Abstract
MicroRNAs (miRs) contribute to cancer development and progression by acting as oncogenes and tumor suppressor genes. miR-9 family members (miR-9s), including miR-9-1, 9-2 and 9-3, have been shown to be oncogenically involved through the downregulation of E-cadherin expression, which promotes the epithelial-mesenchymal transition. Tumor suppressive roles of miR-9s have also been reported to silence miR-9 through methylation, which is associated with an shortened overall survival (OS) period in several types of cancer. In this study, the impact of miR-9s methylation on non-small cell lung cancers (NSCLC) was investigated. In total, 293 resected NSCLC samples were examined and the miR-9s methylation status was determined using a combined bisulfite restriction analysis. miR-9 expression was analyzed by in situ hybridization. Methylation of miR-9-1, 9-2 and 9-3 was present in 20 (7%), 33 (11%) and 34 (12%) of the cases, respectively. Methylation of any miR-9s (miR-9s methylation) was observed in 76 of the cases (26%), and miR-9 expression was silenced in cases with miR-9s methylation. Logistic regression analysis demonstrated that male gender [odds ratio (OR), 2.0; 95% confidence interval (95% CI), 1.1–3.6; P=0.01] and pathologically negative lymph node metastasis (OR, 4.8; 95% CI, 1.4–17.2; P=0.002) were independent relative factors for miR-9s methylation. Additionally, miR-9s methylation [hazard ratio (HR), 4.2; 95% CI, 1.2–27.0; P=0.026] and early pathological stage (HR, 8.3; 95% CI, 2.1–28.6; P=0.004) were found to be independent predictive factors for prolonged OS time by the Cox proportional hazard test. miR-9s methylation which induces expression silencing is common in NSCLC cases without lymph nodal metastasis, suggesting that miR-9s are oncogenically involved in NSCLC carcinogenesis through the promotion of tumor metastasis.
Keywords: lung cancer, non-small cell lung cancer, microRNA, miR-9, methylation, in situ hybridization
Introduction
Lung cancer is a leading cause of cancer-related mortality worldwide (1). Although previous studies have demonstrated the molecular pathogenesis of lung cancer, little progress has been made in terms of its prevention, diagnosis and treatment (2). Thus, further investigation of its molecular pathogenesis is required to reduce the mortality rate caused by lung cancer.
MicroRNAs (miRs) are a group of non-coding small RNAs mostly regulating their target mRNAs by post-transcriptional repression (3). They bind to the target mRNA transcript at the 3′-UTR, generating destabilization that inhibits protein translation (4). Intensive studies have been carried out investigating the involvement of miRs in human malignant tumors due to their ability to regulate several genes involved in multiple cancer pathways (5). miRs are highly involved in various cell processes, including proliferation, differentiation, apoptosis and development, by simultaneously controlling the expression levels of hundreds of genes (6,7). In cancer cells, both oncogenic and tumor suppressive miRs have been reported to contribute to cancer development and progression (7,8). Notably, certain miRs, such as miR-200s, have a biphasic role as oncogenes or tumor suppressor genes (TSGs) (9).
miR-9, including the family members miR-9-1, 9-2 and 9-3, is encoded by three chromosomal loci: miR-9-1 on chromosome (Chr) 1q22, miR-9-2 on Chr 5q14.3 and miR-9-3 on Chr 15q26.1. miR-9 has been reported to have both oncogenic and tumor suppressive roles. In its oncogenic function, miR-9 induces epithelial-mesenchymal transition via the direct suppression of E-cadherin mRNA (8), which downregulates β-catenin to promote vascular endothelial growth factor expression, leading to angiogenesis and tumor metastasis. Moreover, miR-9 is able to repress the expression of TSGs such as FOX1 (10) and CDX2 (11). By contrast, in its tumor suppressive role, the loss of miR-9 expression through methylation is associated with metastasis or poor prognosis in several types of cancer (12–14).
In this study, we determined the miR-9s methylation status in surgically resected non-small cell lung cancers (NSCLC) cases to investigate its role and impact on NSCLC.
