (21)] predicted a signal sequence at the N terminus of LST-1L (Fig. 1D), but not in LST-1S or SYGL-1; algorithms designed to identify low complexity [SEG (22)] or intrinsically disordered sequences [MFDp2 (23)] predict that both proteins harbor multiple low complexity sequences and are largely disordered (Fig. 1D). We conclude that lst-1 and sygl-1 encode novel proteins with a central role in stem cell regulation.
lst-1 and sygl-1 Function Redundantly to Promote GSC Self-Renewal.
To investigate more rigorously the effects of lst-1 and sygl-1 on GSC self-renewal, we obtained two deletion mutants: lst-1(ok814) and sygl-1(tm5040) (Fig. 1C and Fig. S2). The lst-1 deletion removes most exons and introns unique to the longer lst-1 isoform; the sygl-1 deletion removes the first exon, including the ATG translational start codon. These two mutants are likely loss-of-function alleles, because they reproducibly mimic the strongest defect observed with RNAi (see below).
Both lst-1 and sygl-1 single deletion mutants possessed germ lines comparable in size and organization to wild type (Fig. 2 A–C and F), and they were both self-fertile as hermaphrodites and cross-fertile as males. By contrast, all lst-1 sygl-1 double deletion mutants displayed a dramatic Glp defect, regardless of sex (Fig. 2 D–F and Fig. S3 A–J). When lst-1 or sygl-1 single mutants were targeted with RNAi against the other [e.g., lst-1(ok814) sygl-1(RNAi)], the resultant germ lines were also Glp (Table S3). We conclude that lst-1 and sygl-1 function redundantly to promote GSC self-renewal in larvae of both sexes.
We observed no obvious somatic defect in either single mutant or the double mutant. For example, all were viable with normal body shape. Nonetheless, we note that lst-1 plays roles in neurons and the developing vulva, indicating that at least lst-1 likely functions in tissues other than the germ line (17, 24).
lst-1 sygl-1 Glp Defect Comparable to glp-1 Glp Defect.
Mutants lacking the GLP-1/Notch receptor make only four to eight germ cells, which differentiate precociously to produce 16–32 sperm (each germ cell makes 4 sperm) (9). To ask whether lst-1 sygl-1 Glp sterility mimicked that of a glp-1 mutant, we used a germ-line marker (PGL-1) to count germ cell number (Fig. 2F and Fig. S3 K–N) and also asked whether germ cells differentiated precociously during development. Newly hatched lst-1 sygl-1 L1 larvae contained two germ cells, as did wild-type controls; late lst-1 sygl-1 L1s had made a total of five germ cells on average (n = 10; range, 4–7), and that number was essentially unchanged in early L3s, which contained only six germ cells on average (n = 9; range, 4–8). By contrast, wild-type early L3 larvae contained ∼35 germ cells on average, as did lst-1 and sygl-1 single mutants (Fig. 2F). Moreover, lst-1 sygl-1 germ cells differentiated as sperm during the third larval stage (L3), which is one stage earlier than normal and therefore precocious (Fig. S4). Importantly, sperm number in double mutants was consistent with the number of PGL-1–positive undifferentiated germ cells, and no evidence for cell death was seen. We conclude that the lst-1 sygl-1 GSC defect is indistinguishable from that of a glp-1 null mutant (9).
