Abstract
Beta-microseminoprotein (MSP)/MSMB is an immunoglobulin superfamily protein synthesized by prostate epithelial cells and secreted into seminal plasma. Variants in the promoter of the MSMB gene have been associated with the risk of prostate cancer (PCa) in several independent genome-wide association studies. Both MSMB and an adjacent gene, NCOA4, are subjected to transcriptional control via androgen response elements. The gene product of NCOA4 interacts directly with the androgen receptor as a co-activator to enhance AR transcriptional activity. Here, we provide evidence for the expression of full-length MSMB-NCOA4 fusion transcripts regulated by the MSMB promoter. The predominant MSMB-NCOA4 transcript arises by fusion of the 5′UTR and exons 1–2 of the MSMB pre-mRNA, with exons 2–10 of the NCOA4 premRNA, producing a stable fusion protein, comprising the essential domains of NCOA4. Analysis of the splice sites of this transcript shows an unusually strong splice acceptor at NCOA4 exon 2 and the presence of Alu repeats flanking the exons potentially involved in the splicing event. Transfection experiments using deletion clones of the promoter coupled with luciferase reporter assays define a core MSMB promoter element located between –27 and –236 of the gene, and a negative regulatory element immediately upstream of the start codon. Computational network analysis reveals that the MSMB gene is functionally connected to NCOA4 and the androgen receptor signaling pathway. The data provide an example of how GWAS-associated variants may have multiple genetic and epigenetic effects.
Introduction
Human prostate cancer (PCa) is the most common cancer affecting males in developed countries. Unlike many cancers, it is often indolent, multifocal, and genetic studies have failed to find a consistent high penetrance locus in its causation. PCa is a complex disease that involves interaction between genetic susceptibility and environmental factors, environment and socioeconomic status (Crawford 2003). MSP (beta-microseminoprotein), encoded by the MSMB gene, is secreted at high levels by the prostate, and variation in MSP levels can be easily detected in both the serum and semen via a validated immunoassay (Valtonen-Andre et al. 2008). MSP has potential utility as a diagnostic tool in detecting PCa, with several aspects of its molecular biology suggesting that it might be more specific for PCa than prostate-specific antigen (PSA) (Bjartell et al. 2007; Reeves et al. 2006). Nuclear receptor coactivator 4 (NCOA4, also known as 70 kDa androgen receptor coactivator or ARA70) is a ligand-dependent AR-associated protein that enhances the transcriptional activity of androgen receptor (AR) in human PCa cells in the presence of dihydrotestosterone or testosterone (Yeh and Chang 1996). As a potential facilitator of PCa progression, ARA70-induced AR transactivation may result in decreased apoptosis and increased cell proliferation in PCa cells via a PSA-mediated mechanism (Niu et al. 2008). In addition, overexpression of an alternatively spliced 35 kDa ARA70 variant, termed ARA70-beta, promoted cellular invasion in an AR-independent manner (Peng et al. 2008). ARA70 was first identified as a gene fused to an oncogene and subsequently as a co-activator for AR (Peng et al. 2008).
The MSMB promoter variant rs10993994 was identified in two independent genome-wide association studies (GWAS) to be significantly associated with the risk of PCa (Eeles et al. 2008; Thomas et al. 2008). Now that this region has been extensively re-sequenced, additional variants close to rs10993994 have been investigated. It has been shown that a variant located in the neighboring gene, NCOA4, may play a role in PCa (Chang et al. 2009), and may involve alleles of rs10993994 that differentially influence the expression of MSMB and NCOA4 genes in prostate tissue (Chang et al. 2009; Lou et al. 2009; Nacu et al. 2011; Pomerantz et al. 2010).
Among the genetic alterations that characterize many cancers is gene fusion, which often results in the production of a fusion protein that may have a new or altered function (Rabbitts 1994; Rowley 2001). Interestingly, about 80 % of all known gene fusions have been associated with bone and soft tissue sarcomas, leukemias, and lymphomas, which account for only 10 % of all human cancers (Mitelman et al. 2004). In contrast, common epithelial cancers, which account for 80 % of cancer-related deaths, have been associated with only 10 % of known recurrent gene fusions (Kumar et al. 2009; Mitelman et al. 2005). However, the recent discovery of a recurrent gene fusion, TMPRSS2-ERG, in a majority of prostate cancers (Tomlins et al. 2007) and EML4-ALK in non-small-cell lung cancer (NSCLC) (Soda et al. 2007), has provided impetus for a search for gene fusions in epithelial cancers (Choi et al. 2008; Koivunen et al. 2008; Perner et al. 2008; Rikova et al. 2007; Tomlins et al. 2005).
Intergenic trans-splicing, the joining of exons from distinct genes into one mature mRNA, has been shown to occur in mammalian cells and model organisms (Horiuchi and Aigaki 2006) and high-throughput sequencing reveals that trans-splicing and cis-splicing events are widespread in human cells (Al-Balool et al. 2011). Cis-splicing is a mechanism to generate hybrid proteins in adjacent genes, but its role in cancer is unclear. There is, however, precedent for investigating trans-splicing in cancer. Li et al. (2008) described intra-chromosomal trans-splicing of JAZF1 and JJAZF1 in normal endometrial stromal cells that mimics the aberrant hybrid transcript generated by the t(7;17)(p15;q21) translocation found in about 50 % of endometrial sarcomas, and speculated that the trans-splice product is pro-neoplastic. Similarly, MDS1/EVI1 fusion transcripts were found in normal cells that mimic cancer-associated gene fusion products (Fears et al. 1996). In fact, intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family (Fears et al. 1996). More recently, SLC45A3/ELK4 trans-splicing was identified in benign prostate lesions (Rickman et al. 2009). These findings form the basis for an emerging concept that alternative splicing events may generate fusion protein expression that becomes a pivotal event in multi-step carcinogenesis.
The work described herein was stimulated by a series of observations: (1) MSMB and NCOA4 are located less than 10 kb apart; (2) computational network analysis revealed that the MSMB gene is functionally connected with the NCOA4 gene via the AR signaling pathway; (3) three reported expressed sequence tags (EST), DB215804, DB233878 and DB240089, indicate that MSMB and NCOA4 form a complex locus in which mRNA transcripts utilize exons from both genes (Thierry and Thierry 2006), and (4) MSMB-NCOA4 fusion transcripts were identified in a recent study of read-through gene fusions in PCa (Nacu et al. 2011). The high relevance of these two genes to prostate tissue suggests a potential functional role for this fusion transcript in the development of PCa.
Results
Identification of MSMB-NCOA4 fusion transcripts
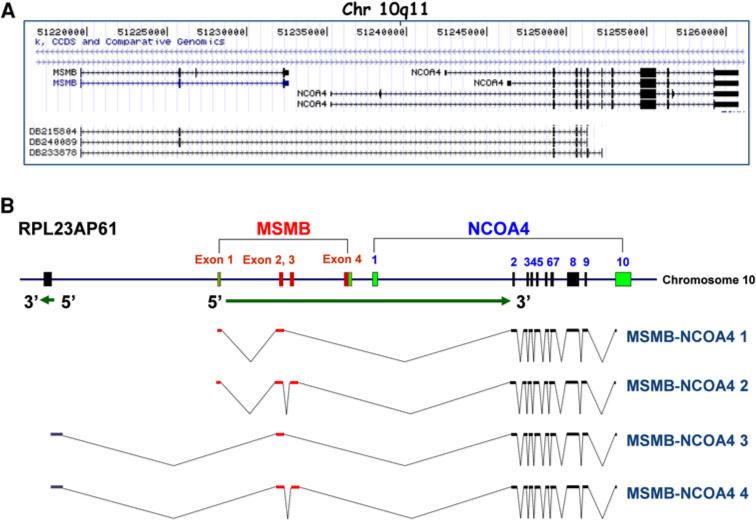
MSMB and NCOA4 are separated by ~3 kb on human chromosome 10q11 and three EST clones containing MSMB and NCOA4 exons have been reported (Fig. 1a). To investigate transcription from that region, we designed primers corresponding to several different positions in known MSMB and NCOA4 exons (Table S1). These were intended to identify fusion transcripts and determine their transcription start sites. A 2,027 bp MSMB-NCOA4 fusion transcript was amplified using primers from the 5′UTR of MSMB exon 1 (RT-MSMB-NCOA4 F1) and the 3′UTR of NCOA4 exon 10 (RT-MSMB-NCOA4 Rev1) (Fig. 1; S1). The MSMB-NCOA4 fusion transcript, containing the full sequence of MSMB exons 1–2 followed by complete sequences of NCOA4 exons 2–10 (E2–E2) was consistently amplified from prostate tissues, normal trachea tissue, and five prostate cancer cell lines (see Fig. 1b).
