Abstract
Objectives
To locate the acquired antibiotic resistance genes, including the amikacin resistance transposon TnaphA6, in the genome of an Australian isolate belonging to Acinetobacter baumannii global clone 1 (GC1).
Methods
A multiply antibiotic-resistant GC1 isolate harbouring TnaphA6 was sequenced using Illumina HiSeq, and reads were used to generate a de novo assembly and determine multilocus sequence types (STs). PCR was used to assemble the AbaR chromosomal resistance island and a large plasmid carrying TnaphA6. Plasmid DNA sequences were compared with ones available in GenBank. Conjugation experiments were conducted.
Results
The A. baumannii GC1 isolate G7 was shown to include the AbaR3 antibiotic resistance island. It also contains an 8.7 kb cryptic plasmid, pAb-G7-1, and a 70 100 bp plasmid, pAb-G7-2, carrying TnaphA6. pAb-G7-2 belongs to the Aci6 Acinetobacter plasmid family. It encodes transfer functions and was shown to conjugate. Plasmids related to pAb-G7-2 were detected in further amikacin-resistant GC1 isolates using PCR. From the genome sequence, isolate G7 was ST1 (Institut Pasteur scheme) and ST231 (Oxford scheme). Using Oxford scheme PCR-based methods, the isolate was ST109 and this difference was traced to a single base difference resulting from the inclusion of the original primers in the gpi segment analysed.
Conclusions
The multiply antibiotic-resistant GC1 isolate G7 carries most of its resistance genes in AbaR3 located in the chromosome. However, TnaphA6 is on a conjugative plasmid, pAb-G7-2. Primers developed to locate TnaphA6 in pAb-G7-2 will simplify the detection of plasmids related to pAb-G7-2 in A. baumannii isolates.
Keywords: A. baumannii, conjugative resistance plasmids, aphA6
Introduction
Aminoglycosides are one of the antibiotic classes that can be used to treat carbapenem-resistant Acinetobacter baumannii infections. However, many isolates are resistant to one or more members of this class. Several genes, each conferring resistance to different combinations of aminoglycosides, have been reported in this species.1–6 These genes occur singly or in combinations leading to a variety of resistance phenotypes. In Australian A. baumannii isolates, only five aminoglycoside resistance genes have been identified to date, namely aacC1 (conferring resistance to gentamicin), aadB (conferring resistance to gentamicin, kanamycin and tobramycin), aphA1 (conferring resistance to kanamycin and neomycin), aphA6 (conferring resistance to amikacin, kanamycin and neomycin) and aadA1 (conferring resistance to streptomycin and spectinomycin).7,8 The aacC1 and aadA1 genes are in cassettes in a class 1 integron, and this integron is usually associated with the aphA1b gene in Tn6020 and located in a genomic resistance island in the chromosome of most global clone 1 (GC1) strains8 and some global clone 2 (GC2) strains.9 However, the aadB gene cassette is often in a small plasmid,6 and the location of the aphA6 gene is not known.
The aphA6 gene, which encodes an aminoglycoside (3′) phosphotransferase, was first identified in a large plasmid from a clinical A. baumannii isolate prior to 1988.10,11 The plasmid was shown to be transferable to other Acinetobacter species, but not to Escherichia coli,10 suggesting that its host range may be restricted to the Acinetobacter genus. Since then, the aphA6 gene has largely been found in Acinetobacter species,2,7,12 and in one study it was concluded that a gene rather than a plasmid or strain was responsible for the spread of resistance to amikacin.12 Supporting this view, we previously identified a transposon TnaphA6 (GenBank accession number JF343537) that consists of the aphA6 gene flanked by directly oriented copies of ISAba125, and found that this transposon is present in all of the Australian amikacin-resistant isolates belonging to GC2.7 The aphA6 gene was also found to be in TnaphA6 in three GC1 isolates, two of which, D78 and D81, were recovered in 2010 at a Sydney hospital and one of which, G7, was recovered in 2002 at a Melbourne hospital.13
Here, the whole genome sequence of isolate G7 was determined and used to identify the antibiotic resistance genes and the plasmids present, and to determine the location of TnaphA6. The AbaR-type genomic island was also assembled, and the sequence type (ST) of this isolate was determined and compared with that determined using standard methods.