GLP-1/Notch signaling is also required to maintain GSCs throughout larval development and in adults (9). To ask whether lst-1 and sygl-1 were similarly required in later development, we treated wild-type L4 larvae with RNAi and then scored their germ lines as adults (2 d later) (Fig. S5A). Features diagnostic of GSCs were scored as follows: (i), presence of a mitotic zone (25); (ii), presence of mitotic marker REC-8 (26) in distal germ cells (Fig. 2 G and H); and (iii), absence of meiotic marker HIM-3 (27) in the same germ cells (Fig. S5 C and D). When treated with either control RNAi (empty vector), lst-1 RNAi, or sygl-1 RNAi (each mixed 1:1 with empty vector), all adult distal germ lines maintained a mitotic zone (Fig. 2G and Fig. S5 B and C). However, when treated with lst-1 sygl-1 double RNAi, adult germ lines lost their mitotic zone and instead contained only differentiating germ cells in the meiotic cell cycle (Fig. 2H and Fig. S5B). In addition, these lst-1 sygl-1 double RNAi adult germ lines lost REC-8 staining and gained HIM-3 staining to the distal end (Fig. 2H and Fig. S5D). We conclude that lst-1 and sygl-1 function in adults to promote GSC self-renewal. Therefore, like GLP-1/Notch signaling, lst-1 and sygl-1 are essential for GSC self-renewal and function in larvae, adults, and both sexes.
lst-1 sygl-1 Is Epistatic to glp-1(gf) but Not gld-2 gld-1 Germ-Line Tumors.
If lst-1 and sygl-1 are critical GLP-1/Notch targets for GSC self-renewal, they should act downstream of GLP-1/Notch signaling and upstream of GLD regulators promoting meiotic differentiation (Fig. S6A). We first explored these predictions genetically, asking whether the lst-1 sygl-1 Glp phenotype was epistatic to the germ-line tumorous phenotype of two key mutants. First was the constitutively active gain-of-function allele glp-1(oz112gf), which encodes an unregulated GLP-1/Notch receptor and drives germ-line tumors independently of signaling ligand (28). All glp-1(gf) mutants made germ-line tumors when treated with control RNAi (Fig. S6 B and F), but >90% of glp-1(gf); lst-1(RNAi) sygl-1(RNAi) animals produced tiny germ lines with only sperm (Fig. S6 C and F). Because lst-1 and sygl-1 are required for glp-1(gf) to drive tumor formation, they likely do not act upstream, but instead act either downstream or in parallel to GLP-1/Notch signaling. Second was the gld-2 gld-1 double null mutant, which makes germ-line tumors because of a failure to enter the meiotic cell cycle and differentiate (29). Like the gld-2 gld-1 double mutant, all gld-2 gld-1 lst-1 sygl-1 quadruple mutants formed germ cell tumors (Fig. S6 D–F). Therefore, lst-1 and sygl-1 likely work upstream of gld-1 and gld-2. These epistasis results are consistent with the idea that lst-1 and sygl-1 are critical GLP-1/Notch targets for GSC self-renewal.
GLP-1/Notch Signaling Activates lst-1 and sygl-1 Expression Within the Niche.
To test the idea that lst-1 and sygl-1 function downstream of GLP-1/Notch signaling, we asked whether GLP-1/Notch signaling activates their expression. When assayed by mRNA in situ hybridization, both lst-1 and sygl-1 mRNAs localized to the distal-most germ line, within the GSC niche (Fig. 3 A and B, arrows); both genes are also expressed more proximally in developing oocytes (Fig. 3 A and B), which became useful as a control in later experiments. To ask whether their niche expression relies on GLP-1/Notch signaling, we took advantage of the gld-2 gld-1 double mutant, which generates a tumorous germ line independently of GLP-1/Notch signaling (29) and also independently of lst-1 and sygl-1 (Fig. S6 D–F). When GLP-1/Notch signaling was normal and active, lst-1 and sygl-1 mRNAs were both expressed in germ cells within the niche of gld-2 gld-1 double mutants, but when GLP-1/Notch signaling was eliminated, both mRNAs became undetectable (Fig. 3 C and D). Therefore, lst-1 and sygl-1 expression in the GSC niche depends on GLP-1/Notch signaling.
Fig. 3.