Materials and methods
Tissue samples
Between January, 2005 and August, 2009, 556 NSCLC cases underwent complete pulmonary resection at the Department of Cancer and Thoracic Surgery, in the Okayama University Hospital, Japan. Of these, 293 specimens were made available for this study, while 20 corresponding non-malignant lung tissues from these patients were also used for this study. Institutional review board permission and informed consent were obtained for the cases.
Detection of miR-9s methylation by combined bisulfite restriction analysis (COBRA)
COBRA is a semiquantitative method used to measure methylation at specific methylation-sensitive restriction sites (15). Briefly, genomic DNA was isolated from tissue samples by proteinase K digestion followed by phenol-chloroform (1:1) extraction and ethanol precipitation (16). Genomic DNA (200 ng) was subjected to bisulfite treatment using the EZ DNA Methylation Gold™ kit (Zymo Research, Irvine, CA, USA). Primers designed for the predicted bisulfite-modified sequences were based on the nucleotide sequence submitted to GenBank (accession numbers: NR_029691 for miR-9-1, NR_030741 for miR-9-2, and NR_029692 for miR-9-3) and were as follows: miR-9-1, forward: GGGGTTTGGTTGTTTATTT and reverse: TCCACTACCCTTCTCTAAAAA: miR-9-2, forward: GGA ATAAATTTTGAAGGTAATAGATTT and reverse: CAC AAACCTCTATCTTCCTCTTACC; miR-9-3, forward: GTTTGTTTATTTTTTTTGGTTTTT and reverse: AACCT CCCTTAACCAATACC. PCR was carried out in 25 μl reaction mixtures under the following conditions: an initial denaturation step at 95°C for 12 min, followed by 45 cycles of 94°C for 30 sec, annealing at 54°C (miR-9-1), 58°C (miR-9-2) or 55°C (miR-9-3) for 60 sec and extension at 72°C for 60 sec., with a final extension step at 72°C for 7 min with the GeneAmp PCR System 9700 (Applied Biosystems, Carlsbad, CA, USA). PCR products (5 μl) were digested for 9 h with BstUI (New England Biolabs, Beverly, MA, USA) that recognizes sequences unique to methylated and bisulfite-converted alleles ( CG|CG site). The restricted products were electrophoresed on 2.2% agarose gels [3:1 mixture of agarose (Sigma-Aldrich Corp., St. Louis, MO, USA) and NUSieve® GTG® agarose (Cambrex Bio Science Rockland, Inc., Rockland, ME, USA)] and stained with ethidium bromide.
In situ hybridization (ISH) of miR-9
Sections (10 μm) for ISH analysis and 5 μm sections stained with hematoxylin and eosin (H&E) were generated from paraffin-embedded tissue blocks. The pathological classification was determined from an H&E-stained 5 μm slide section based on the 7th World Health Organization criteria of lung cancer (17). Sections (10 μm) were deparaffinized in xylene, then dehydrated in an ethanol series (100, 90, 70 and 50%). After blocking endogenous peroxidase with 3% hydrogen peroxide, sections were digested with 1 μg/ml proteinase K (Roche Diagnostics, Indianapolis, IN, USA) at 37°C for 15 min. The LNA-modified probe, complementary to miR-9 (100 nM; Exiqon, Woburn, MA, USA), diluted with NTE buffer (ISHR 7; Nippon Gene Co., Ltd., Tokyo, Japan) was hybridized with the sections for 16 h at 37°C. Post-hybridization washes were performed three times for 20 min in 2X standard saline citrate (SSC)/25% formamide and three times for 10 min in 0.1X SSC at 37°C. The probe signal was amplified using Tyramide Signal Amplification Plus DNP kits (PerkinElmer, Inc., Waltham, MA, USA) and color reactions were developed in 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium solution. Nuclear counterstaining was performed with 0.2% methylgreen.