Materials and methods
Isolates and isolate characterization
The GC1 isolates G7, D78 and D81 have previously been described.13 The ST was determined using standard procedures for the Oxford-based multilocus sequence typing (MLST) scheme (http://pubmlst.org/abaumannii/).
DNA sequencing and sequence analysis
Genomic DNA was isolated from G7 and sequenced using Illumina HiSeq at the Wellcome Trust Sanger Institute. Paired-end reads of 100 bp were assembled using Velvet,14 yielding 109 contigs with an average read depth of 76-fold. The presence of plasmids within the G7 assembly that were related to previously sequenced ones found in Acinetobacter strains were identified by a Stand alone BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of publicly available sequences included on the European Bioinformatics Institute plasmid list (http://www.ebi.ac.uk/genomes/plasmid.html). Contigs containing regions related to parts of the sequenced plasmids were retrieved using BLAST searches. Junctions between contigs predicted by comparison with related plasmids or AbaR were amplified using PCR and published primers8 or primers designed specifically for the purpose (Table 1), and the amplicons were sequenced as described previously. The complete AbaR island and plasmid sequences were assembled using Sequencher 5.1 (Gene Codes Corporation, Ann Arbor, MI, USA). Reading frames were predicted using ORF Finder (www.ncbi.nlm.nih.gov/projects/gorf/) and annotated manually. In addition, STs were determined from the reads using short read sequence typing15 in conjunction with the databases of the Oxford (http://pubmlst.org/abaumannii/) and Institut Pasteur (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html) schemes.
Table 1.
Primers used for assembling pAb-G7-2
| Primer name | Primer sequence (5′–3′) | Amplicon length (bp) |
|---|---|---|
| RH1398 | TTTGACGTTGCTCTTGTTGC | 991 |
| RH1399 | TTCTCCCAAGTGGTCAGGTC | |
| RH1394 | TGGTTGGCAGAACAAGATGA | 1372 |
| RH1395 | TCAAACGATGCAATGGAAGA | |
| RH1503 | GAAGATCCAGAAGCGGGATA | 1575 |
| RH1397 | CCATGTTCTTTTCCACATGC | |
| RH1501 | CTTGAGGAAGGGATGGTTGA | 1930 |
| aphA6R | GGACAATCAATAATAGCAAT | |
| aphA6F | ATACAGAGACCACCATACAGT | 2540 |
| RH1502 | TTGCTTTAATCGGTGGTTCC |
Conjugation
A spontaneous rifampicin-resistant mutant of A. baumannii ATCC 17978, which is resistant only to sulphonamides,16 was isolated for use as a recipient. Equal amounts of overnight cultures of the donor and recipient cells were mixed and incubated on an L agar plate overnight. Cells were resuspended and diluted in 0.9% saline, and transconjugants were selected by plating on L agar plates containing rifampicin (100 mg/L) and kanamycin (20 mg/L). To ensure that rare transconjugants were not spontaneous rifampicin-resistant derivatives of the donor, potential transconjugants were purified and screened for growth on L agar containing ceftazidime (25 mg/L), to which the donor was resistant and the recipient susceptible. Transconjugants identified in this way were tested for resistance to aminoglycoside antibiotics using disc diffusion with discs (Oxoid) containing amikacin (30 μg), gentamicin (10 μg), kanamycin (30 μg), neomycin (30 μg), tobramycin (10 μg) or netilmicin (30 μg).
GenBank accession numbers
The sequence of plasmid pAb-G7-2 was deposited in GenBank under accession number KF669606.