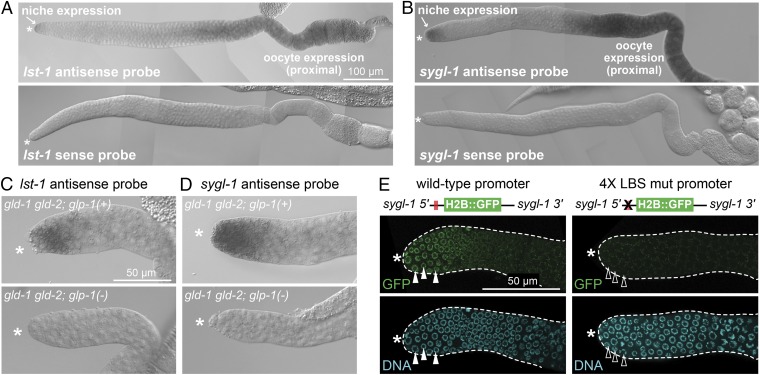
lst-1 and sygl-1 are targets of GLP-1/Notch activation. (A and B) lst-1 and sygl-1 mRNA expression in wild-type young adult gonads. (C and D) Expression of lst-1 and sygl-1 in GSCs within the niche requires GLP-1/Notch. Shown are L4 gonads. GLP-1(+): lst-1, 97% positive (n = 37); sygl-1, 97% positive (n = 36). GLP-1(-): lst-1, 0% positive (n = 28); sygl-1, 0% positive (n = 33). (E) Wild-type sygl-1 promoter drives reporter expression in germ cells within the niche (100% GFP-positive; n = 45), whereas 4× LBS mut sygl-1 promoter does not (0% distal positive; n = 46). Filled white triangles mark nuclei positive for H2B::GFP; empty white triangles mark nuclei negative for H2B::GFP. Because reporters were occasionally silenced, only germ lines expressing GFP proximally were scored (SI Materials and Methods). (A–E) Asterisk marks distal end.
LAG-1 Binding Sequences in sygl-1 Promoter Required for Expression Within Niche.
GLP-1/Notch signaling activates transcription via the LAG-1/CSL DNA-binding protein and its binding to the LAG-1 binding sequence (LBS) (30). The lst-1 LBSs were previously shown to mediate Notch activation in the soma (17), and, given the reliance of lst-1 expression on GLP-1/Notch signaling in the distal germ line (Fig. 3C), it seemed likely that the lst-1 LBSs would also be critical for Notch activation in the distal germ line. We therefore turned to sygl-1 and asked whether its LBSs were critical for expression within the niche. For this experiment, we generated a Mos1-mediated single copy insertion of a Histone 2B::GFP reporter transgene driven by the sygl-1 promoter, which harbors a cluster of four LBSs (Figs. 1C and 3E) (Materials and Methods). The wild-type sygl-1 promoter drove expression of Histone 2B::GFP in the distal-most germ cells (Fig. 3E), but that expression was eliminated when all four LBSs were mutated from the consensus RTGGGAA to RACGGAA, a sequence that LAG-1 cannot bind in vitro (30) (Fig. 3E). The lack of distal expression was not attributable to gene silencing, as the LBS mutant reporter supported proximal expression (Fig. S7). Therefore, the sygl-1 LBSs are responsible for expression distally in the niche but not proximally in oocytes.
lst-1 and sygl-1 Are Not Required for GLP-1/Notch Signaling Per Se.
One potential caveat to the model that lst-1 and sygl-1 function downstream of GLP-1/Notch to regulate GSCs might have been that they encode essential components of Notch signaling rather than encoding key downstream stem cell regulators. To explore this concern, we asked whether GLP-1/Notch signaling remains functional in the lst-1 sygl-1 double mutant.