Statistical analysis
Differences in statistical significance among the categorized groups were compared using the Chi-square test. The logistic regression analysis using the variables with the univariate analysis was used to further explore statistically significant differences observed and to identify baseline factors that may independently correlate with miR-9s methylation. A univariate analysis of overall survival (OS) was performed using the Kaplan-Meier method with log-rank testing, while a multivariate analysis was performed using the Cox proportional hazard model. The data were analyzed using the JMP software version 9.0.0 (SAS Institute, Inc., Cary, NC, USA). P<0.05 was considered to indicate a statistically significant difference for each analysis.
Results
Patient characteristics
Patient characteristics are provided in Table I. The median age of the patients was 66 years (range, 29–87). Younger patients were defined as ≤66 (n=147) years old, while elderly patients were >66 years old (n=146). The cases were divided into two groups of never (n=114) and ever (n=179) smokers. Never smokers were defined as individuals with a lifetime exposure to ≤100 cigarettes, while ever smokers had a lifetime exposure to >100 cigarettes. The early pathological stage of NSCLC was defined as stage I NSCLC (n=205), while the advanced as stages II and III NSCLC (n=88).
Table I.
Clinicopathological parameters and miR-9s methylation (n=293).
| Any miR-9s methylation (n=76)
|
|||
|---|---|---|---|
| Subsets | n | % | P-value |
| Agea (n) | |||
| ≤66 (147) | 29 | 20 | 0.03 |
| >66 (146) | 47 | 32 | |
| Gender (n) | |||
| Male (172) | 54 | 31 | 0.01 |
| Female (121) | 22 | 18 | |
| Smoking (n) | |||
| Never (114) | 27 | 24 | NS |
| Ever (179) | 49 | 27 | |
| Histology (n) | |||
| AD (221) | 54 | 24 | NS (AD vs. SQ and others) |
| SQ (62) | 16 | 26 | |
| Others (10) | 6 | 60 | |
| Stage (n) | |||
| I (205) | 60 | 29 | (I vs. 210 II–III) |
| II (35) | 6 | 17 | 0.047 |
| III (53) | 10 | 19 | |
| p-N (n) | |||
| Negative (220) | 67 | 30 | 0.002 |
| Positive (73) | 9 | 12 | |
Median, 66 years; range, 29–87 years. AD, adenocarcinoma; SQ, squamous cell carcinomas; others include 5 adenosquamous cell carcinomas and 5 large cell carcinomas; p-N, pathological lymph node metastasis; NS, not significant.
miR-9s methylation by COBRA and miR-9 expression
A representative example of COBRA is shown in Fig. 1A. A malignant pleural mesothelioma cell line, NCI-H290, was used as the positive control for the methylation of miR-9-1, 9-2 and 9-3. The methylation of miR-9-1, 9-2 and 9-3 was present in 20 (7%), 33 (11%) and 34 (12%) of 293 NSCLC cases, respectively. The methylation of any miR-9s (miR-9s methylation) was observed in 76 of 293 NSCLC cases (26%) (Fig. 1B). Of these 76 cases, two harbored methylation of the three (1%), seven harbored methylation of two (2%) and 67 (23%) harbored methylation of one miR-9s. miR-9 expression was determined by ISH assay in eight NSCLC cases. Representative examples are shown in Fig. 3. The five miR-9-methylated cases, including the two harboring methylation in miR-9-1 and 9-2, but not in miR-9-3, and the three harboring methylation in either miR-9-1, 9-2 or 9-3, showed a negative miR-9 expression, whereas the other three miR-9s unmethylated cases showed a positive miR-9 expression. None of the 20 corresponding non-malignant lung tissues harbored any miR-9s methylation.
Figure 1.