Results and discussion
G7 includes AbaR3
G7 is resistant to the aminoglycosides amikacin, gentamicin, kanamycin, neomycin, streptomycin and spectinomycin, as well as to tetracycline, ceftazidime and cefotaxime. It is susceptible to imipenem, meropenem and ticarcillin, and to nalidixic acid and ciprofloxacin. All of the antibiotic resistance genes found in the Tn6019-based complex transposon known as AbaR3, which is found in the comM gene of most GC1 isolates, were found in the G7 genome. The complete AbaR island was assembled from 11 contigs that matched AbaR3 using PCR primers published previously to link the contigs. The G7 genomic resistance island was found to be identical to AbaR3,17 and includes the aacC1-orfP-orfQ-aadA1 cassette configuration in a class 1 integron with the small deletion in the intI1 gene that has recently been identified as being characteristic of the AbaR3 lineage.18 Genes in AbaR3 account for resistance to gentamicin (aacC1), kanamycin and neomycin (aphA1b) and streptomycin and spectinomycin (aadA1) and the tet(A) determinant accounts for tetracycline resistance.
Plasmid pAb-G7-2
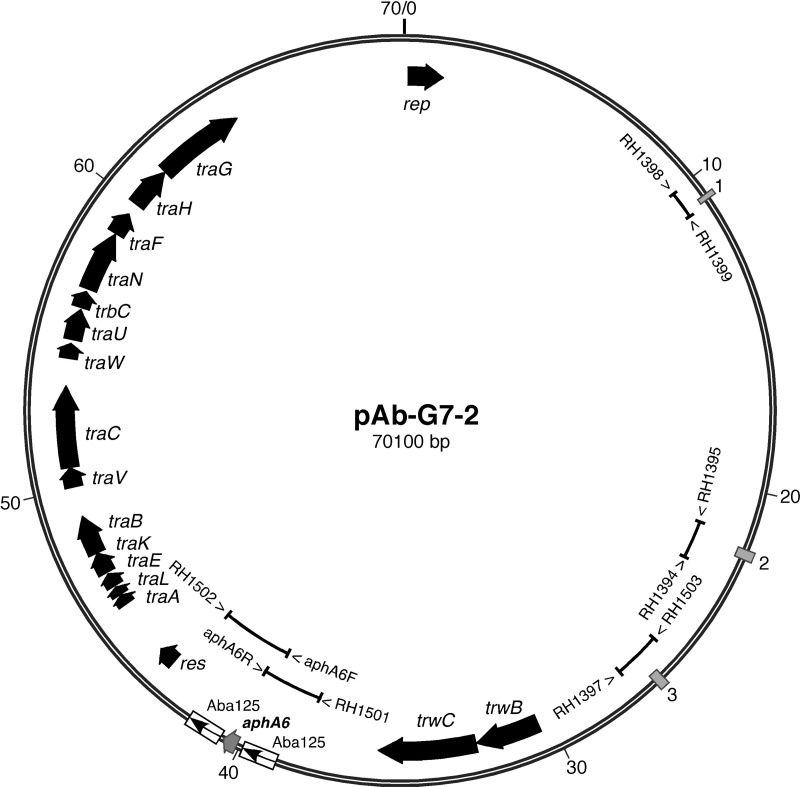
The resistance of G7 to amikacin has been shown to be due to the aphA6 gene in TnaphA6,13 but the transposon was not found in the AbaR3 island. G7 contains two plasmids. One, pAB-G7-1, of 8731 bp, is almost identical to pAB0057 (GenBank accession number CP001183), a cryptic plasmid found in AB0057.17 The second plasmid, named pAb-G7-2, includes TnaphA6. The complete 64 366 bp plasmid pACICU2 (GenBank accession number CP000865), which was reported as a cryptic plasmid in the GC2 isolate ACICU2,19 most closely matched pAb-G7-2, with over 99.9% identity over the full length of pACICU2. However, pAb-G7-2, at 70 100 bp, was larger and contained two additional segments (Figure 1). The first of these is TnaphA6 (3072 bp), which, in pAb-G7-2, replaces a single copy of ISAba125 in pACICU2 (GenBank accession number CP000865). TnaphA6 is surrounded by a duplication of a 3 bp sequence (AGC) that is present only once in the related cryptic plasmids p2ABTCDC0715 (70 894 bp; GenBank accession number CP002524) and ABKp1 (74 451 bp; GenBank accession number CP001922) that do not include TnaphA6 or ISAba125 at this location. The second segment in pAb-G7-2, p2ABTCDC0715 and ABKp1 that was not found in pACICU2, is 4.7 kb long and lies between two copies of a 423/424 bp segment (the boxes numbered 2 and 3 in Figure 1). The 423/424 bp segment is found only once in pACICU2. A third copy of 229 bp from one end of the 423 bp segment (numbered 1 in Figure 1) is also present in all plasmids in this group.