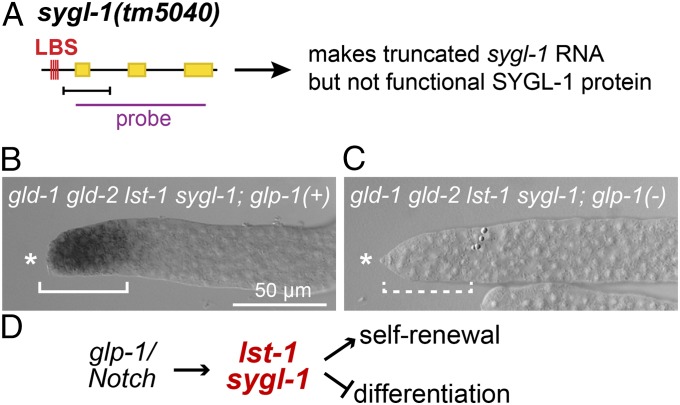
Our assay took advantage of the gld-2 gld-1 lst-1 sygl-1 germ-line tumor (Fig. S6E), which carries an ideal reporter of GLP-1/Notch signaling: the sygl-1(tm5040) deletion mutant retains its LBS cluster and generates a truncated sygl-1(tm5040) mRNA that lacks its translational start codon and lacks wild-type SYGL-1 activity (Figs. 1C and 4A and Fig. S2B). The gld-2 gld-1 lst-1 sygl-1 quadruple mutant expressed this GLP-1/Notch reporter in germ cells within the niche as expected (Fig. 4B). More importantly, that reporter expression required GLP-1/Notch signaling: when signaling was eliminated, reporter expression was no longer detected in the niche (Fig. 4C). Therefore, GLP-1/Notch signaling is active and functional in animals harboring both lst-1 and sygl-1 deletions. We conclude that lst-1 and sygl-1 deletions do not affect GLP-1/Notch signaling per se even though their double-mutant phenotype mimics a complete loss GLP-1/Notch signaling. Therefore, these two pivotal stem cell regulators are targets of GLP-1/Notch signaling and function downstream of GLP-1/Notch signaling to promote GSC self-renewal (Fig. 4D).
Fig. 4.
lst-1 and sygl-1 do not affect GLP-1/Notch signaling. (A) GLP-1/Notch reporter assay exploits sygl-1(tm5040) deletion mutant (see Results). Conventions same as Fig. 1C. (B and C) L4 hermaphrodite gonads were probed for GLP-1/Notch reporter by in situ hybridization. Shown are representative images of distal gonads. Both mutants have tumorous germ lines that appear the same. Asterisk marks distal end. (B) The gld-2 gld-1 lst-1 sygl-1 quadruple mutant stains positively for the GLP-1/Notch reporter (solid bracket) (91% positive; n = 35). (C) The gld-2 gld-1 lst-1 sygl-1; glp-1 quintuple mutant does not stain positively for the GLP-1/Notch reporter (dashed bracket) (0% positive; n = 27). (D) Model for lst-1 and sygl-1 function in the GSC self-renewal pathway.
Discussion
A central question in stem cell biology is how niche signaling maintains stem cells. Here, we identify two genes, lst-1 and sygl-1, which are activated by GLP-1/Notch signaling and function redundantly to maintain the stem cell state. Remarkably, these two genes fully account for the stem cell function of GLP-1/Notch niche signaling. Both lst-1 and sygl-1 encode novel proteins and therefore may represent a new class of stem cell regulators. Here, we place this discovery in context and discuss its implications.
The DTC Niche and GLP-1/Notch Signaling Maintain GSCs by Activation of Two Redundant Genes, lst-1 and sygl-1.
The DTC niche was discovered decades ago and its use of GLP-1/Notch signaling to maintain GSCs was found soon thereafter (see Introduction). A longstanding question has been how the DTC niche and GLP-1/Notch signaling promote GSC self-renewal. A partial answer is fbf-2 activation (11), but FBF only maintains GSCs in adults, whereas the DTC niche and GLP-1/Notch signaling maintain them in both larvae and adults (9, 13). Therefore, a larval GSC-promoting activity was predicted to exist—an activity that would complement the previously found adult GSC-promoting activity of FBF. Our search for this additional GSC-promoting activity was guided by several predictions.