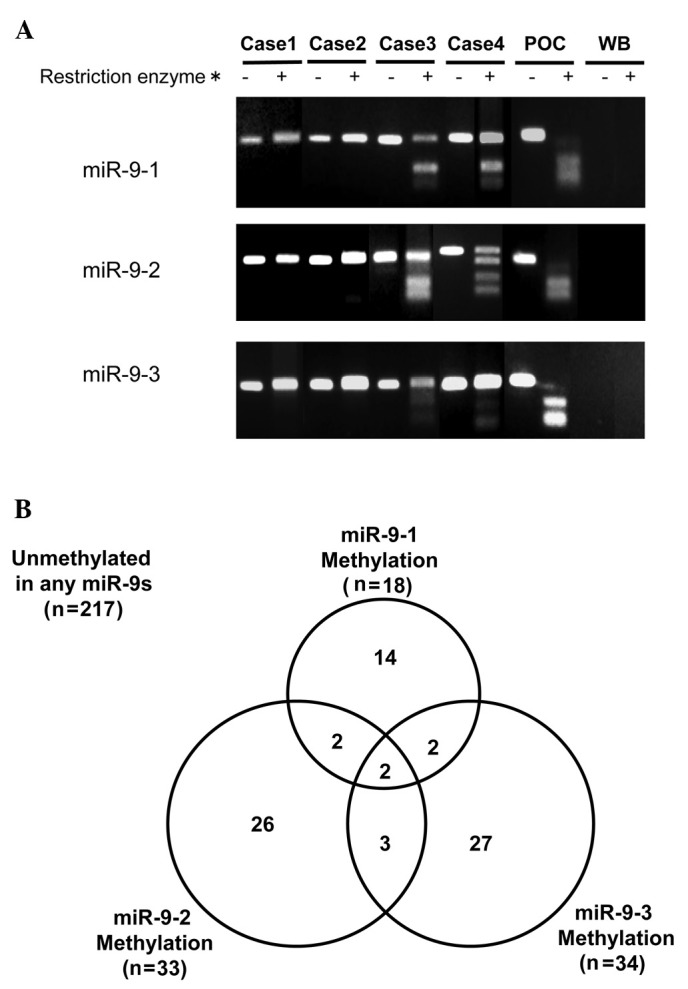
Detection of miR-9s methylation by combined bisulfite restriction analysis (COBRA) is shown. (A) Representative example of COBRA is shown. Cases 1 and 2 were unmethylated in the three miR-9 genes, while the miR-9s were methylated in Cases 3 and 4. POC, positive control (a mesothelioma cell line, NCI-H290); WB, water blank; *, BstUI. (B) Interrelationship between methylation of miR-9-1, 9-2 and 9-3 is shown.
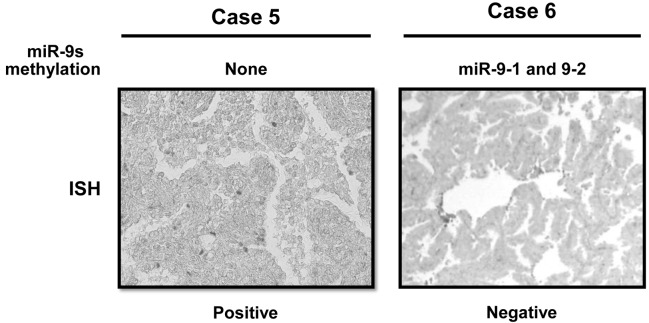
Figure 3.
Representative examples of in situ hybridization of miR-9 are shown. Case 5, in which none of the miR-9s was methylated, shows a positive expression of miR-9. By contrast, Case 6, harboring miR-9-1 and 9-2 methylations shows a negative expression of miR-9.
miR-9s methylation and clinicopathological factors
The associations between miR-9s methylation and clinicopathological factors are shown in Table I. miR-9s methylation was significantly more frequent in elderly [32 vs. 20% (younger patients): P=0.03], male patients [31 vs. 18% (female): P=0.01], in early pathological stage [29 vs. 18% (advanced stage): P=0.047] and pathologically lymph node-negative patients [30 vs. 12% (lymph nodal metastasis positive): P=0.002]. Logistic regression analysis of the significant univariate analysis factors showed that male gender [odds ratio (OR), 2.0; 95% confidence interval (95% CI), 1.1–3.6; P=0.01] and pathologically negative lymph node metastasis (OR, 4.8; 95% CI, 1.4–17.2; P=0.002) were independent relative factors for miR-9s methylation.