Fig. 1.
Characterization of the MSMB gene and MSMB-NCOA4 fusion transcripts. a UCSC genome browser view of MSMB and part of NCOA4. Three transcripts, DB215804, DB240089 and DB233878 span the MSMB and NCOA4 genes. The region is shown from (51,217,500) to (51,261,000) on Chromosome 10q11. b Schematic organization of MSMB and MSMB-NCOA4 fusion transcripts. The exons are indicated as boxes with corresponding exon numbers. Sequences derived from MSMB are shown in red, from NCOA4 are shown in dark blue, and from RPL23AP61 is shown in gray. Green boxes represent untranslated regions. The structures of four fusion transcripts identified in this study by 5′ RACE assay and sequencing, MSMB-NCOA4 1 to 4 are shown. Transcription orientations are indicated by arrows (color figure online)
Mapping the fusion gene transcription initiation site
To characterize the 5′ start sites of the MSMB-NCOA4 fusion transcripts, we performed 5′ RACE experiments. Four novel MSMB-NCOA4 transcripts were identified in this study from RNA of human prostate tissues, and normal trachea tissue (Fig. 1b). The first transcript, MSMB-NCOA4 1, represented a fusion of the complete exons 1–2 of MSMB with exons 2–10 of NCOA4 (E2-E2) as described previously (see Figure S1). The second transcript, MSMB-NCOA4 2, contains exons 1–3 of MSMB fused with exons 2–10 of NCOA4 (E3-E2). The third and fourth transcripts, MSMB-NCOA4 3 and 4, contain 5′ sequence from the upstream RPL23AP61 pseudogene fused to either exon 2 or exons 2–3 of MSMB followed by exons 2–10 of NCOA4 at the 3′ end (Fig. 1b; Figure S1). The MSMB-NCOA4 3 and 4 transcripts lack MSMB exon 1 sequence compared with MSMB-NCOA4 1 and 2. Sequencing of the first MSMB-NCOA4 transcript revealed an open reading frame that creates a fusion protein. The other transcripts were only detected in tumor tissue by cloning and sequencing, without a visible band by RT-PCR (Fig. 2).
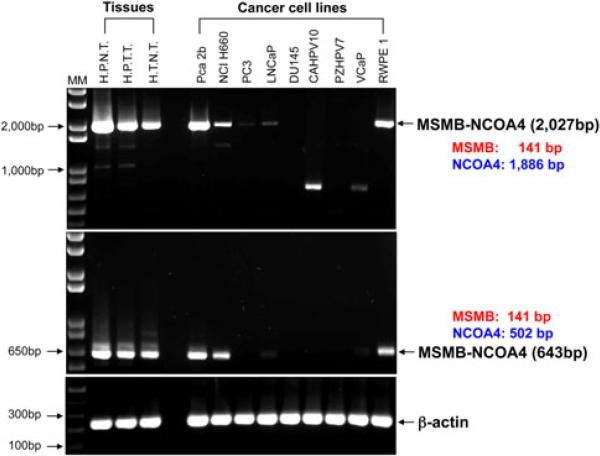
Fig. 2.
RT-PCR of MSMB-NCOA4 fusion transcripts in human tissues and cancer cell lines. cDNA prepared from human prostate normal tissue (HPNT), human prostate tumor tissue (HPTT), human trachea normal tissue (HTNT) and 9 prostate cell lines was subjected to RT-PCR using two primer sets, the 5′MSMB exon1 primer (forward)/3′NCOA4 exon10 (3′UTR, reverse) (2,027 bp), and the 5′MSMB exon 1 primer/3′NCOA4 exons 5–6 (reverse) (643 bp)
MSMB-NCOA4 fusion protein expression in vitro and in cell lines
Sequence analysis of MSMB-NCOA4 1 (E2-E2) revealed a 2,027 bp cDNA including 32 bp of the 5′UTR of MSMB and 27 bp of the 3′UTR of NCOA4, generating an ORF of 656 aa, putatively encoding an ~73 kDa protein from the fusion transcript. This structure of MSMB-NCOA4 has the potential to encode a protein with two distinct domains, MSMB at its amino (N) terminus and NCOA4 at its carboxyl (C) terminus.
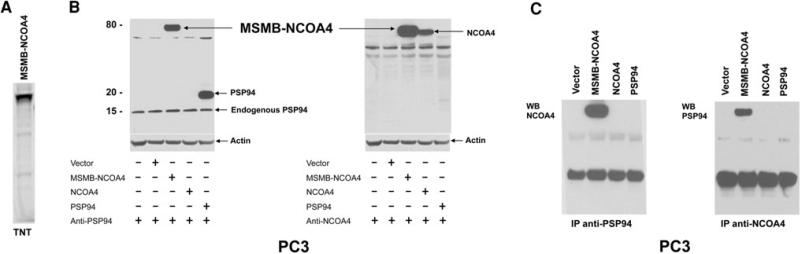
To determine if the MSMB-NCOA4 fusion protein can be expressed, we performed in vitro transcription/translation of the MSMB-NCOA4 cDNA. As shown in Fig. 3a, a specific signal was observed at 73 kDa. PC3 cells were then transiently transfected for 72 h with the pcDNA3.1+-MSMB-NCOA4 vector carrying the full MSMB-NCOA4 fusion transcript. Protein expression was verified by western blot analysis of cell lysates using a polyclonal anti-MSMB antibody affinity-purified on a peptide encompassing amino acids 21–144 of the MSMB N-terminus, and a monoclonal anti-NCOA4 antibody on a peptide encompassing amino acids 505–615 of NCOA4 C-terminus (Fig. 3b). The anti-MSMB antibody recognized the endogenous (15 kDa) and recombinant (18 kDa) MSMB proteins in MSMB transfectants containing a Myc-DDK-tag (Fig. 3b). The anti-NCOA4 C terminus antibody did not react with lysates of MSMB transfectants, but showed a specific signal with NCOA4 transfectants (Fig. 3b). The lysates of PC3 transfected with the MSMB-NCOA4 expression vector showed a specific band at 73 kDa, when probed with either anti-MSMB or anti-NCOA4 antibodies. To further confirm the identity of the MSMB-NCOA4 fusion protein, we performed immunoprecipitation with either the anti-MSMB N-terminal or the anti-NCOA4 C-terminal antibody. Western blotting of immunoprecipitated protein with either anti-NCOA4 (Fig. 3c, left panel) or anti-MSMB (Fig. 3c, right panel) showed a specific signal around 73 kDa.
Fig. 3.
