Abstract
We investigated the effect of activating a competing, artificially generated, neural representation on encoding of contextual fear memory. We used a cfos based transgenic approach to introduce the hM3Dq DREADD receptor into neurons based on their natural activity patterns. Neural activity can then be specifically and inducibly increased in the hM3Dq expressing neurons by an exogenous ligand. When an ensemble of neurons for one context (ctxA) was artificially activated during conditioning in a distinct context (ctxB), animals formed a hybrid memory representation. Reactivation of the artificially stimulated network within the conditioning context was required for retrieval of the memory. The memory was specific for the spatial pattern of neurons artificially activated during learning while similar stimulation impaired recall when not part of the initial conditioning.
Direct electrical stimulation can be used to define functional domains in the brain, can elicit stereotyped behavioral responses, drive self-stimulation behavior, and serve as CS or US in conditioning paradigms (1-4). This type of stimulation has typically been focal, using either microelectrodes, or more recently, genetically encoded mediators of neural excitability such as channelrhodopsin (5, 6). While this discrete, temporally coordinated, focal stimulation can drive behavior, we know much less about the effects of stimulating broadly distributed neural networks. In the mammalian cortex there is significant, non-random, spontaneous neural activity that is internally generated rather than arising from sensory inputs, and this activity influences the processing of natural sensory stimuli (7-10). How does this internally generated activity influence the formation of a new memory representation?
To investigate this question we used transgenic mice (Fig 1A) in which the hM3Dq receptor is expressed in an activity dependent manner by a cfos promoter driven tTA transgene (hM3Dqfos mouse) (11, 12). hM3Dq is a Gq coupled receptor that responds specifically to clozapine-N-oxide (CNO) and produces strong depolarization and spiking in pyramidal neurons (12). Transgenic animals exposed to a particular environmental stimulus will express hM3Dq in those neurons that are sufficiently active to induce the cfos promoter, and this naturally occurring neural ensemble can be subsequently reactivated artificially in the transgenic mice by delivery of CNO. Artificial activity induced in this manner will retain the spatial character of the neural ensemble, but will not preserve the temporal dynamics achieved by natural-stimuli.
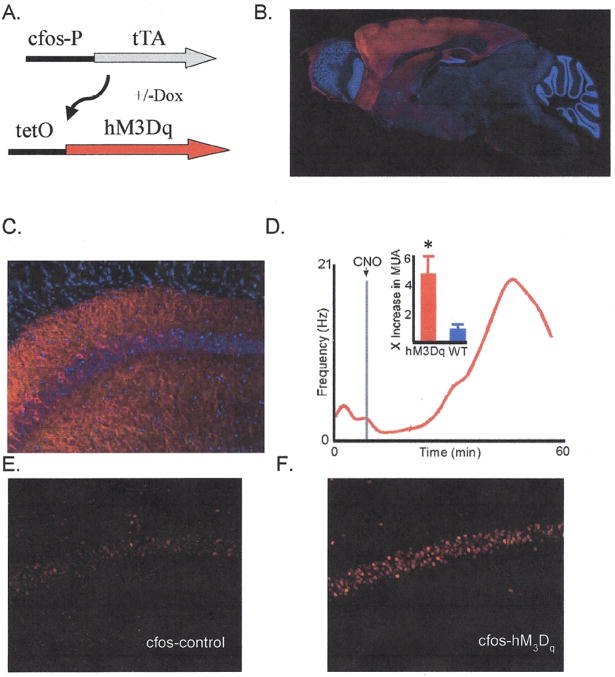
Figure 1. Expression and activation of the hM3Dq transgene.