miR-9s methylation and clinical outcome
Over a median follow-up period of 32 months, the OS rates of the cases (n=293), miR-9-methylated (n=76) and unmethylated cases (n=217) were 91.8, 97.3 and 90%, respectively. miR-9-methylated cases had a significantly more prolonged OS period compared to miR-9-unmethylated cases (P=0.049) (Fig. 2). Female gender, never smoker, adenocarcinoma histology, early pathological stage and pathologically lymph node-negativity were additional significant predictive factors for a prolonged OS period in the univariate analysis (Table II). The multivariate analysis of the significant univariate analysis demonstrated that miR-9s methylation [hazard ratio (HR) 4.2, 95% CI: 1.2–27.0, P=0.026] and early pathological stage (HR 8.3, 95% CI: 2.1–28.6, P=0.004) were independent predictive factors for a prolonged OS period.
Figure 2.
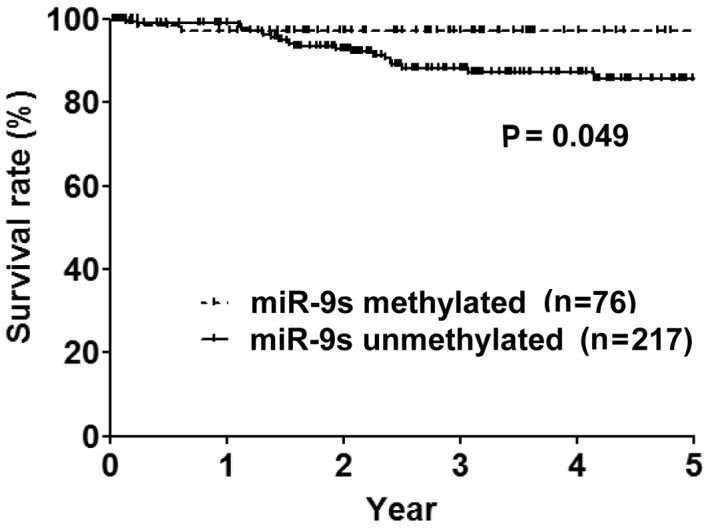
Overall survival curves based on miR-9s methylation status are shown.
Table II.
Impact of miR-9s methylation on overall survival.
| Factors | Median OS (month) | P-value |
|---|---|---|
| miR-9s methylation | 0.049 | |
| Positive | 33.3 | |
| Negative | 31.1 | |
| Age (years) | 0.3 | |
| ≤66 | 36.4 | |
| >66 | 28.3 | |
| Gender | 0.013 | |
| Male | 29.1 | |
| Female | 34.9 | |
| Smoking history | 0.006 | |
| Never | 35.5 | |
| Ever | 28.3 | |
| Histology | 0.017 | |
| AD | 33.1 | |
| Non-AD | 25.2 | |
| Stage | <0.0001 | |
| I | 33.0 | |
| II and III | 27.0 | |
| p-N | 0.002 | |
| Negative | 32.6 | |
| Positive | 29.9 |
AD, adenocarcinoma; OS, overall survival; p-N, pathological lymph nodal metastasis.
Discussion
miRs are capable of acting as oncogenes or TSGs and the observed widespread alteration in miR expression patterns is highly correlated with various human malignancies (18). Regarding NSCLC, miR alterations also have clinical implications: reduced let-7 expression (19) and miR-155 over-expression (20) reportedly correlate with improved or worse clinical outcomes, respectively. In the present study, miR-9 families were demonstrated to frequently methylate, especially in lymph node-negative cases, and to silence miR-9 expression in cases with miR-9s methylation. These findings suggest that miR-9 is oncogenically involved in NSCLC.
Methylation in CpG islands of promoter regions is an important epigenetic mechanism of gene silencing that is often observed in cancer, leading to the inhibition of gene transcription (21). Similar to protein coding genes, miRs are downregulated through epigenetic mechanisms (11,22,23). Although methylation typically affects TSGs, it also occurs in oncogenes, such as COX219, telomerase reverse transcriptase and epidermal growth factor receptor, silencing their expression (24–27). miR-9 is reportedly involved in an oncogenic as well as a tumor-suppressing manner in human malignancies.