Two straightforward predictions were that this gene would be a direct target of GLP-1/Notch signaling and that its RNA would be expressed in the distal-most GSCs within their niche. Another prediction, which was based on the striking failure to find this gene despite extensive mutant screens, was that it likely functioned redundantly with at least one other gene. A final and more risky prediction was that it might be an FBF target, a highly speculative prediction that turned out to be crucial. A comparison of candidate Notch and FBF targets identified 15 in common, one of which was expressed in germ cells within the niche according to an extensive in situ hybridization database; RNAi directed against that 1 gene plus the 14 others led to identification of lst-1 and sygl-1, two genes required for GSC self-renewal in larvae. Follow-up experiments demonstrated that a double mutant harboring deletions of both lst-1 and sygl-1 abolished GSC self-renewal in a manner indistinguishable from a glp-1 null mutant. However, removal of either gene alone, using either RNAi or a deletion mutant, did not affect GSCs, confirming the prediction of redundancy for genes driving the larval GSC-promoting activity.
Surprisingly, lst-1 and sygl-1 are not larval-specific but instead are essential for the GSC state in both larvae and adults. Thus, in addition to the dramatic GSC loss found upon deletion of the two genes, adult GSCs fail to self-renew when treated later in development with RNAi against both lst-1 and sygl-1. The lst-1 gene was previously identified as a target of Notch signaling in the soma (17). Here, we show that lst-1 and sygl-1 are also regulated by GLP-1/Notch signaling in GSCs within the niche and that lst-1 and sygl-1 function downstream of GLP-1/Notch signaling rather than as an integral component of signaling per se. Together these findings demonstrate that the niche and GLP-1/Notch signaling maintain GSCs by activation of lst-1 and sygl-1 and that these two genes are essential effectors of niche signaling for GSC self-renewal. We do not yet know whether overexpression of lst-1 or sygl-1 is sufficient for GSC self-renewal, because transgenes driving them in an unregulated fashion could not be generated. Nonetheless, their dramatic loss-of-function phenotype demonstrates unambiguously that lst-1 and sygl-1 are critical downstream targets of niche signaling that are essential for all GSC self-renewal.
How Might LST-1 and SYGL-1 Promote GSC Maintenance?
A major question emerging from this work is how LST-1 and SYGL-1 proteins promote GSC self-renewal. Few clues come from their amino acid sequences. LST-1 harbors a single C-terminal Nanos-like zinc finger, suggesting RNA binding activity. However, LST-1 is not a canonical Nanos protein, which possesses two zinc fingers in tandem (31); moreover, zinc fingers can perform many molecular functions in addition to RNA binding (32). The LST-1L protein also harbors a predicted signal sequence, implying a possible membrane association or secretion of this isoform. The only other features of note are low complexity sequences (LCSs) in both LST-1 and SYGL-1. In other proteins, LCSs can promote assembly of RNA-protein granules (33), consistent with the possibility of a role in RNA regulation. However, ideas for LST-1 and SYGL-1 molecular activities remain highly speculative at the current time.
The lst-1 and sygl-1 amino acid sequences are not conserved outside of nematodes. As such, these two key stem cell regulators may have evolved recently and represent a new mechanism for stem cell regulation, or they may represent a broadly conserved mechanism of stem cell regulation that relies on proteins whose amino acid sequences are not recognizable across evolution. Instructive cases of proteins not recognizable by amino acid sequence similarity have been found in other pathways. For example, C. elegans SYS-1 encodes a β-catenin whose amino acid sequence bears little resemblance to canonical homologs; its discovery therefore relied on genetic phenotype and X-ray crystal structure rather than amino acid similarity (34). Here, discovery of LST-1 and SYGL-1 required a well-defined niche/stem cell system in a genetically tractable organism, sophisticated knowledge of Notch and FBF targets and the motivation to probe for redundant functions. To our knowledge, similar searches have not been applied broadly. Although not yet definitive, we favor the idea that LST-1 and SYGL-1 are pioneers for a previously unidentified class of stem cell regulators.
