Abstract
The association between the polymorphism of cyclin D1 (CCND1) and esophageal carcinogenicity has been widely examined, however, it remains controversial. To evaluate the importance of CCND1 G870A polymorphism with regard to the risk of esophageal cancer, a meta-analysis was carried out that reviewed the available literature in order to clarify the controversies. This meta-analysis included 1,154 cases and 1,678 controls for CCND1 G870A polymorphism from seven published case-control studies. The odds ratio (OR) and 95% confidence interval (CI) were calculated using the Stata software version 11.1. The results were pooled using a dominant model to appropriately reflect a biological model of the genetic effect. No significant association was observed in the Caucasian (OR=1.64; 95% CI, 0.84–3.20) or the Asian populations (OR=1.30; 95% CI, 0.65–2.62), while no significant association was found in esophageal squamous cell carcinoma (ESCC) (OR=1.74; 95% CI, 0.79–3.81) or esophageal adenocarcinoma (EADC) (OR=1.18; 95% CI, 0.77–1.81). However, the comparison of A vs. G in CCND1 G870A showed significant differential susceptibility to esophageal cancer (OR=1.26; 95% CI, 1.00–1.59). These findings suggested that the CCND1 G870A polymorphism has no association with esophageal cancer risk in ethnicity and histology, respectively. Further studies are required to assess these associations in greater detail.
Keywords: cyclin D1, G870A, polymorphism, esophageal cancer, meta-analysis
Introduction
Esophageal cancer is a global health problem that ranked eighth in terms of incidence and sixth in terms of mortality in 2002 (1). Esophageal tumors usually lead to dysphagia, pain and other symptoms and are diagnosed via a biopsy. Esophageal cancer is divided into squamous cell carcinoma and adenocarcinoma. Squamous cell carcinoma arises from the cells that line the upper part of the esophagus. Adenocarcinoma arises from glandular cells that are present at the junction of the esophagus and stomach (2). Genetic as well as environmental factors play a role in the carcinogenesis of esophageal cancer (3,4). Individual variations in cancer risk have been associated with specific polymorphisms of various genes that are present in a significant proportion of the normal population.
Cyclin D1 (CCND1) is a mitogenic sensor for the cell cycle mechanism and cellular oncogene (5). High activity of cyclin D1 can lead to chemoresistance due to the dual roles of cyclin D1 in promoting cell proliferation and inhibiting drug-induced apoptosis (6). Overexpression of cyclin D1 contributes to immature cell passage through the G1-S transition, induces propagation of unrepaired DNA damage and the accumulation of genetic errors and a selective growth advantage for the altered cells (7). Cyclin D1 are important regulators of the cell cycle and apoptosis and each contains functional single nucleotide polymorphisms (SNPs) (CCND1 G870A) that are involved in the susceptibility and outcome of various human malignancies (8).
Poor statistical power has been controversial for data from individually published studies. The aim of this study was to evaluate the association between CCND1 G870A and esophageal cancer risk in a large-scale case-control study. However, we analyzed two subgroups based on ethnicity and histology, in which the variant alleles potentially exert a stronger biologic effect.
Materials and methods
Search strategy
Relevant studies were selected by searching PubMed, EMBASE and HuGENet databases prior to July 1, 2012. A highly sensitive search strategy was used to identify relevant articles exploring the following terms or their combination: ‘CCND1’, ‘cyclin D1’, ‘PRAD1’, ‘polymorphism’, ‘G870A’, ‘rs603965’ and ‘esophageal cancer’. Reference lists of identified studies and related reviews and proceedings of international meetings were reviewed to ensure identification of all potentially eligible studies.
Eligibility criteria
Identified studies included in our meta-analysis had to be: i) originally on CCND1 G870A poly morphism and esophageal cancer risk; ii) a study to obtain G870A genotypes and the genotype distributions being reported separately; iii) case-control or cohort studies; iv) on a control population without severe respiratory disease and malignant tumor patients and v) a clear information source of the population.
Data extraction
From each eligible study, we extracted the following information: first author, year of publication, country of origin, ethnicity, sum of cases and controls with genotypes, diagnostic standards and genotype methods.
Statistical analysis
Funnel plots, together with the Begg’s rank correlation test, were used to assess publication bias. Additionally, a subgroup analysis of the Caucasian and Asian populations was performed to account for the subpopulation structure. The effect of these polymorphisms on esophageal cancer risk was estimated using a logistic regression analysis to obtain pooled estimates of the odds ratio (OR) and a 95% confidence interval (CI). The most plausible gene effect model was determined without assuming a priority genetic model to avoid multiple comparisons.