Expression of MSMB-NCOA4 fusion protein in the transfected PC3 cell line. a Expression of the MSMB-NCOA4-encoded protein in vitro using the TNT system. b Western blotting analysis of the fusion construct. PC3 cells transfected with the pcDNA3.1+ empty vector, the pcDNA3.1+-MSMB-NCOA4 expression vector, pCMV6-NCOA4 and pCMV6-PSP94 were analyzed by western blot using polyclonal anti-MSMB antibody (left panel) and monoclonal anti-NCOA4 antibody (right panel). c Immunoprecipitation (IP) using anti-MSMB raised against the N-terminus of MSMB (left panel) or anti-NCOA4 against the C-terminus of NCOA4 (right panel)
The expression of MSMB-NCOA4 in transfected prostate cancer cell lines
To confirm expression of the fusion protein, we amplified the full-length MSMB-NCOA4 1 (E2-E2) transcript from normal prostate tissue and inserted it into the expression vectors, pcDNA3.1+ and pEGFPC1, respectively. These expression vectors and commercially available pCMV6-NCOA4 and pCMV6-PSP94 vectors were transiently transfected into PC3, LNCaP and DU145 cells (Fig. 4a).
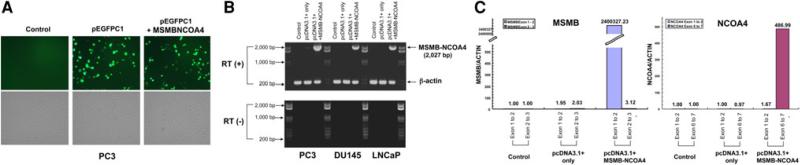
Fig. 4.
Transfection with expression vectors carrying the MSMB-NCOA4 fusion transcript. a Transfection efficiency was shown by GFP-positive cells. The extent of EGFP expression in PC3 was determined following transfection with the pEGFPC1-MSMBNCOA4 plasmid using fluorescence microscopy. b RT-PCR amplification of the MSMB-NCOA4 fusion gene using RT-MSMB-NCOA4 F1 and RT-MSMB-NCOA4 Rev1 primer pairs in PC3, DU145 and LNCaP cells transfected with pcDNA3.1+-MSMB-NCOA4 and pcDNA3.1+ vector only. RT (+) is shown in the top panel, and RT (–) is shown in the bottom panel. c Expression level of the MSMB-NCOA4 fusion transcript in transfected PC3 cells by TaqMan real-time RT-PCR using primer and probe sets corresponding to different exons of MSMB and NCOA4 genes (Table S2). Overexpression was confirmed using primer/probe sets containing MSMB exons 1–2 and NCOA4 exons 6–7, but no overexpression was observed when primer/probe sets containing MSMB exons 2–3 and NCOA4 exons 1–2 were used. Relative quantities of the fusion transcript were normalized by beta-actin, and calibrated to control sample (PC3 only)
MSMB-NCOA4 mRNA was overexpressed in three cell lines transfected with an expression vector carrying MSMB-NCOA4 fusion transcript, compared with the vector-only control (Fig. 4b). Of them, PC3 showed the highest expression level of MSMB-NCOA4. We developed two TaqMan real-time PCR primer and probe sets to target different regions of MSMB (MSMB I across exons 1–2 and MSMB II across exons 2–3 of MSMB) and NCOA4 (NCOA4 I across exons 1–2 and NCOA4 II across exons 6–7 of NCOA4) (Table S1). The results show high expression of the fusion transcript in transfected PC3 cells using MSMB I and NCOA4 II assays (Fig. 4c).
Co-regulation of the expression of MSMB and MSMB-NCOA4 fusion transcripts in human tissue and cancer cell lines
To determine whether the expression of the fusion transcript is regulated by the MSMB promoter, we analyzed its expression in normal prostate tissue, prostate tumor tissue, trachea tissue, and prostate cancer cell lines using RT-PCR followed by agarose gel electrophoresis. A 2,027 bp RT-PCR product was identified in tissues from 5 of 9 prostate tumor cell lines, including PCa2b, NCI H660, PC3, LNCaP and RWPE1, using a forward RT-MSMB-NCOA4 F1 primer located in the 5′UTR of MSMB and a reverse primer, RT-MSMB-NCOA4 Rev1, located in the 3′UTR of NCOA4 (Table S1; Fig. 2, top panel). Using another reverse primer, RT-MSMB-NCOA4 Rev2, we detected a 643-bp RT-PCR product, with exactly the same tissue and cell distribution as the products obtained using the RT-MSMB-NCOA4 Rev1 primer (Fig. 2, middle panel). While all tissues expressed the MSMB-NCOA4 fusion transcripts, normal prostate tissue showed the highest expression level.
To further survey for the presence of MSMB-NCOA4 fusion transcripts in tissues and cell lines, we developed a TaqMan real-time RT-PCR assay using an absolute quantitation method (see “Materials and methods” for details). Using this assay, the MSMB-NCOA4 fusion transcript was detected in 14 of 26 tumor cell lines and all eight human tissues (Table S2), and, again, was expressed at the highest level in normal human prostate tissue compared with other tissues and cells. We also examined levels of MSMB, NCOA4 and AR mRNA expression in relation to the fusion transcripts (Table S2). Expression of the MSMB-NCOA4 fusion transcript was positively correlated with MSMB expression in tissues and cancer cell lines in a statistically significant manner. The Pearson correlation for the value of MSMB-NCOA4 transcript I and MSMB transcription is 0.70, yielding a two-tailed P value of ~6.6 × 10–6.
Functional analysis of MSMB promoter activity by deletion mapping
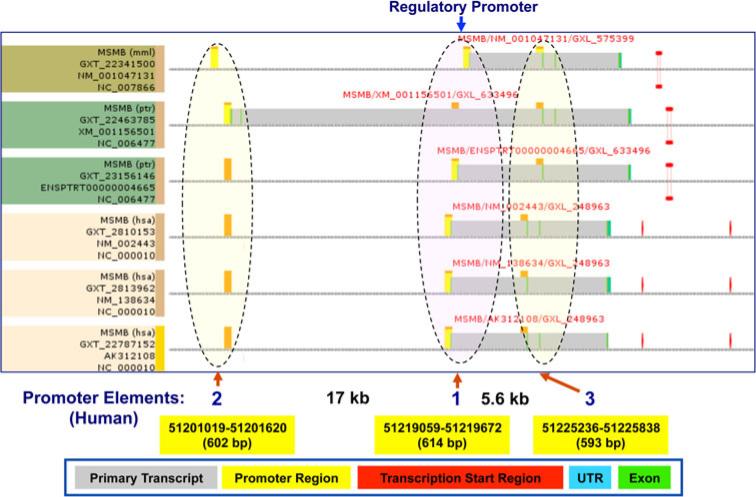
In silico promoter region prediction and gene analysis were performed using the ElDorado and Gene2Promoter online programs. The potential promoter region upstream of the MSMB gene was predicted by interspecies comparison. Multiple promoter elements are located immediately upstream of MSMB exon 1, and likely play a role in the regulation of MSMB expression (Fig. 5).
Fig. 5.