Notch Target Genes and Stem Cell Self-Renewal.
In addition to C. elegans GSCs, other types of stem cells also depend on Notch signaling for self-renewal (see Introduction) (reviewed in ref. 8). Critical downstream targets have also been identified, the most well-studied being genes encoding the Hes/Hey family of bHLH transcription factors. For example, mouse embryos with a conditional brain-specific CSL knockout lose virtually all neural stem cells (NSCs) (35) and Hes1; Hes3; Hes5 triple-mutant embryos similarly lose NSCs in many parts of the developing brain (36), consistent with the idea that these transcription factors are key targets of niche signaling. However, a brain-specific CSL knockout has a broader effect on NSCs than found in the Hes1; Hes3; Hes5 triple mutant (35, 37), and a muscle-specific CSL knockout similarly has a more dramatic defect than a mutant lacking key Hes genes (38, 39). Therefore, removal of known downstream Notch targets in brain and muscle stem cells does not fully recapitulate the effect of removing the DNA-binding protein responsible for most Notch signaling. This discrepancy may be explained by additional targets or by a lack of congruence between Notch signaling and CSL activity (reviewed in ref. 40). Regardless, our work reports two GLP-1/Notch targets that do in fact fully recapitulate the effect of this canonical signaling pathway on stem cell maintenance. Indeed, lst-1 and sygl-1 provide a previously unidentified example of Notch target genes that fully explains the effects of Notch signaling on stem cells of a particular tissue. A fascinating possibility is that similarly pivotal targets of niche signaling exist in other stem cell systems.
Materials and Methods
Nematode Culture and RNAi.
Strains were maintained at 20 °C following standard protocols (41), except for H2B::GFP reporter transgenic lines, which were maintained at 25 °C. Wild type was the N2 Bristol strain. See SI Materials and Methods for a full list of strains and alleles used in this study. RNAi feeding experiments were carried out following established protocols (42). For multiple gene knockdowns, HT115 bacteria containing lst-1, sygl-1, and empty (pL4440) RNAi vectors were grown separately in overnight cultures and then seeded to RNAi plates in equal volumes.
Immunocytochemistry and mRNA in Situ Hybridization.
Antibody staining of dissected gonads was carried out as described (15), and staining of whole animals was carried out as described (43). See SI Materials and Methods for protocols. mRNA in situ hybridization was performed on dissected gonads from either adults grown to 24 h post L4 stage or L4 larvae, using digoxigenin-labeled DNA probes (44). See SI Materials and Methods for protocols.
Transgenic C. elegans.
Transgenes were inserted into the genome using the Mos1-mediated single copy insertion method (45) (SI Materials and Methods). The presence of H2B::GFP was scored in unfixed dissected gonads in PBS plus 0.1% Tween-20, 0.25 mM Levamisole, and Hoechst 33342 (Invitrogen) diluted 1:10,000 and visualized using the Zeiss Axioimager microscope.
Supplementary Material
Acknowledgments
We thank the C. elegans knockout consortia for deletions, the Caenorhabditis Genetics Center [funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (Grant P40 OD010440)] for strains, Susan Strome for PGL-1 and SP56 antibodies, the Developmental Studies Hybridoma Bank for the anti–DAO-5 antibody, and Monica Zetka for anti–HIM-3 antibodies. We thank the members of the J.K. and Wickens laboratories for insightful comments. J.K. was supported by NIH Grant GM069454 and is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401861111/-/DCSupplemental.