First, the Pearson’s Chi-square test (P≥0.05) (9) was used to check for deviations from the Hardy-Weinberg equilibrium (HWE) in the controls in each study. The measurement of the overall inconsistency index (I2) (10) was then carried out to describe the proportion of total variation caused by heterogeneity (11). Meta-regression was performed to explore potential heterogeneity in various types of study designs. The I2 test detected mainly moderate heterogeneity for the SNPs, thus a logistic regression was performed to evaluate gene effects via the random-effects model.
The parameters θ2 and θ3 were calculated using the formula: log it (πij)= αi+θ2 zi2+θ3 zi3, and ORAB/AA= exp (θ2), ORBB/AA= exp (θ2), αi are indicators of the study-specific fixed-effects, and θ2 and θ3 are dummy variables of genotypes AB and BB. The appropriate genetic model was identified using the criteria (12): i) no association: θ2=θ3 (ORAB/AA=ORBB/AA=1); ii) dominant model: θ2≠0, θ3≠0 and θ2=θ3 (ORAB/AA=ORBB/AA≠1); iii) recessive model: θ2=0 (ORAB/AA=1) and θ3≠0 (ORBB/AA≠1); iv) co-dominant model: θ2≠0, θ3≠0 and 2θ2=θ3 (OR2AB/AA=ORBB/AA).
Metagen (http://bioinformatics.biol.uoa.gr/~pbagos/metagen/) was used by selecting the genetic model. Statistical analyses were carried out using the Stata software version 11.1 (Stata Corporation, College Station, TX, USA). The statistical evaluations were carried out using a two-sided test with a significance level of 0.05.
Results
Study inclusion and characteristics
Based on the inclusion criteria, we searched the PubMed, EMBASE and HuGENet databases by reading titles and abstracts. Ten original study articles concerning the association between CCND1 G870A polymorphism and esophageal cancer risk were retrieved. The full text of these articles was reviewed intensively and three articles without the control group were excluded. Thus, seven case-control studies (1,154 esophageal cancer cases and 1,678 controls) on the association between CCND1 G870 polymorphism and esophageal cancer risk were included in this meta-analysis (13–19). Of these included articles, four studies were on Caucasians (15,16,18,19) and three studies were on Asians (13,14,17).
The eligible studies were consistent with HWE. Their characteristics are shown in Table I, and frequency of CCND1 G870A polymorphism in various populations are listed in Table II. Casson et al (15), Jain et al (17) and Hussain et al (19) showed a significant association between this polymorphism and esophageal cancer. However, four other studies demonstrated no significant association between this polymorphism and esophageal cancer (13,14,16,18).
Table I.
Characteristics of all eligible studies in this meta-analysis.
| Eligible subjects | Selective characteristics of cases and controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Authors | Year | Country | Ethnicity | Cases | Controls | Cases | Controls | Source of controls | Method | Ref. |
| Yu, et al | 2003 | China | Asian | 321 | 345 | Most of the cases and controls have been characterized in a molecular epidemiological study | Population controls were accrued from a database of the nutritional survey conducted in the same regions | Population | PCR-RFLP | (13) |
| Zhang, et al | 2003 | China | Asian | 120 | 183 | Histological tumor typing was carried out on the basis of the resection specimens in the Department of Pathology of the same hospital | The healthy controls were recruited from blood donors | Population | PCR | (14) |
| Casson, et al | 2005 | Canada | Caucasian | 56 | 95 | Cases that underwent diagnostic esophagogastroscopy were asked to provide three additional biopsy samples, in addition to the usual number of biopsies, which were placed in formalin for immunohistochemistry and routine histopathologic diagnosis | Patients who attended for consultation regarding unrelated, benign conditions were screened for GERD-related symptoms, a history of ‘hiatus hernia’, ‘dyspepsia’, antacid use; if negative, patients were asked to participate as controls | Hospital-based | PCR | (15) |
| Geddert, et al | 2005 | Germany | Caucasian | 56 | 253 | Tumors with their epicenter in the esophagus were regarded as esophageal in origin | The healthy controls were recruited from blood donors | Population | PCR | (16) |
| Jain, et al | 2007 | India | Asian | 151 | 201 | Histologically confirmed esophageal cancer patients of squamous cell carcinoma or adenocarcinoma were inducted into this study. | During the same period, blood samples of 201 unrelated controls were randomly selected from the outpatient clinics | Hospital-based | PCR-RFLP | (17) |
| Liu, et al | 2010 | USA | Caucasian | 299 | 450 | Patients with histologically confirmed EADC | Healthy unrelated age-, gender-, and gender-matched visitor controls with no history of cancer or gastroesophageal reflux disease were recruited from the same institutions | Hospital-based | TaqMan assays | (18) |
| Hussain, et al | 2011 | India | Caucasian | 151 | 151 | This study included a total of 151 histologically confirmed, untreated squamous cell carcinoma patients | The controls were recruited from a healthy Indian population | Population | PCR-RFLP | (19) |
PCR-RFLP, polymerase chain reaction-restriction fragment length; PCR, polymerase chain reaction; GERD, gastroesophageal reflux disease; EADC, esophageal adenocarcinoma.