A) Transgenic mice used in this study carry the 2 transgenes shown allowing Dox regulated and neural activity dependent expression of the hM3Dq receptor. B) Overall spatial expression profile of the hM3Dq transgene in mice off dox maintained in the homecage. Immunofluorescence was strong in hippocampus, basalateral amygdala, and throughout the cortex. Fluorescence was also observed to a small extent in the pontine nucleus and in brainstem. C) Expression in the CA1 region of the hippocampus showing sparse and distributed expression of the hM3Dq transgene. D) CNO injection causes increased neural activity in hM3Dqfos mice. Red curve shows multi unit activity (MUA) recorded from dorsal CA1of an anesthetized hM3Dqfos mouse over time. Inset gives fold increase in MUA (4.76 for hM3Dqfos vs. .9 for WT, mean 30-40 minutes post-injection/mean pre-injection baseline. n=6 and 6, *=Wilcoxon signed-rank: P<0.01). E & F) cfos induction 1.5 hours after CNO administration in a control (left) and hM3Dqfos (right) mouse. hM3Dqfos mice showed on average a 2.5-fold increase in cfos expression in the hippocampal CA1 region compared to control mice (see supplementary table 1 hM3Dqfos n = 10, control, n = 10, T-test p <.02).
The expression of hM3Dq is widely distributed in the brain of hM3Dqfos double transgenic mice in the absence of Doxycycline (Dox), to allow tTA driven transcription (Fig. 1 B&C). Within a given brain area expression is limited to a fraction of excitatory neurons based on neural activity driving the cfos promoter. Dox can be used to control the specific time window in which active neurons are genetically tagged with hM3Dq by modulating tTA driven transcription (11, 13). To test the kinetics of CNO based neural activation in these animals we performed in vivo recording in the hippocampus of anesthetized animals. Following CNO injection we found an increase in neuronal activity that reached a maximum intensity between 30 and 40 minutes post CNO injection (Fig 1D). In order to examine more broadly the increase in neural activity we used endogenous cfos expression as an indicator of neural activity (Fig 1E&F). We found significant increases in cfos labeling across multiple brain regions (ranging from 2-20 fold) in CNO injected hM3Dqfos transgenic vs. control animals (Table S1). Labeling for cfos was found in both hM3Dq positive and negative neurons with 91±2% of hM3Dq positive neurons in CA1 co-labeled with cfos (Fig. S2).
In standard contextual fear-conditioning animals develop a memory for the conditioning chamber in which they receive a foot-shock. The ability to form the context association is dependent on the hippocampus, which participates in encoding a representation of the environment (14, 15). To test the effects of competing circuit activation on formation of a memory trace we designed the fear-conditioning protocol outlined in fig. 2. On day 1 hM3Dfos mice were exposed to a novel context (ctxA) in order to drive expression of the hM3Dq transgene into neurons activated in that context. On day 2 animals were injected with Dox to inhibit further hM3Dq receptor expression and with CNO to stimulate activity in the pattern of neurons that expressed the receptor. The mice were then fear conditioned in a distinct context (ctxB), and 24 hours later, memory performance was tested in the absence and presence of CNO. Thus, we are in effect firing the neurons active in ctxA while the animals are fear conditioned in ctxB.
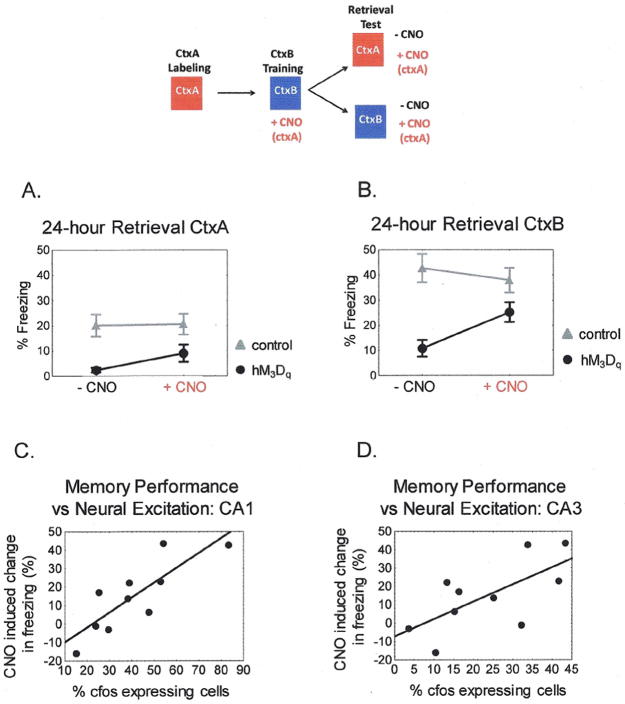
Figure 2. Incorporation of Synthetic Neural Activity into a 24-hr Memory Representation.