Identification of promoter elements of the MSMB gene using EIDorado. Three potential promoter regions surrounding the MSMB gene are shown. Promoter element 1 is located adjacent to the MSMB 5′UTR region. The promoter elements 2 and 3 are located 17 kb upstream and 5.3 kb downstream of the MSMB transcription start site, respectively
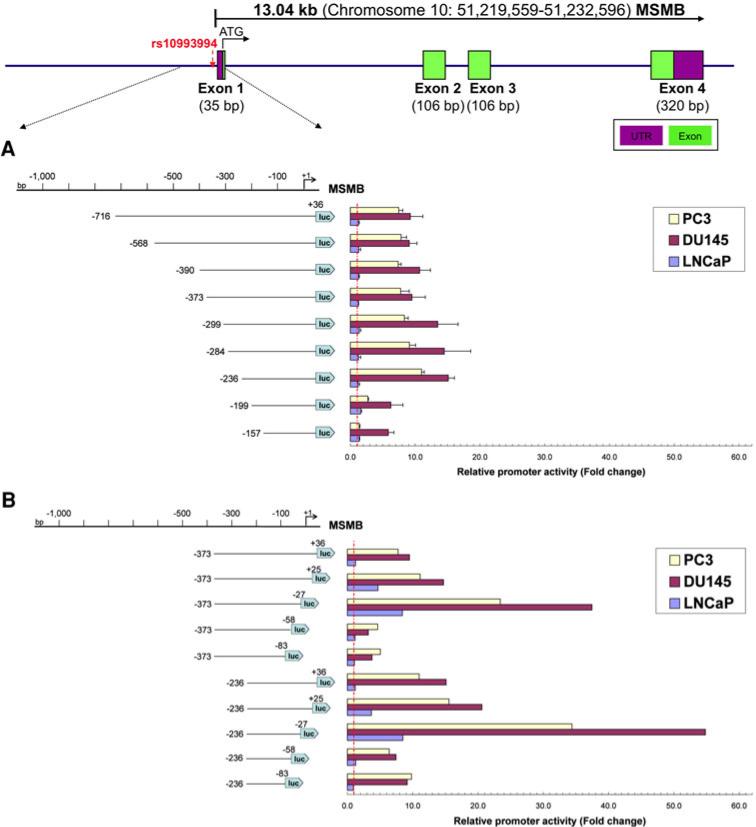
To determine the minimal sequences required for promoter function and identify cis-acting elements controlling MSMB promoter activity, a series of truncated luciferase constructs were generated by progressive deletions at the 5′ end of a 752 bp fragment (–716/+36) comprising the putative promoter region, to produce nine constructs (Fig. 6a). Plasmids containing the MSMB gene fragments were transiently transfected into three prostate cancer cell lines and the promoter activity was measured as described in the “Materials and methods”.
Fig. 6.
Functional localization of the MSMB promoter. A schematic of the MSMB gene structure is shown above. Four MSMB exons are indicated by the numbered green rectangles. A schematic diagram of the 1,000 bp 5′-flanking region of MSMB exon 1 and 5′ serial (a) or 3′ serial (b) truncation constructs of the MSMB promoter and their corresponding luciferase activities in different cell types are shown. Serial deletions at the 5′ and the 3′ ends of the promoter fragment of MSMB are shown on the left. The promoter activities measured after transfection into PC3, DU145 and LNCaP cells are shown on the right. The relative size and position of fragments cloned into the pGL3 vector are indicated by the lines below the schematic, and the numbers in parentheses on either side of each fragment indicate the distance in nucleotides upstream from the MSMB start codon of the 5′ and 3′ ends of each fragment. The luciferase activity of the pGL3 constructs is shown as fold-increase of corrected light units relative to an empty pGL3 vector control. Values represent the mean, and error bars indicate the SEM of at least three independent experiments (color figure online)
As shown in Fig. 6a, luciferase activity was not diminished by deleting DNA up to position –236; rather, an increase in promoter activity was observed for this construct when compared with the original PGL3–716/+36 reporter in PC3 and DU145 cell lines. The highest promoter activity was observed in the construct PGL3–236/+36. Further deletions decreased promoter activity significantly. However, no promoter activity change was found in LNCaP cells (Fig. 6a).
The above data suggest that sequences contained between nucleotides (nt)–236 and +36 in PGL3–716/+36 construct bear the cis-acting elements required for maximal MSMB transcriptional activation, and this reporter vector as well as PGL3–373/+36 was used for the following 3′ deletion analysis. It should be noted, however, that the regions from nt –373 to –284 might contain additional negative cis-regulatory elements, relevant in the context of prostate cancer PC3 and DU145 cells.
To further identify the 3′ boundary of the core promoter, a series of plasmids were constructed sharing the same 5′ boundaries at either position –373 or –236, and variable 3′ ends from +36 to –83. The deletion constructs were transiently transfected in PC3, DU145 and LNCaP human prostate cancer cell lines, and luciferase activities were measured. As shown in Fig. 6b, the deletion from +36 to +25 within the MSMB core promoter region resulted in a 5-fold increase in luciferase activity, and the deletion from +25 to –27 resulted in a 19-fold and 24-fold increase in PC3 and DU145 cells, respectively (Fig. 6b). In contrast to the results from the 5′ deletion analysis, luciferase activity in LNCaP cells transfected with the 3′ deletion constructs showed 3.5-fold to 7-fold increases with the +25 to –27 deletions. Further deletions (–58, –83) dramatically reduced promoter activity, indicating that the core promoter is contained within the –236 to –27 region. These results suggested that sequences contained in the 210 bp proximal to the transcription initiation site are necessary and sufficient for MSMB promoter activity, and inhibitory elements were identified in both distal and proximal areas of the MSMB promoter. These two regulatory elements in the distal and proximal regions of the MSMB promoter were thus confirmed (Fig. 7). The 210 bp core promoter had the highest transcription activity in all the prostate cancer cell lines we tested, indicating that this fragment is responsible for basal MSMB promoter activity. Because deletion of the immediate 5′UTR results in significantly enhanced transcription activity in the prostate cancer cell lines, we suggest that there are negative cis-acting elements in this region.
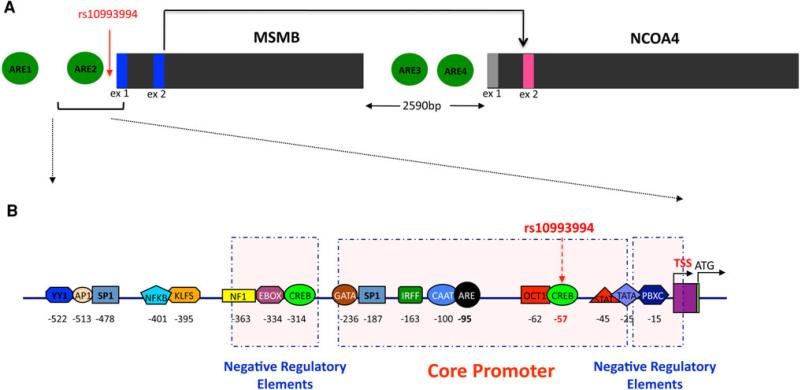
Fig. 7.
Schematic model of regulatory binding sites 5′ to the MSMB and NCOA4 genes. a Potential androgen response elements (AREs) upstream of MSMB and in between MSMB and NCOA4 in the top 15 sequences examined. Scores were derived from a frequency matrix built based on (Denayer et al. 2010). The sequences for the potential AREs are GGGTCACAAAGTTCT, CACTCAATGTGTTCT, GGTTCAGGCAGTTCT, and AGAGAACCCTGTTCT for ARE1, ARE2, ARE3, and ARE4, respectively. b A schematic of predicted transcription factor binding modules in the proximal MSMB promoter region. Predictions are organized into three groups, one core regulatory element group and two negative regulatory element groups
Computational analysis of splice sites participating in MSMB-NCOA4 fusion transcripts
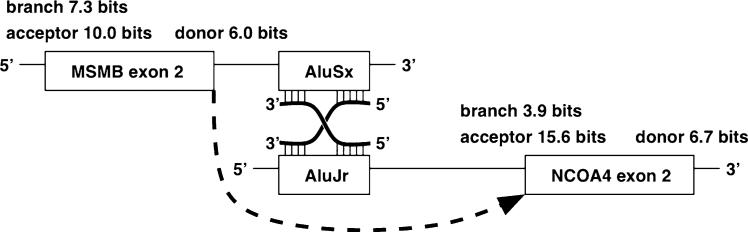
To begin to explore the mechanism of generation of the hybrid MSMB-NCOA4 fusions, we characterized the splice sites, flanking sequences and branch point surrounding the exons involved using information theory-based methods (Rogan et al. 1998). We identified that the MSMB exon 2 participating in the major trans-splice transcript contains a strong splice donor and the splice acceptor for exon 2 of NCOA4 is extremely strong (Fig. 8). At 15.6 bits the acceptor of exon 2 of NCOA4 is in the strongest ~4 % of acceptors in the genome. We also identified Alu sequences immediately downstream of MSMB exon 2 and upstream of NCOA4 exon 2 that may contribute to splicing (Fig. 8, see “Discussion”).