Table II.
Heterogeneity test showing frequency of CCND1 G870A polymorphism in various populations.
| Cases (%)
|
Controls (%)
|
P-value for HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Authors | Year | Ethnicity | GG | GA | AA | GG | GA | AA | Refs. | |
| Yu, et al | 2003 | Asian | 21.2 | 48.9 | 29.9 | 16.8 | 51.3 | 31.9 | 0.354 | (13) |
| Zhang, et al | 2003 | Asian | 9.2 | 61.7 | 29.2 | 20.8 | 55.7 | 23.5 | 0.118 | (14) |
| Casson, et al | 2005 | Caucasian | 21.4 | 48.2 | 30.4 | 36.8 | 54.7 | 8.4 | 0.063 | (15) |
| Geddert, et al | 2005 | Caucasian | 28.6 | 46.4 | 25.0 | 24.9 | 53.8 | 21.3 | 0.224 | (16) |
| Jain, et al | 2007 | Asian | 14.6 | 50.3 | 35.1 | 18.4 | 55.2 | 26.4 | 0.114 | (17) |
| Liu, et al | 2010 | Caucasian | 26.4 | 51.5 | 22.1 | 28.4 | 47.8 | 23.8 | 0.369 | (18) |
| Hussain, et al | 2011 | Caucasian | 13.2 | 65.6 | 21.2 | 37.1 | 47.7 | 15.2 | 0.986 | (19) |
CCND1, cyclin D1; HWE, Hardy-Weinberg equilibrium.
Meta-analysis results
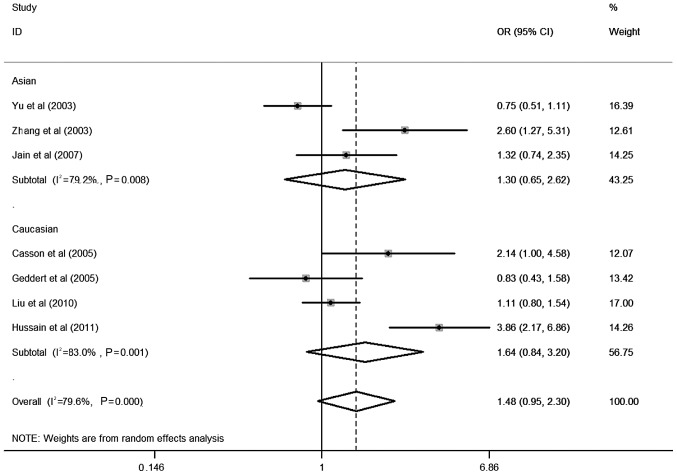
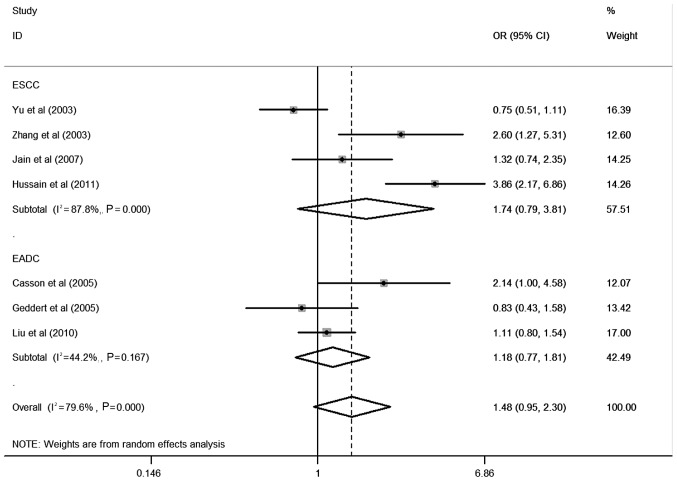
Table II shows the detailed results of the heterogeneity test, the most appropriate genetic model and the association between CCND1 G870A polymorphism and esophageal cancer risk evaluated using OR with 95% CI. In the overall analysis, using the random-effects model, significant associations were detected in the comparison of A vs. G in CCND1 G870A polymorphism and esophageal cancer risk were observed in dominant models (OR=1.26; 95% CI, 1.00–1.59). Moreover, in stratified studies based on ethnicity, no significant association was found in Caucasian (OR=1.64; 95% CI, 0.84–3.20) or Asian populations (OR=1.30; 95% CI, 0.65–2.62) (Fig. 1), and no significant association was found in esophageal squamous cell carcinoma (ESCC) (OR=1.74; 95% CI, 0.79–3.81) and esophageal adenocarcinoma (EADC) (OR=1.18; 95% CI, 0.77–1.81) (Fig. 2).