A) Freezing in ctxA 24-hours after conditioning in ctxB. hM3Dqfos n = 14, control n = 13. hM3Dqfos mice freeze significantly less than control mice in ctxA in the absence and presence of CNO. Repeated measures ANOVA main effect of genotype F(1,26) = 10.96, p <.005. CNO has no significant effect on freezing in either group. Post hoc Bonferroni hM3Dfos p = 0.192, control p = 1.00. B) Transgenic hM3Dqfos mice show impaired 24-hour memory for ctxB that is rescued by injection of CNO. Repeated measures ANOVA genotype x CNO interaction F(1.25) = 10.15, p <.005. Post hoc Fisher’s LSD found that hM3Dqfos mice were freezing significantly less than control mice in ctxB in the absence of CNO, p < 0.001, but were statistically similar in ctxB in the presence of CNO, p = 0.117, and showed a significant increase in freezing in ctxB with CNO compared to ctxB alone, p < 0.001. C and D) Correlation between the difference in freezing scores in the presence and absence of CNO and endogenous cfos expression 1 hour after memory testing in hippocampal area CA1, D, r = 0.8276, p <.005 and CA3, E, r = 0.6742, p <.05.
We anticipated 3 potential outcomes. The strong synthetic activation of ctxA neurons could be dominant and serve as a CS to produce an associative fear memory. This would lead to a fear response to CNO or possibly even a fear response to ctxA itself if the artificial and natural activation of the neurons were sufficiently similar. This was not observed as the level of freezing in ctxA was not significant in transgenic animals either with or without CNO injection (Fig 2A). A protocol in which ctxA neurons were activated by CNO and animals were shocked immediately in ctxB (to prevent formation of a ctxB representation (13)) also failed to produce a CNO dependent memory (Fig. S3). Similarly, if the neurons active during conditioning itself were tagged with the hM3D transgene, CNO did not produce significant freezing (Fig. S5). Thus the synthetic activity alone could not serve as a CS in fear conditioning. A second possibility was that the natural sensory experience in ctxB would dominate and transgenic animals would show normal conditioning to ctxB. The hM3Dfos animals displayed a severe deficit in freezing to ctxB suggesting that the CNO induced activity was interfering with normal encoding of memory for ctxB (Fig. 2B). A third possibility was that animals would form a hybrid representation, incorporating elements of both the CNO induced artificial stimulation and the natural sensory cues from ctxB. This appears to be the case as the transgenic animals showed a significant increase in freezing in response to CNO delivered in the ctxB setting during the 24-hour memory test (Fig 2B). We observed similar results in 2 separate experiments when a different contextual set-up for ctxA neural labeling was used (Fig. S1, S4). The requirement for reactivation of the transgene expressing neurons during memory retrieval suggests that their activity was incorporated into the memory trace. Consistent with this idea, we found a correlation between freezing during memory retrieval and the degree of neural activation, assessed by cfos expression in the hippocampus (Fig. 2 C&D).