References
- 1.Morrison SJ, Spradling AC. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander AD, et al. What does the concept of the stem cell niche really mean today? BMC Biol. 2012;10:19. doi: 10.1186/1741-7007-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81(2):208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- 4.Xie T, Spradling AC. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 2000;290(5490):328–330. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]
- 5.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294(5551):2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 6.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopan R. Notch signaling. Cold Spring Harb Perspect Biol. 2012;4(10):pii: a011213. doi: 10.1101/cshperspect.a011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140(4):689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 9.Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51(4):589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 10.Morgan DE, Crittenden SL, Kimble J. The C. elegans adult male germline: Stem cells and sexual dimorphism. Dev Biol. 2010;346(2):204–214. doi: 10.1016/j.ydbio.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7(5):697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Kershner A, et al. Germline stem cells and their regulation in the nematode Caenorhabditis elegans. Adv Exp Med Biol. 2013;786:29–46. doi: 10.1007/978-94-007-6621-1_3. [DOI] [PubMed] [Google Scholar]
- 13.Crittenden SL, et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417(6889):660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 14.Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science. 2001;291(5506):1055–1058. doi: 10.1126/science.1055642. [DOI] [PubMed] [Google Scholar]
- 15.Lee M-H, Hook B, Lamont LB, Wickens M, Kimble J. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans. EMBO J. 2006;25(1):88–96. doi: 10.1038/sj.emboj.7600901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell. 2006;17(7):3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303(5658):663–666. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 18.Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci USA. 2010;107(8):3936–3941. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumsta C, Hansen M. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS ONE. 2012;7(5):e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 21.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 22.Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Comput Chem. 1993;17(2):149–163. [Google Scholar]
- 23.Mizianty MJ, Peng Z, Kurgan L. MFDp2: Accurate predictor of disorder in proteins by fusion of disorder probabilities, content and profiles. Intrinsically Disord Proteins. 2013;1(1):e24428. doi: 10.4161/idp.24428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh K, et al. C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr Biol. 2011;21(10):825–834. doi: 10.1016/j.cub.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crittenden SL, Troemel ER, Evans TC, Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994;120(10):2901–2911. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- 26.Hansen D, Hubbard EJA, Schedl T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol. 2004;268(2):342–357. doi: 10.1016/j.ydbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 27.Zetka MC, Kawasaki I, Strome S, Müller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13(17):2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry LW, Westlund B, Schedl T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development. 1997;124(4):925–936. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]
- 29.Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125(10):1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- 30.Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H) Development. 1996;122(5):1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 31.Curtis D, et al. A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J. 1997;16(4):834–843. doi: 10.1093/emboj/16.4.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laity JH, Lee BM, Wright PE. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11(1):39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 33.Kato M, et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips BT, Kimble J. A new look at TCF and β-catenin through the lens of a divergent C. elegans Wnt pathway. Dev Cell. 2009;17(1):27–34. doi: 10.1016/j.devcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30(9):3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatakeyama J, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131(22):5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 37.Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135(15):2531–2541. doi: 10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- 38.Vasyutina E, et al. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci USA. 2007;104(11):4443–4448. doi: 10.1073/pnas.0610647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukada S, et al. Hesr1 and Hesr3 are essential to generate undifferentiated quiescent satellite cells and to maintain satellite cell numbers. Development. 2011;138(21):4609–4619. doi: 10.1242/dev.067165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson JE, Macdonald RJ. Notch-independent functions of CSL. Curr Top Dev Biol. 2011;97:55–74. doi: 10.1016/B978-0-12-385975-4.00009-7. [DOI] [PubMed] [Google Scholar]
- 41.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahringer J (2006) Reverse genetics. WormBook (The C. elegans Research Community). Available at www.wormbook.org.
- 43. Duerr JS (2006) Immunohistochemistry. WormBook (The C. elegans Research Community). Available at www.wormbook.org.
- 44. Lee M-H, Schedl T (2006) RNA in situ hybridization of dissected gonads. WormBook (The C. elegans Research Community). Available at www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 45.Frøkjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40(11):1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.