Figure 1.
Forest plot (random-effects analysis) of esophageal cancer risk associated with CCND1 G870A polymorphism based on ethnicity using a dominant genetic model.
Figure 2.
Forest plot (random-effects analysis) of esophageal cancer risk associated with CCND1 G870A polymorphism based on histology using a dominant genetic model.
Publication bias
The Begg’s rank correlation test and funnel plots were performed to access the publication bias. No evident asymmetry was observed in the shapes of the funnel plots, indicating no evidence of publication bias in this meta-analysis (t=1.55, P=0.182).
Discussion
The aim of this study was to elucidate the association between the CCND1 G870A polymorphism and esophageal cancer risk. A meta-analysis published in 2011, showed that the CCND1 G870A genotype exhibited a statistically significant risk for cancers of the digestive tract (20). In the present study we explored the association between CCND1 G870A polymorphism and the risk of esophageal cancer based on various ethnicities and histology in the most appropriate genetic model and detected no significant associations in Caucasians and Asians. Additionally, no significant association was found in ESCC and EADC, whereas the comparison of A vs. G in CCND1 G870A showed a significant differential susceptibility to esophageal cancer. However, the reason for the results of the CCND1 polymorphism is unclear. Environmental differences and genetic backgrounds play a role in the association between the CCND1 G870A polymorphism and the risk for esophageal cancer. Notably, the results of our meta-analysis were inconsistent with the findings of Chen et al (20), likely due to the different genetic model selected. We identified the most appropriate genetic model using the method described by Bagos et al (11), which allows for the assessment of hetero geneity, thus our result is likely to be more accurate. Additionally, we considered the association between CCND1 G870A polymorphism and the risk of esophageal cancer rather than cancers of the digestive tract.
Heterogeneity is inevitable in a meta-analysis (21,22), and was therefore also evident in our meta-analysis. Sources of heterogeneity may be derived from various channels: studies included in this meta-analysis cover various ethnicities and environments. Moreover, various methodologies including source of the controls, diagnostic criteria and genotypic methods may lead to heterogeneity.
Additional possible limitations should be taken into account and contributed to the poor statistical power of this meta-analysis. For example, we only selected articles published electronically from databases in English and excluded articles published in other languages, in print or not published at all. Although the Begg’s funnel plot and the Egger’s test did not detect it, publish bias may exist. Moreover, since not all the included articles contain complete data, a subgroup analysis should be conducted.
In conclusion, the present meta-analysis has demonstrated that the comparison of A vs. G in the CCND1 G870A polymorphism may increase the risk of esophageal cancer. Considering the limitations of this meta-analysis, additional large-scale investigations, as well as well-designed and more accurate methods of genotypic are required to confirm the association between CCND1 G870A and esophageal cancer risk under the complex landscape of the cell cycle and cancer risk.