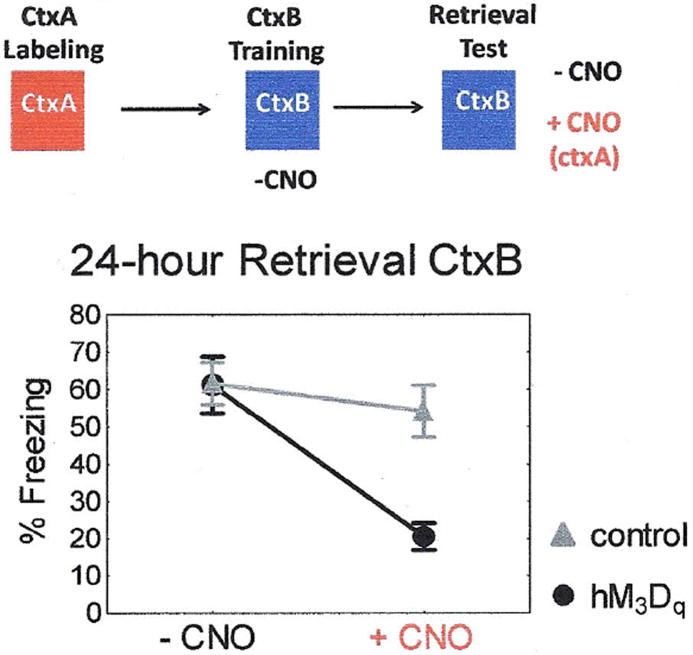
Retrieval of a memory representation likely involves the reactivation of some neurons that were active during the initial learning (11, 16-18). To test the susceptibility of this spatial code to competing neural network activation, hM3Dqfos mice were exposed to ctxA to allow expression of the hM3Dq transgene but then conditioned in ctxB without CNO stimulation of the ctxA neural ensemble (Fig.3). As expected, these animals developed wild-type levels of freezing to ctxB 24-hours after conditioning. Now, however, activation of the hM3Dq expressing neurons impaired memory performance during retrieval in ctxB. This suggests that CNO induced activation of a competing neural network interferes with the learned spatial code and degrades recognition if this activity was not present during the initial training. This is not surprising given that even limited focal hippocampal stimulation has been shown to disrupt spatial memory (19).
Figure 3. Disruption of Memory Retrieval by Synthetic Neural Activation.
Transgenic hM3Dfos mice develop a normal 24-hr context memory when conditioned in the absence of CNO. This memory is disrupted by CNO injection to activate the competing ctxA representation. hM3Dqfos n = 12, control n = 12. Repeated measures ANOVA main effect of genotype F(1, 22) = 5.3, p <.05, CNO F(1, 22) = 28.6, p < 0.001, and genotype × CNO interaction F(1, 22) = 13.5, p = 0.001. Post-hoc Fisher LSD revealed that hM3Dfos mice were freezing significantly less in the presence of CNO compared to before CNO administration p < 0.001, and were freezing significantly less that control mice in the presence of CNO p < 0.001.
Does the hybrid fear memory formed by hM3Dqfos mice incorporate the specific pattern of ctxA neurons activated by CNO during learning or are the animals responding to a less specific alteration in brain state? To distinguish between these possibilities we conditioned animals in the presence of CNO induced firing of ctxA labeled neurons but then placed the animals on Dox to allow turnover of the hM3Dq receptor. Two days later we removed Dox from the animals’ diet, and placed them in a new home cage to allow de novo expression of the hM3Dq receptor in a distinct group of neurons (ctxC). Fourteen days after initial conditioning, we tested memory performance as assessed by freezing scores in ctxB in the absence and presence of CNO induced synthetic activation. We found no increase in freezing in hM3Dqfos mice in response to CNO (Fig 4A), demonstrating a requirement for reactivation specifically of the learned, ctxA, neural ensemble rather than a generalized change in brain state caused by CNO induced activity.
Figure 4. Memory performance during synthetic reactivation is network specific.