Fig. 8.
A conceptual model for splicing of the MSMB/NCOA4 transcripts. A reverse transcript of an Alu element may base pair with the two independent mRNA transcripts or within a single transcript to form a helix with a crossover as in a Holliday junction. The junction would be in the middle of the repeat element sequences. A prediction of this model is that there would be a reverse promoter driving either one or both of the Alu element copies. If they are strong and local, they would match one sequence perfectly and match the other well enough to more frequently bring the two exons together. An alternative hypothesis is that the RNA polymerase reading the AluSx dislodges and starts reading AluJr since the sequences are similar. Then the splicing proceeds as before to fuse MSMB exon 2 to NCOA4 exon 2. In this case there will be a continuous mRNA with a transition somewhere inside the Alu sequences. Alternatively, stabilization of lariat structures required by cis-splicing can be enhanced through the presence of these Alu sequences in the primary transcript
In silico identification of potential cis regulatory elements of the MSMB promoter
The MSMB –236/–27 promoter fragment exhibited the highest transcriptional activity in the luciferase reporter assay. We therefore performed a computational analysis of the MSMB core promoter sequence to identify potential cis regulatory elements using the MatInspector and TFSEARCH programs. Analysis revealed putative binding sites for various transcription factors (Fig. 7b) and, of these, CREB, MYOD, STAT, OCT, PBXC, ETS, SP1, TBP, CAAT and TATA had high match scores. We have already published the observation that the GWAS-associated variant rs10993994 is located in the core –236/–27 MSMB promoter region and affects CREB binding (18); however, the impact of the risk haplotype on other transcription factors identified here not yet been assayed in vitro.
Androgen-induced gene regulation typically occurs through AR interaction with specific DNA sequences known as androgen response elements (AREs). Regulation through positive and negative AREs (pARE and nARE) has been reported for AR-target genes, including prostate-specific antigen (PSA) and transforming growth factor β (TGFβ) (Fabre et al. 1994; Qi et al. 2012). Sequence analysis revealed two putative ARE-like elements located upstream of MSMB and two ARE-like elements in between the MSMB and NCOA4 genes. These elements are designated ARE1 to ARE4 in Fig. 7a. The sequences for MSMB and NCOA4 AREs are very similar to either consensus AREs or some natural ARE motifs, as shown in Figure S2. ARE2 localized to the core MSMB promoter region containing the sequence CACTCAATGTGTTCT; this is similar to the ARE structure found at the proximal promoter region of TGFβ (Qi et al. 2012).
MatInspector was also used to identify putative functional frameworks in the regulatory regions of the MSMB promoter. We searched for common modules containing two transcription factor-binding sites at a distance between 5 and 50 bp. The modules EBOX–CREB, GATA–SP1, BRNF–RXRF, IRFF–CREB, CAAT–CAAT, OCT1– PBXC, STAT–ETS were identified within the 752 bp MSMB promoter sequence (Fig. 7b).
To investigate the regulatory and functional connection of MSMB with NCOA4, we performed a literature search combined with TF binding site analysis using Genomatix BiblioSphere and MatInspector. Transcripts which showed co-citations with transcription factors, functional co-citations, and co-citations with other genes were selected. Based on these criteria, twenty-one TFs which showed functional co-citations at GFG (gene-for-gene) level 3 were identified. Notably, the MSMB gene is functionally connected with the NCOA4 gene in relation to the androgen receptor signaling pathway and the beta-catenin signaling pathway (Figure S3).
Discussion
Large numbers of genetic loci have been identified through GWAS studies of cancer (Chung and Chanock 2011), but relatively few have begun to be understood at a molecular level. In addition, the effect of associated variants on both transcriptional regulation as well as alternative splicing has been proposed but rarely demonstrated. While alternative splicing is known to be a major source of protein diversity in mammalian cells, a role for transcripts joining exons from two genes (transcription-induced cis- or trans-splicing) is also becoming increasingly clear (Akiva et al. 2006; Gray et al. 1999; Kaye 2009; Parra et al. 2006). For adjacent genes, read-through transcription has been demonstrated in a few cases, but trans-splicing has also been documented. Remarkably, a comprehensive analysis of the 5′ transcription start site of 399 annotated protein coding genes (ENCODE pilot project) using pooled 5′ RACE products to hybridize with a high-density genome tiling array showed that approximately 50 % of the genes had distal 5′ exons that often spanned adjacent genes at distances up to 200 kb (Denoeud et al. 2007). Fusion transcripts are often pathognomonic in hematopoetic malignancies and Ewing's sarcoma. The recent discovery of recurrent gene fusions in prostate (Tomlins et al. 2005) and lung cancer (Choi et al. 2008; Koivunen et al. 2008; Perner et al. 2008; Rikova et al. 2007) points to their role in common epithelial tumors as well.
In this paper, we document a chimeric transcript, present in both normal and tumor cells, with relevance to prostate cancer. A SNP upstream of MSMB (rs10993994) has been implicated by multiple groups in prostate cancer risk by GWAS (Eeles et al. 2008; Pomerantz et al. 2010; Thomas et al. 2008) and we previously reported that MSMB transcription was significantly up-regulated in normal prostate tissue compared to prostate tumor tissue and cell lines derived from metastatic prostate cancers (Lou et al. 2009). These observations suggest that MSMB, which encodes MSP, and its neighboring gene NCOA4, an androgen receptor co-activator, may play an important role in multistep carcinogenesis. Here, we report the existence of common novel fusion transcripts resulting from these two genes. We identified four MSMB-NCOA4 fusion transcripts in human prostate and trachea tissues by RT-PCR and 5′ RACE, all of which incorporated canonical RNA splice sites from both genes. The most abundant chimeric spliced transcript, occurring in 80 % of the events, removes several exons of the upstream gene and the first exon of the downstream gene (Akiva et al. 2006). The four MSMB-NCOA4 fusion transcripts described in this study fall within this category of common chimeric transcripts.
MSMB and NCOA4 genes map to human chromosome 10q11, and are situated about 3 kb apart. The predominant fusion transcript, MSMB-NCOA4 1 (E2-E2), contains the ATG initiation codon of MSMB fused in-frame to NCOA4 exons 2–10. NCOA4 is a chromosomal fusion partner of the RET proto-oncogene, a transmembrane receptor of the tyrosine kinase family, that is frequently activated in papillary thyroid carcinoma (Santoro et al. 1994). NCOA4 interacts with the AR to enhance AR transcriptional activity, through hydrophobic side chain interactions, and NCOA4 may function as a linker protein between the nuclear receptors and the basal transcription machinery, thereby recruiting or stabilizing the pre-initiation complexes on the promoter (Alen et al. 1999).
Analysis of the splice sites participating in the major fusion transcript revealed that they are canonical splice sites, and in fact, the acceptor in front of NCOA4 exon 2 is extremely strong. This fusion resembles the 80 % of such fusions that join to the second exon of the downstream gene (Akiva et al. 2006) and the two genes are separated by only 3 KB. This favors the model of cis-read-through for adjacent genes (Akiva et al. 2006). However, there is an AluSX element just 3′ of the MSMB exon 2, and an AluJr element 5′ to NCOA4 exon 2. It is possible therefore that these elements facilitate a trans-splicing event between hnRNAs of the two genes (Fig. 8). The mechanism proposed by which Alu elements mediate alternative splicing is independent of cis- trans-distinctions; however, it may be required for trans-splicing. It is also interesting that the expression of the fusion transcript is correlated with the expression of MSMB and with the genotype 5′ to the locus. Allele-specific expression of trans-splicing products has been observed, and therefore additional genetic variants might participate in such epigenetic events. For example, another variant strongly associated with prostate cancer is rs10486567, located in intron 2 of the JAZF1 gene, another gene known to participate in trans-splicing (Li et al. 2008; Thomas et al. 2008).