Acknowledgments
This study was supported by a grant from the Science Technology Research and New Product trial of the Guangxi Zhuang Autonomous Region (to Mingwu Chen) (no. 10124001A-47).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 3.Corley DA, Buffler PA. Oesophageal and gastric cardia adenocarcinomas: analysis of regional variation using the Cancer Incidence in Five Continents database. Int J Epidemiol. 2001;30:1415–1425. doi: 10.1093/ije/30.6.1415. [DOI] [PubMed] [Google Scholar]
- 4.Hiyama T, Tanaka S, Shima H, Kose K, Kitadai Y, Ito M, Sumii M, Yoshihara M, Shimamoto F, Haruma K, Chayama K. Somatic mutation of mitochondrial DNA in Helicobacter pylori-associated chronic gastritis in patients with and without gastric cancer. Int J Mol Med. 2003;12:169–174. [PubMed] [Google Scholar]
- 5.Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 6.Biliran H, Jr, Wang Y, Banerjee S, Xu H, Heng H, Thakur A, Bollig A, Sarkar FH, Liao JD. Overexpression of cyclin D1 promotes tumor cell growth and confers resistance to cisplatin-mediated apoptosis in an elastase-myc transgene-expressing pancreatic tumor cell line. Clin Cancer Res. 2005;11:6075–6086. doi: 10.1158/1078-0432.CCR-04-2419. [DOI] [PubMed] [Google Scholar]
- 7.Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 8.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 9.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagos PG, Nikolopoulos GK. A method for meta-analysis of case-control genetic association studies using logistic regression. Stat Appl Genet Mol Biol. 2007;6 doi: 10.2202/1544-6115.1281. Article17. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Ha TC, Tai BC. XRCC1 gene polymorphisms and breast cancer risk in different populations: a meta-analysis. Breast. 2009;18:183–191. doi: 10.1016/j.breast.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Yu C, Lu W, Tan W, Xing D, Liang G, Miao X, Lin D. Lack of association between CCND1 G870A polymorphism and risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2003;12:176. [PubMed] [Google Scholar]
- 14.Zhang J, Li Y, Wang R, Wen D, Sarbia M, Kuang G, Wu M, Wei L, He M, Zhang L, Wang S. Association of cyclin D1 (G870A) polymorphism with susceptibility to esophageal and gastric cardiac carcinoma in a northern Chinese population. Int J Cancer. 2003;105:281–284. doi: 10.1002/ijc.11067. [DOI] [PubMed] [Google Scholar]
- 15.Casson AG, Zheng Z, Evans SC, Geldenhuys L, van Zanten SV, Veugelers PJ, Porter GA, Guernsey DL. Cyclin D1 polymorphism (G870A) and risk for esophageal adenocarcinoma. Cancer. 2005;104:730–739. doi: 10.1002/cncr.21229. [DOI] [PubMed] [Google Scholar]
- 16.Geddert H, Kiel S, Zotz RB, Zhang J, Willers R, Gabbert HE, Sarbia M. Polymorphism of p16 INK4A and cyclin D1 in adenocarcinomas of the upper gastrointestinal tract. J Cancer Res Clin Oncol. 2005;131:803–808. doi: 10.1007/s00432-005-0021-4. [DOI] [PubMed] [Google Scholar]
- 17.Jain M, Kumar S, Lal P, Tiwari A, Ghoshal UC, Mittal B. Role of BCL2 (ala43thr), CCND1 (G870A) and FAS (A-670G) polymorphisms in modulating the risk of developing esophageal cancer. Cancer Detect Prev. 2007;31:225–232. doi: 10.1016/j.cdp.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Liu G, Cescon DW, Zhai R, Zhou W, Kulke MH, Ma C, Xu W, Su L, Asomaning K, Heist RS, et al. p53 Arg72Pro, MDM2 T309G and CCND1 G870A polymorphisms are not associated with susceptibility to esophageal adenocarcinoma. Dis Esophagus. 2010;23:36–39. doi: 10.1111/j.1442-2050.2009.00960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain S, M Y, Thakur N, Salam I, Singh N, Mir MM, Bhat MA, Siddiqi MA, Das BC, Bharadwaj M. Association of cyclin D1 gene polymorphisms with risk of esophageal squamous cell carcinoma in Kashmir Valley: a high risk area. Mol Carcinog. 2011;50:487–498. doi: 10.1002/mc.20732. [DOI] [PubMed] [Google Scholar]
- 20.Chen B, Cao L, Yang P, Zhou Y, Wu XT. Cyclin D1 (CCND1) G870A gene polymorphism is an ethnicity-dependent risk factor for digestive tract cancers: a meta-analysis comprising 20,271 subjects. Cancer Epidemiol. 2012;36:106–115. doi: 10.1016/j.canep.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Higgins JP. Commentary: heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2008;37:1158–1160. doi: 10.1093/ije/dyn204. [DOI] [PubMed] [Google Scholar]
- 22.Groenwold RH, Rovers MM, Lubsen J, van der Heijden GJ. Subgroup effects despite homogeneous heterogeneity test results. BMC Med Res Methodol. 2010;10:43. doi: 10.1186/1471-2288-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]