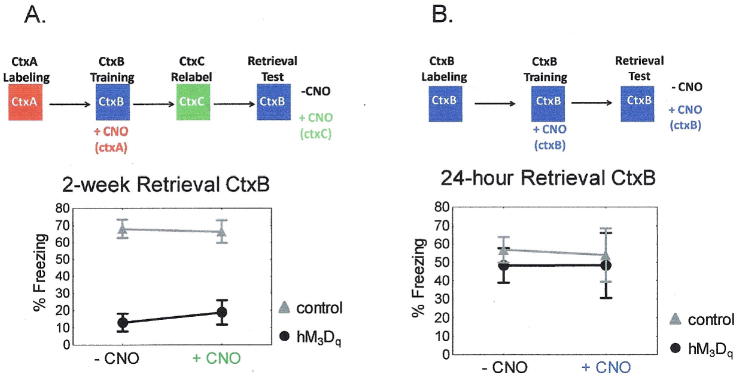
A) When CNO induced synthetic activation does not occur in identical neural populations during memory formation and memory retrieval, a memory deficit is observed. hM3Dfos mice show significantly less freezing than control mice in ctxB both in the absence and presence of CNO. hM3Dfos n = 14, control = 17. Repeated measures ANOVA main effect of genotype F(1, 23) = 51.15, p < 0.001. B) When hM3Dqfos mice are exposed to ctxB off of dox to induce hM3Dq expression and then fear conditioned on dox after CNO injection in ctxB, synthetic activation by CNO is not necessary for memory recall in ctxB. ctxB: hM3Dqfos n = 9, control n = 10, ctxBcno: hM3Dqfos n = 5, control n = 6. Repeated measures ANOVA F(2, 18) = 0.0474, p = 0.954.
To further address the issue of ensemble specificity we preexposed animals to the fear conditioning context (ctxB) on day 1 to express the hM3Dq receptor in neurons that are activated in that context. We reasoned that the synthetic activation of this pattern of neurons would more likely overlap with the natural activity during learning in ctxB and should therefore not interfere with the production of a normal ctxB representation. When animals were fear conditioned following injection of CNO to artificially activate the ctxB ensemble during learning they developed wild-type levels of 24-hour context fear memory that was independent of CNO stimulation (Fig 4B). This is in contrast to the deficit produced in animals pre-exposed to the novel ctxA and further supports the contention there must be a match in the spatial pattern of neural activity at learning and retrieval.
Several recent studies have suggested flexibility in the specific neurons incorporated into a fear memory trace in the amygdala through a selection mechanism in which more excitable neurons are preferentially incorporated into the trace (16-18). The current results do not appear to be due to this type of selection as the reactivation of the neurons with CNO is required for retrieval while in the previous studies the stimulated neurons were part of a representation that could be naturally retrieved. This difference may be due to different requirements for forming simple associations in the amygdala vs more complex representations in the hippocampus and cortex.
In the current study the artificially stimulated neural ensembles become incorporated into the memory and there must be a match between the pattern of activity at the time of learning and the time of retrieval. In one recent study, ChR2 stimulation of a random population of neurons in the piriform cortex combined with odorant during conditioning found that either the artificial stimulation or the odorant alone could produce recall, suggesting independent and non interfering representations (20). In this study we found that the CNO activation alone could not act as an independent cue. While these studies differed in a variety of parameters including anatomy and size of the artificially stimulated ensembles, one critical difference may be that the activity induced by hM3Dq is not temporally coordinated in response to the inducing stimulus (CNO), as is the case with ChR2 driven stimulation by light. In that case the sensory input during conditioning and retrieval in ctxB may coordinate the activity of the CNO depolarized cells to provide some degree of temporal coordination to the CNO driven neurons and account for the requirement for the compound stimulus. Alternately, it is possible that the uncoordinated CNO based stimulus could serve as a CS if it was limited to a discrete primary sensory area as in the previous study.
Current views of sensory processing recognize the role of internally generated (spontaneous) neural activity in generating a representation from a given sensory input (8). This activity is not random but has spatial and temporal structure that is thought to represent defined ensembles formed through previous learning related plasticity. Moreover, in psychology the idea of a schema as a preexisting framework of relationships which modulates learning suggests that new memories are not produced de novo from experience but interact with existing circuit activity (21, 22). While the CNO based stimulation does not replicate the temporal dynamics of this naturally occurring internal activity, the approach allows the activation of a distributed spatial pattern of neurons recruited during a specific experience (ctxA exposure). The results demonstrate that this spatial pattern of activity at the time of learning and retrieval must match for appropriate recall. The results imply a strong spatial component to coding in this form of learning and support the idea that the internal dynamics of the brain at the time of learning contribute to memory encoding.