Alignment of the predicted human and mouse MSMB promoter regions demonstrates a high degree of similarity and conservation. This suggests that transcription of MSMB may be regulated by similar mechanisms across mammalian species. The minimal MSMB core promoter region was identified as a –236/–27 fragment relative to the previously reported transcription start site. In addition, two negative regulatory elements in the distal and proximal regions of the MSMB promoter were confirmed (Fig. 7b). The 210 bp core promoter had the highest transcription activity in all the prostate cancer cell lines we tested, indicating that this fragment is responsible for basal MSMB promoter activity. Deletion of the immediate 5′UTR results in significantly enhanced transcription activity in the PCa cell lines, suggesting that negative cis-acting elements are in this region.
Through in silico analysis, the putative transcription factor binding sites CREB, MYOD, STAT, OCT, PBXC, ETS, SP1, TBP, CAAT and TATA, and modules CAAT–CAAT, OCT1–PBXC, STAT–ETS were identified within the core MSMB fragment –236/–27. The 3′ deletion assay identified 31 bp (–27 to –58) as necessary for the repressor effect (Fig. 6b). These results suggest that repression of the MSMB gene requires recruitment of other transcription factors to the 3′ end of the promoter sequence. The further deletion from –27 to –58 abrogated promoter activity, demonstrating that the region surrounding rs10993994, located at –57, is critical for MSMB promoter activation. Several putative transcription factor binding sites were identified between –27 and –58, including E4F, EGRF, HIC, GATA and CREB. Mutation at rs10993994 destroys a potential binding site for CREB, leading to a sharp decrease of MSMB promoter activity. Therefore, the transcription factor binding sites and modules identified in this study are likely to be key controlling elements in the regulation of the MSMB gene. These findings provide a basis for further delineating the precise function of each transcription factor in MSMB regulation, and revealing the molecular basis of MSMB repression.
The androgen receptor regulates the expression of many genes that are essential for male tissue-specific differentiation. The receptor acts by direct binding to AREs found in the promoters of regulated genes. These AREs are organized as two 5′-TGTTCT-3′ like motifs separated by three base pairs. They may vary in structure and be present at considerable distance upstream from regulated promoters. The presence of multiple AREs in the MSMB/NCOA4 region indicates that these genes might be subjected to AR regulation through these AREs. These observations suggest a mechanism in which multiple transcription factors bind to the MSMB promoter and interact with AR through AREs contributing to MSMB expression. In addition, the gene product of NCOA4, is an AR co-activator that may specifically regulate its own expression and/or the expression of MSMB. Further characterization of the predicted AREs in the MSMB/NCOA4 genes may provide a greater understanding of AR-mediated PCa progression.
In conclusion, this study confirms the presence of a novel fusion transcript between MSMB and NCOA4 expressed in PCa cells and surrounding tissues, giving further insight into the role of this complex locus in PCa susceptibility. The data provide an understanding of the MSMB 5′ regulatory response elements controlling MSMB expression that also probably control the expression of the fusion transcript, and a mechanism through which the fusion transcript may arise. The identification of the MSMB-NCOA4 fusion transcript and the functional connections between MSMB, NCOA4, and androgen receptor suggests new markers and new therapeutic targets for PCa.
Materials and methods
Cell lines
The benign immortalized prostate cell line RWPE1 and the PCa cell lines PC3, DU145, LNCaP, PCa2b, NCIH660, PZHPV7, CAHPV10, VCaP, 22RV1, WPMY-1, WPE-Stem and breast cancer cell line SKBR3 and stomach cell line AGS were obtained from American Type Culture Collection (ATCC). Breast cancer cell line MCF10F was purchased from the University of Michigan, SUM229, SUM159, and SUM149 were purchased from Asterand, Inc. Cells were maintained according to the supplier's instructions.
RNA isolation and cDNA synthesis and RT-PCR
Total RNAs were extracted and purified as described (18). One microgram of total RNA was reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche) with oligonucleotide (dT)18 primer according to the manufacturer's instructions. The primers designed for RT-PCR and 5′ RACE assays corresponding to regions across the MSMB and NCOA4 genes are listed in Table S1. RT-PCR was carried out in a volume of 50 μl containing 100 ng of cDNA and 20 pmol of each primer using Platinum® TaqGold HIF Kit (Invitrogen). Thermal cycling conditions included 10 cycles of 94 °C for 30 s, 58 °C for 45 s and 68 °C for 45 s, and 28 cycles of 94 °C for 30 s, 56 °C for 45 s and 68 °C for 45 s. After RT-PCR, products were separated on a 1.5 % agarose gel.
5′ RACE assays
5′ RACE assays were carried out with both First Choice RLM RACE kit (Ambion) and 5′/3′ RACE kit, 2nd Generation (Roche) to confirm the transcription starting site (TSS) of the MSMB-NCOA4 fusion transcripts, according to the manufacturer's directions.
For sequence analysis, the RT-PCR and 5′ RACE products were subcloned into a pCR2.1 vector using a TOPO TA Cloning kit (Invitrogen) according to the manufacturer's instructions. At least twelve clones were sequenced in an ABI 3730 DNA Sequencer (Applied Biosystems) for each product tested. Each experiment was done at least two times.
Construction of expression vectors and transfection
The expression constructs of the full-length MSMB-NCOA4 transcript were generated by subcloning the corresponding cDNA fragment into pcDNA 3.1+ (Invitrogen) and pEGFPC1 (Clontech) vectors. The MSMB-NCOA4 cDNAs were obtained by RT-PCR with the RT-MSMB-NCOA4 F1 forward primer and the reverse RT-MSMB-NCOA4 Rev1 primer (Table S1). The MSMB-NCOA4 expression construct contained full-length MSMB-NCOA4 fusion cDNA, including 141 bp of MSMB and 1,886 bp of NCOA4, starting at the transcription start site of MSMB part of the fusion. The constructs were verified by sequencing.
We have compared three different transfection reagents, FuGENE 6, FuGENE HD and HilyMax. High transfection efficiency was detected in all cell lines when using HilyMax; therefore, we selected HilyMax for transfection experiments in this study. The pcDNA 3.1+-MSMB-NCOA4 and pEGFPC1-MSMB-NCOA4 plasmid DNAs were transfected into PC3, DU145 and LNCaP cells using the HilyMax transfection reagent (Dojindo) according to the manufacturer's instructions. Briefly, 24 h before transfection, 1 × 105 to1 × 106 cells were plated on 100 mm dishes. Equal amounts (3 μg) of pcDNA 3.1+-MSMB-NCOA4 and pcDNA 3.1+ alone were individually transfected into each plate. After 24 h of transfection, cells were harvested and total RNA was extracted and purified as described (18). To establish stable cell lines, the PC3 cells transfected with pcDNA 3.1+-MSMB-NCOA4 were selected with 650 μg/ml G418 in complete RPMI 1640 medium. Transfection efficiency was measured by analysis of the percentage of GFP-positive cells in the PC3 cell line transfected with pEGFPC1-MSMB-NCOA4 plasmid DNA.
The expression level of the MSMB-NCOA4 fusion transcript in the transfected cells was examined by RTPCR using RT-MSMB-NCOA4 F1 and RT-MSMBN-COA4 Rev1 primers (Table S1), and by the TaqMan real-time RT-PCR method. To distinguish between fusion and endogenous transcripts, two primers/probe sets for MSMB or NCOA4 were used. Hs00738230_m1 and Hs00159303_m1 corresponded to MSMB exons 1–2 and exons 2–3, respectively, whereas NCOA4 II primer probe sets (Table S1) and Hs00428328_m1 corresponded to NCOA4 exons 6–7 and exons 1–2, respectively. The expression of the beta-actin gene (Hs99999903_m1) was used to normalize results.