Figure 1.
Map of pAb-G7-2. The open boxes represent the ISAba125, with internal arrows indicating the orientation of the tnp gene, and the filled boxes represent the repeat units. The broad arrows represent genes; open reading frames of unknown function are not shown. The location of the PCR primers used to map the junctions of TnaphA6 with the backbone and to link across the repeat units are shown within the circle.
All four plasmids carry the same rep gene, which encodes the predicted replication initiation protein, and belong to the Acinetobacter plasmid group recently designated as repAci6.20 In a recent survey performed using PCR to detect several different repA genes, plasmids in this group appeared to be widespread as they were found to be present in 93 of 96 diverse European A. baumannii isolates.21 However, the aminoglycoside resistance profiles of these isolates were not reported and it would be of interest to determine whether the aphA6 gene is present in any members of that collection.
Plasmid pAb-G7-2 is conjugative
The four sequenced repAci6 plasmids also carry genes involved in conjugal transfer in two separate regions. One region contains a cluster of mating pair formation (MPF) or type IV secretion system genes (tra genes in Figure 1) that is of the type recently classified as MPFF22 and the second region contains the genes for mobilization (trwC and trwB in Figure 1). This indicated that they might be conjugative, and pAb-G7-2 was shown to transfer amikacin, kanamycin and neomycin resistance into an aminoglycoside-susceptible, rifampicin-resistant A. baumannii recipient strain, ATCC 17978rifR.
Location of TnaphA6 in D78 and D81
The location of transposons can represent simple, useful markers for specific plasmids. Primers that amplify a region that completely includes the left- or right-hand copies of ISAba125 only when TnaphA6 is in the location found in pAb-G7-2 (primers are listed in Table 1 and locations shown in Figure 1) were used to examine two additional GC1 isolates, D78 and D81, that had been shown previously to carry TnaphA6. The transposon was found to be in exactly the same location in a plasmid carried by these isolates. The 4.7 kb segment flanked by the two copies of the 423 bp sequence was also shown to be present using primers shown in Figure 1. Together, these data suggest that they also carry a plasmid identical or closely related to pAb-G7-2.
Sequence typing
The ST of G7 was determined from the genome sequence and compared with ones determined previously by standard methods. G7 was ST1 (Institut Pasteur MLST scheme), consistent with the assignment to GC1. However, the ST under the Oxford scheme determined using the procedure described in the MLST web site was ST109 (10-12-4-11-4-9-5) compared with ST231 (10-12-4-11-4-98-5) when the genome sequence data were used. This difference was traced to a single base difference in the gpi allele caused by inclusion of the original primer in the region analysed. Differences in this region are revealed in genome sequences, making ST109 and ST231 the same ST.
Conclusions
The AbaR3 resistance island found on the chromosome carries most of the acquired antibiotic resistance genes in G7, including the aacC1 gentamicin resistance gene. The conjugative ability of the amikacin resistance plasmid pAb-G7-2 should allow it to become widespread, and the PCRs developed will allow it to be tracked globally.
Funding
This study was supported by NHMRC Project Grant APP1026189 and Wellcome Trust grant number 098051. M. H. was supported by a University of Sydney Postgraduate Research Award. K. E. H. was supported by an NHMRC PostDoctoral Fellowship (no. 628930).
Transparency declarations
None to declare.