In the former case, miR-9 expression levels were elevated in primary tumors in breast cancer patients with metastasis compared to metastasis-free patients (28), while other reports showed the oncogenic involvement of miR-9 based on basic experiments (8) and clinical findings (29). In their study, Tsai et al demonstrated that the tumor-suppressive miR-9 was silenced by promoter methylation in gastric cancer (30), while Heller et al found that patients with miR-9-3 methylation showed shorter OS duration among 101 NSCLC patients (31). In this study, the methylation status of the miR-9 family members was determined using COBRA in a larger patient set compared to previous reports, and NSCLC patients with methylation of any miR-9 family member were found to be an independent factor associated with a prolonged OS period. Moreover, miR-9-3 methylation showed no prognostic impact (data not shown).
In conclusion, miR-9s methylation, which silences miR-9 expression, is associated with early-stage and pathologically lymph node-negative cases of NSCLC, suggesting miR-9 to be oncogenically involved in NSCLC in promoting metastasis and tumor aggressiveness.
Acknowledgments
The authors would like to thank Ms. Chikako Isobe for preparing the sections from paraffin-embedded tissue blocks.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Toyooka S, Mitsudomi T, Soh J, et al. Molecular oncology of lung cancer. Gen Thorac Cardiovasc Surg. 2011;59:527–537. doi: 10.1007/s11748-010-0743-3. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31:809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 8.Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korpal M, Ell BJ, Buffa FM, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myatt SS, Wang J, Monteiro LJ, et al. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann U, Hasemeier B, Christgen M, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 13.Hildebrandt MA, Gu J, Lin J, et al. Hsa- miR-9s methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29:5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- 14.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrmann BG, Frischauf AM. Isolation of genomic DNA. Methods Enzymol. 1987;152:180–183. doi: 10.1016/0076-6879(87)52018-3. [DOI] [PubMed] [Google Scholar]
- 17.Venneti S, Boateng LA, Friedman JR, et al. MiR-9 and MiRNA-200a distinguish hemangioblastomas from metastatic clear cell renal cell carcinomas in the CNS. Brain Pathol. 2012;22:522–529. doi: 10.1111/j.1750-3639.2011.00551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 19.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 20.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Toyooka S, Toyooka KO, Miyajima K, et al. Epigenetic down-regulation of death-associated protein kinase in lung cancers. Clin Cancer Res. 2003;9:3034–3041. [PubMed] [Google Scholar]
- 22.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 23.Toyota M, Suzuki H, Sasaki Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 24.Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–6090. [PubMed] [Google Scholar]
- 25.Santini V, Kantarjian HM, Issa JP. Changes in DNA methylation in neoplasia: pathophysiology and therapeutic implications. Ann Intern Med. 2001;134:573–586. doi: 10.7326/0003-4819-134-7-200104030-00011. [DOI] [PubMed] [Google Scholar]
- 26.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 27.Scartozzi M, Bearzi I, Mandolesi A, et al. Epidermal growth factor receptor (EGFR) gene promoter methylation and cetuximab treatment in colorectal cancer patients. Br J Cancer. 2011;104:1786–1790. doi: 10.1038/bjc.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khew-Goodall Y, Goodall GJ. Myc-modulated miR-9 makes more metastases. Nat Cell Biol. 2010;12:209–211. doi: 10.1038/ncb0310-209. [DOI] [PubMed] [Google Scholar]
- 29.Tsai KW, Liao YL, Wu CW, et al. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6:1189–1197. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravgaard KH, Lyng MB, Laenkholm AV, et al. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat. 2012 doi: 10.1007/s10549-012-1969-9. [DOI] [PubMed] [Google Scholar]
- 31.Heller G, Weinzierl M, Noll C, et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin Cancer Res. 2012;18:1619–1629. doi: 10.1158/1078-0432.CCR-11-2450. [DOI] [PubMed] [Google Scholar]