Real-time quantitative RT-PCR
TaqMan® Gene expression assay primer and probe (FAM-labeled) sets (Applied Biosystems) and custom-designed TaqMan MGB-labeled primers and probes were used for detection of the mRNA expression levels of MSMB-NCOA4 fusion, MSMB (PSP94), NCOA4 and AR genes in human tissues and other cancer cell lines (Table S1). Two primer/probe sets were designed to measure fusion transcript levels, with the same forward primer and probe but different reverse primer sequences. The reverse primer of the first MSMB-NCOA4 set, Assay ME2NE2 I, consisted of 22 nucleotides with 3 bp of the MSMB exon 2 sequence at the 3′ end of the primer and 19 bp of the NCOA4 exon 2 sequence. The reverse primer of the second MSMB-NCOA4 set, Assay ME2NE2 II, was 21 bp in length, containing 10 bp of the MSMB exon 2 sequence at the 3′ end of the primer (Table S1).
Standard curve construction
Standard curves were generated using a dilution series of plasmids containing full-length cDNA of MSMB (GenBank Accession Number BC005257.1) (ATCC), NCOA4 (Gen-Bank Accession Number NM_005437), AR (GenBank Accession Number BC132975), and beta-actin (GenBank Accession Number BC061604) (Open Biosystems). The pcDNA 3.1+-MSMB-NCOA4 construct was used for the standard curve to measure levels of fusion transcript in tissues and cancer cell lines. The copy number of plasmid cDNA was calculated by optical density according to the exact molar mass determined from the sequences. Serial dilutions were made to obtain 101–107 copies. The slope and intercept were calculated for each run using linear regression analysis of log copy number versus threshold cycle (Ct) value for both target genes and beta-actin standard curves. The linear dynamic range and inter-intra assay precision were examined from three separate experiments. The relative mRNA expression level of the target genes was normalized for human tissue and cancer cell lines by the following formula: (copy number of target gene)/(copy number of beta-actin) × 105 (Table S2).
Protein expression
In vitro expression construct
The MSMB-NCOA4 fusion protein was expressed using the in vitro TNT Quick Coupled Transcription/Translation System (Promega) in the presence of [35S] methionine, according to the manufacturer's protocol (Amersham Pharmacia Biotech).
Expression of proteins in vivo
Whole cell lysates were prepared from the PC3 cell line 72 h after transfection with pcDNA3.1+-MSMB-NCOA4. Seventy-five micrograms of whole cell protein were separated in a 4–12 % NuPAGE Bis–Tris gel, transferred to PVDF membrane (Invitrogen). The primary antibodies used were polyclonal goat-anti-PSP94 (R&D system) at a dilution of 1:200, and mouse monoclonal anti-NCOA4 (Abnova) at a dilution of 1:500 to analyze the MSMB-NCOA4 fusion protein. Beta-actin was used as an internal control. HRP-conjugated anti-goat IgG, anti-mouse IgG (1:3,000, Cell Signaling Technology MA, USA) and anti-rabbit IgG were used as secondary antibodies.
To further confirm the identity of the MSMB-NCOA4 fusion protein, the proteins were immunoprecipitated using the Immunoprecipitation Kit—Dynabeads Protein G (Invitrogen) according to the manufacturer's protocol. Five microgram of the polyclonal goat-anti-PSP94 antibody was pre-incubated with 50 μl of Dynabeads Protein G, and then 200 μl of cell lysate was added (200–500 μg protein) to the resuspended Dynabeads–Ab complex, incubated for 30 min, then washed three times with 200 μl of wash buffer The immunprecipitate was electrophoresed and transferred to PVDF membranes as described above. The membranes were blotted with mouse monoclonal anti-NCOA4 antibody against the NCOA4 C-terminal region. We also used the anti-NCOA4 C-terminal antibody to immunoprecipitate protein, and then performed a western blot using the goat-anti-PSP94 antibody.
In silico analysis of the MSMB regulatory region
A 40 kb sequence surrounding the MSMB gene was obtained through the Ensembl database and the UCSC genome browser for human, mouse and chimpanzee. Identification of putative TF-binding sites and modules in the regulatory region upstream of the transcription start site of the MSMB gene was performed online at Genomatix using ElDorado, PromoterInspection, MatInspector, BiblioSphere and FrameWorker programs (Genomatix Software, Germany) and at the TFSEARCH website (http://www.cbrc.jp/research/db/TFSEARCH.html).
Generation of luciferase reporter plasmids
A promoter fragment was generated by PCR using gene-specific forward primers starting at –716 and reverse primers starting at +36 relative to the transcription start site of the MSMB gene from MCF10A (breast epithelial cell) genomic DNA. The PCR product was cloned into the TOPOTA vector (Invitrogen), and the presence of the C nucleotide polymorphism at both rs10993994 and rs12770171 was confirmed by sequencing. A series of truncated MSMB promoter constructs, including 9 deletions from the 5′ side and 10 deletions on the 3′ side, were created by PCR using the primers shown in Table S3. Luciferase reporter plasmids were generated as described (Lou et al. 2009).
Cell transfection and luciferase assays
Three prostate cancer cell lines, PC3, LNCaP and DU145, were used for the analysis of promoter constructs. Cells were transfected with plasmid DNA and luciferase assay was performed as described (Lou et al. 2009).
Supplementary Material
Acknowledgments
The authors would like to thank Allison Bierly and Wei Tan for critical reading of the manuscript and helpful comments. The project was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and from SAIC-Frederick under contract #NO1-CO-12400. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- ARE
Androgen response element
- AR
Androgen receptor
- GWAS
Genome-wide association studies
- PCa
Prostate cancer
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-012-1182-2) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest.
Ethical standards The experiments comply with the current laws of the United States of America.
Contributor Information
Hong Lou, Human Genetics Section, Basic Research Program, SAIC-Frederick Inc., National Cancer Institute-Frederick, Frederick, MD 21702, USA.
Hongchuan Li, Molecular Immunology Section, Basic Research Program, SAIC-Frederick Inc., National Cancer Institute-Frederick, Frederick, MD 21702, USA.
Meredith Yeager, Core Genotyping Facility, Advanced Technology Program, SAIC-Frederick, Inc., National Cancer Institute-Frederick, Frederick, MD 21702, USA.
Kate Im, Cancer and Inflammation Program, Laboratory of Experimental Immunology, Center for Cancer Research, National Cancer Institute-Frederick, Frederick, MD 21702, USA.
Bert Gold, Cancer and Inflammation Program, Laboratory of Experimental Immunology, Center for Cancer Research, National Cancer Institute-Frederick, Frederick, MD 21702, USA.
Thomas D. Schneider, Gene Regulation and Chromosome Biology Laboratory, Molecular Information Theory Group, Frederick, MD 21702, USA
Joseph F. Fraumeni, Jr., Division of Cancer Epidemiology and Genetics, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA
Stephen J. Chanock, Division of Cancer Epidemiology and Genetics, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA
Stephen K. Anderson, Molecular Immunology Section, Basic Research Program, SAIC-Frederick Inc., National Cancer Institute-Frederick, Frederick, MD 21702, USA Cancer and Inflammation Program, Laboratory of Experimental Immunology, Center for Cancer Research, National Cancer Institute-Frederick, Frederick, MD 21702, USA.
Michael Dean, Cancer and Inflammation Program, Laboratory of Experimental Immunology, Center for Cancer Research, National Cancer Institute-Frederick, Frederick, MD 21702, USA.