Supplementary Material
Acknowledgments
We would like to thank Kiriana Cowansage for helpful discussions. This work was supported by grants from the NIMH and NIDA (MM), the NIMH and the Michael Hooker Distinguished Chair in Pharmacology (BLR), a graduate fellowship from the California Institute for Regenerative Medicine (AG).
References and Notes
- 1.Doty RW. Annu Rev Psychol. 1969;20:289. doi: 10.1146/annurev.ps.20.020169.001445. [DOI] [PubMed] [Google Scholar]
- 2.Shinkman PG, Swain RA, Thompson RF. Behav Neurosci. 1996 Oct;110:914. doi: 10.1037//0735-7044.110.5.914. [DOI] [PubMed] [Google Scholar]
- 3.Romo R, Hernandez A, Zainos A, Salinas E. Nature. 1998;392:387. doi: 10.1038/32891. [DOI] [PubMed] [Google Scholar]
- 4.Jasper H, Penfield W. Epilepsy and the Functional Anatomy of the Human Brain. 2. Little, Brown and Co.; 1954. [Google Scholar]
- 5.Huber D, et al. Nature. 2008;451:61. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo L, Callaway EM, Svoboda K. Neuron. 2008;57:634. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenet T, Bibitchkov D, Tsodyks M, Grinvald A, Arieli A. Nature. 2003;425:954. doi: 10.1038/nature02078. [DOI] [PubMed] [Google Scholar]
- 8.Fiser J, Chiu C, Weliky M. Nature. 2004;431:573. doi: 10.1038/nature02907. [DOI] [PubMed] [Google Scholar]
- 9.MacLean JN, Watson BO, Aaron GB, Yuste R. Neuron. 2005;48:811. doi: 10.1016/j.neuron.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 10.Ringach DL. Current Opinion in Neurobiology. 2009;19:439. doi: 10.1016/j.conb.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reijmers LG, Perkins BL, Matsuo N, Mayford M. Science. 2007 Aug 31;317:1230. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- 12.Alexander GM, et al. Neuron. 2009;63:27. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuo N, Reijmers L, Mayford M. Science. 2008 Feb 22;319:1104. doi: 10.1126/science.1149967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anagnostaras SG, Gale GD, Fanselow MS. Hippocampus. 2001;11:8. doi: 10.1002/hipo.1051. [DOI] [PubMed] [Google Scholar]
- 15.Frankland PW, Cestari V, Filipkowski RK, McDonald RJ, Silva AJ. Behav Neurosci. 1998;112:863. doi: 10.1037//0735-7044.112.4.863. [DOI] [PubMed] [Google Scholar]
- 16.Han JH, et al. Science. 2007 Apr 20;316:457. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- 17.Han JH, et al. Science. 2009 Mar 13;323:1492. doi: 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, et al. Nat Neurosci. 2009 Nov;12:1438. doi: 10.1038/nn.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girardeau G, Benchenane K, Wiener SI, Buzsaki G, Zugaro MB. Nat Neurosci. 2009 Oct;12:1222. doi: 10.1038/nn.2384. [DOI] [PubMed] [Google Scholar]
- 20.Choi GB, et al. Cell. Sep 16;146:1004. [Google Scholar]
- 21.Tse D, et al. Science. 2011 Aug 12;333:891. doi: 10.1126/science.1205274. [DOI] [PubMed] [Google Scholar]
- 22.Tse D, et al. Science. 2007 Apr 6;316:76. doi: 10.1126/science.1135935. [DOI] [PubMed] [Google Scholar]
- 23.Korzus E, Rosenfeld MG, Mayford M. Neuron . 2004 Jun 24;42:961. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKhann GM, 2, Wenzel HJ, Robbins CA, Sosunov AA, Schwartzkroin PA. Neuroscience. 2003;122:551. doi: 10.1016/s0306-4522(03)00562-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.