References
- 1.Seward RJ, Lambert T, Towner KJ. Molecular epidemiology of aminoglycoside resistance in Acinetobacter spp. J Med Microbiol. 1998;47:455–62. doi: 10.1099/00222615-47-5-455. [DOI] [PubMed] [Google Scholar]
- 2.Nemec A, Dolzani L, Brisse S, et al. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J Med Microbiol. 2004;53:1233–40. doi: 10.1099/jmm.0.45716-0. [DOI] [PubMed] [Google Scholar]
- 3.Hujer KM, Hujer AM, Hulten EA, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother. 2006;50:4114–23. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han HL, Jang SJ, Park G, et al. Identification of an atypical integron carrying an IS26-disrupted aadA1 gene cassette in Acinetobacter baumannii. Int J Antimicrob Agents. 2008;32:165–9. doi: 10.1016/j.ijantimicag.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Yu YS, Zhou H, Yang Q, et al. Widespread occurrence of aminoglycoside resistance due to ArmA methylase in imipenem-resistant Acinetobacter baumannii isolates in China. J Antimicrob Chemother. 2007;60:454–5. doi: 10.1093/jac/dkm208. [DOI] [PubMed] [Google Scholar]
- 6.Hamidian M, Nigro SJ, Hall RM. Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J Antimicrob Chemother. 2012;67:2833–6. doi: 10.1093/jac/dks318. [DOI] [PubMed] [Google Scholar]
- 7.Nigro SJ, Post V, Hall RM. Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J Antimicrob Chemother. 2011;66:1504–9. doi: 10.1093/jac/dkr163. [DOI] [PubMed] [Google Scholar]
- 8.Post V, White PA, Hall RM. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;16:1162–70. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 9.Nigro SJ, Farrugia DN, Paulsen IT, et al. A novel family of genomic resistance islands, AbGRI2, contributing to aminoglycoside resistance in Acinetobacter baumannii isolates belonging to global clone 2. J Antimicrob Chemother. 2013;68:554–7. doi: 10.1093/jac/dks459. [DOI] [PubMed] [Google Scholar]
- 10.Lambert T, Gerbaud G, Courvalin P. Transferable amikacin resistance in Acinetobacter spp. due to a new type of 3′-aminoglycoside phosphotransferase. Antimicrob Agents Chemother. 1988;32:15–9. doi: 10.1128/aac.32.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin P, Jullien E, Courvalin P. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol Microbiol. 1988;2:615–25. doi: 10.1111/j.1365-2958.1988.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 12.Lambert T, Gerbaud G, Bouvet P, et al. Dissemination of amikacin resistance gene aphA6 in Acinetobacter spp. Antimicrob Agents Chemother. 1990;34:1244–8. doi: 10.1128/aac.34.6.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamidian M, Hall RM. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother. 2011;66:2484–91. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 14.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inouye M, Conway TC, Zobel J, et al. Short read sequence typing (SRST): multi-locus sequence types from short reads. BMC Genomics. 2012;13:338. doi: 10.1186/1471-2164-13-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nigro SJ, Hall RM. GIsul2, a genomic island carrying the sul2 sulphonamide resistance gene and the small mobile element CR2 found in the Enterobacter cloacae subspecies cloacae type strain ATCC 13047 from 1890, Shigella flexneri ATCC 700930 from 1954 and Acinetobacter baumannii ATCC 17978 from 1951. J Antimicrob Chemother. 2011;66:2175–6. doi: 10.1093/jac/dkr230. [DOI] [PubMed] [Google Scholar]
- 17.Adams MD, Goglin K, Molyneaux N, et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol. 2008;190:8053–64. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamidian M, Wynn M, Holt KE, et al. Identification of a marker for two lineages within the GC1 clone of Acinetobacter baumannii. J Antimicrob Chemother. 2014;69:557–8. doi: 10.1093/jac/dkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iacono M, Villa L, Fortini D, et al. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother. 2008;52:2616–25. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertini A, Poirel L, Mugnier PD, et al. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54:4168–77. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towner KJ, Evans B, Villa L, et al. Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D β-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55:2154–9. doi: 10.1128/AAC.01661-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smillie C, Garcillán-Barcia MP, Francia MV, et al. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–52. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]