References
- Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Balool HH, Weber D, Liu Y, Wade M, Guleria K, Nam PL, Clayton J, Rowe W, Coxhead J, Irving J, Elliott DJ, Hall AG, Santibanez-Koref M, Jackson MS. Post-transcriptional exon shuffling events in humans can be evolutionarily conserved and abundant. Genome Res. 2011;21:1788–1799. doi: 10.1101/gr.116442.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alen P, Claessens F, Schoenmakers E, Swinnen JV, Verhoeven G, Rombauts W, Peeters B. Interaction of the putative androgen receptor-specific coactivator ARA70/ELE1alpha with multiple steroid receptors and identification of an internally deleted ELE1beta isoform. Mol Endocrinol. 1999;13:117–128. doi: 10.1210/mend.13.1.0214. [DOI] [PubMed] [Google Scholar]
- Bjartell AS, Al-Ahmadie H, Serio AM, Eastham JA, Eggener SE, Fine SW, Udby L, Gerald WL, Vickers AJ, Lilja H, Reuter VE, Scardino PT. Association of cysteine-rich secretory protein 3 and beta-microseminoprotein with outcome after radical prostatectomy. Clin Cancer Res. 2007;13:4130–4138. doi: 10.1158/1078-0432.CCR-06-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BL, Cramer SD, Wiklund F, Isaacs SD, Stevens VL, Sun J, Smith S, Pruett K, Romero LM, Wiley KE, Kim ST, Zhu Y, Zhang Z, Hsu FC, Turner AR, Adolfsson J, Liu W, Kim JW, Duggan D, Carpten J, Zheng SL, Rodriguez C, Isaacs WB, Gronberg H, Xu J. Fine mapping association study and functional analysis implicate a SNP in MSMB at 10q11 as a causal variant for prostate cancer risk. Hum Mol Genet. 2009;18:1368–1375. doi: 10.1093/hmg/ddp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YL, Takeuchi K, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Hamada T, Haruta H, Watanabe H, Kurashina K, Hatanaka H, Ueno T, Takada S, Yamashita Y, Sugiyama Y, Ishikawa Y, Mano H. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- Crawford ED. Epidemiology of prostate cancer. Urology. 2003;62:3–12. doi: 10.1016/j.urology.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Denayer S, Helsen C, Thorrez L, Haelens A, Claessens F. The rules of DNA recognition by the androgen receptor. Mol Endocrinol. 2010;24:898–913. doi: 10.1210/me.2009-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, Dike S, Wyss C, Henrichsen CN, Holroyd N, Dickson MC, Taylor R, Hance Z, Foissac S, Myers RM, Rogers J, Hubbard T, Harrow J, Guigo R, Gingeras TR, Antonarakis SE, Reymond A. Prominent use of distal 5’ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O'Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- Fabre S, Manin M, Pailhoux E, Veyssiere G, Jean C. Identification of a functional androgen response element in the promoter of the gene for the androgen-regulated aldose reductase-like protein specific to the mouse vas deferens. J Biol Chem. 1994;269:5857–5864. [PubMed] [Google Scholar]
- Fears S, Mathieu C, Zeleznik-Le N, Huang S, Rowley JD, Nucifora G. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc Natl Acad Sci USA. 1996;93:1642–1647. doi: 10.1073/pnas.93.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- Kaye FJ. Mutation-associated fusion cancer genes in solid tumors. Mol Cancer Ther. 2009;8:1399–1408. doi: 10.1158/1535-7163.MCT-09-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas R, Lee J, Richards WG, Sugarbaker DJ, Ducko C, Lindeman N, Marcoux JP, Engelman JA, Gray NS, Lee C, Meyerson M, Janne PA. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AR, Li Q, Hudson WA, Chen W, Sam T, Yao Q, Lund EA, Wu B, Kowal BJ, Kersey JH. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- Lou H, Yeager M, Li H, Bosquet JG, Hayes RB, Orr N, Yu K, Hutchinson A, Jacobs KB, Kraft P, Wacholder S, Chatterjee N, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Ma J, Gaziano JM, Stampfer M, Schumacher FR, Giovannucci E, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Anderson SK, Tucker M, Hoover RN, Fraumeni JF, Jr, Thomas G, Hunter DJ, Dean M, Chanock SJ. Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. Proc Natl Acad Sci USA. 2009;106:7933–7938. doi: 10.1073/pnas.0902104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Mertens F, Johansson B. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosom Cancer. 2005;43:350–366. doi: 10.1002/gcc.20212. [DOI] [PubMed] [Google Scholar]
- Nacu S, Yuan W, Kan Z, Bhatt D, Rivers CS, Stinson J, Peters BA, Modrusan Z, Jung K, Seshagiri S, Wu TD. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genomics. 2011;4:11. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Yeh S, Miyamoto H, Li G, Altuwaijri S, Yuan J, Han R, Ma T, Kuo HC, Chang C. Tissue prostate-specific antigen facilitates refractory prostate tumor progression via enhancing ARA70-regulated androgen receptor transactivation. Cancer Res. 2008;68:7110–7119. doi: 10.1158/0008-5472.CAN-07-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigo R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Li CX, Chen F, Wang Z, Ligr M, Melamed J, Wei J, Gerald W, Pagano M, Garabedian MJ, Lee P. Stimulation of prostate cancer cellular proliferation and invasion by the androgen receptor co-activator ARA70. Am J Pathol. 2008;172:225–235. doi: 10.2353/ajpath.2008.070065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Wagner PL, Demichelis F, Mehra R, Lafargue CJ, Moss BJ, Arbogast S, Soltermann A, Weder W, Giordano TJ, Beer DG, Rickman DS, Chinnaiyan AM, Moch H, Rubin MA. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Shrestha Y, Flavin RJ, Regan MM, Penney KL, Mucci LA, Stampfer MJ, Hunter DJ, Chanock SJ, Schafer EJ, Chan JA, Tabernero J, Baselga J, Richardson AL, Loda M, Oh WK, Kantoff PW, Hahn WC, Freedman ML. Analysis of the 10q11 cancer risk locus implicates MSMB and NCOA4 in human prostate tumorigenesis. PLoS Genet. 2010;6:e1001204. doi: 10.1371/journal.pgen.1001204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Gao S, Chu J, Zhou L, Wang Z. Negative androgen-response elements mediate androgen-dependent transcriptional inhibition of TGF-beta1 and CDK2 promoters in the prostate gland. J Androl. 2012;33:27–36. doi: 10.2164/jandrol.110.011999. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- Reeves JR, Dulude H, Panchal C, Daigneault L, Ramnani DM. Prognostic value of prostate secretory protein of 94 amino acids and its binding protein after radical prostatectomy. Clin Cancer Res. 2006;12:6018–6022. doi: 10.1158/1078-0432.CCR-06-0625. [DOI] [PubMed] [Google Scholar]
- Rickman DS, Pflueger D, Moss B, VanDoren VE, Chen CX, de la Taille A, Kuefer R, Tewari AK, Setlur SR, Demichelis F, Rubin MA. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Rogan PK, Faux BM, Schneider TD. Information analysis of human splice site mutations. Hum Mutat. 1998;12:153–171. doi: 10.1002/(SICI)1098-1004(1998)12:3<153::AID-HUMU3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- Santoro M, Dathan NA, Berlingieri MT, Bongarzone I, Paulin C, Grieco M, Pierotti MA, Vecchio G, Fusco A. Molecular characterization of RET/PTC3; a novel rearranged version of the RETproto-oncogene in a human thyroid papillary carcinoma. Oncogene. 1994;9:509–516. [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Thierry MD, Thierry MJ. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12–S14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Valtonen-Andre C, Savblom C, Fernlund P, Lilja H, Giwercman A, Lundwall A. Beta-microseminoprotein in serum correlates with the levels in seminal plasma of young, healthy males. J Androl. 2008;29:330–337. doi: 10.2164/jandrol.107.003616. [DOI] [PubMed] [Google Scholar]
- Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.