Abstract
Pneumococcal surface adhesin A (PsaA) is a multifunctional lipoprotein known to bind nasopharyngeal epithelial cells, and is significantly involved in bacterial adherence and virulence. Identification of PsaA peptides that optimally bind human leucocyte antigen (HLA) and elicit a potent immune response would be of great importance to vaccine development. However, this is hindered by the multitude of HLA polymorphisms in humans. To identify the conserved immunodominant epitopes, we used an experimental dataset of 28 PsaA synthetic peptides and in silico methods to predict specific peptide-binding to HLA and murine MHC class II molecules. We also characterized spleen and cervical lymph node (CLN)-derived T helper (Th) lymphocyte cytokine responses to these peptides after Streptococcus pneumoniae strain EF3030 challenge in mice. Individual, yet overlapping, peptides 15 amino acids in length revealed residues of PsaA that consistently caused the highest interferon-γ, interleukin-2 (IL-2), IL-5 and IL-17 responses and proliferation as well as moderate IL-10 and IL-4 responses by ex vivo re-stimulated splenic and CLN CD4+ T cells isolated from S. pneumoniae strain EF3030-challenged F1 (B6 × BALB/c) mice. In silico analysis revealed that peptides from PsaA may interact with a broad range of HLA-DP,-DQ and-DR alleles, due in part to regions lacking β-turns and asparagine endopeptidase sites. These data suggest that Th cell peptides (7, 19, 20, 22, 23 and 24) screened for secondary structures and MHC class II peptide-binding affinities can elicit T helper cytokine and proliferative responses to PsaA peptides.
Keywords: MHC, peptide vaccine, Streptococcus pneumoniae, T helper type 1/2 cytokine
Introduction
Streptococcus pneumoniae continues to be a major cause of morbidity and mortality among very young, elderly and immunocompromised individuals worldwide.1 More than 1·2 million people per year, including 0·8 million children <5 years of age, succumb to pneumococcal diseases.2 The rapid emergence of multidrug-resistant strains of S. pneumoniae has limited the effectiveness of antibiotics to treat this preventable disease.3 Hence, pneumococcal vaccines are of utmost importance to provide protection against S. pneumoniae infection. Animal experiments have identified several pneumococcal proteins and polysaccharides as promising vaccine candidates.4 Indeed, the 7-valent conjugate vaccine (PCV7) and 23-valent non-conjugated polysaccharide vaccine (Pneumovax-23) are used worldwide to reduce the pneumococcal burden. However, both of these vaccines are limited to certain pneumococcal strains and do not generate robust anti-polysaccharide responses in infants and the elderly.4
Pneumococcal surface adhesin A (PsaA) is a model vaccine antigen, because of its role in pneumococcal pathogenesis and conservation among virulent strains.4–6 The present study describes the characterization of T helper (Th) cell epitopes of a candidate pneumococcal vaccine antigen, PsaA, which is a cell wall-associated surface protein and plays a major role in pneumococcal virulence by binding human lactoferrin and interferes with complement deposition on the bacterial surface.4 PsaA is a divalent metal-ion-binding lipoprotein component of an ATP-binding cassette transport system that has specificity for manganese.7,8 It plays a vital role in bacterial adherence and nasopharyngeal colonization, which further progresses to invasive disease, by crossing natural physical and immunological barriers. PsaA is an immunogenic protein that activates both humoral and cellular branches of the immune system. Murine studies showed that anti-PsaA antibodies confer protection against nasopharyngeal carriage and systemic infection.9 Moreover, this protein was found to be highly conserved in > 90 strains of S. pneumoniae so far reported, including all clinically relevant strains. On these grounds, PsaA has progressed as a promising target for pneumococcal vaccine development. However, vaccine development efforts have been hindered by the limited characterization of immunogenic S. pneumoniae epitope(s) that can bind to multiple HLA alleles. Importantly, CD4+ T cells play a significant role in the clearance of pneumococcal colonization and are required for optimal protective antibody responses to pneumococcal protein PsaA.10 Identification of a suitable CD4+ T-cell epitope(s) will be critical to develop an effective pneumococcal vaccine.
A synthetic peptide designed to induce protective immunity must: (i) show homology to the peptides naturally presented to antigen-presenting cells during infection, (ii) induce an appropriate effector immune response to the pathogen and (iii) be recognized by the majority of the diverse human population.
Indeed, wide-ranging HLA haplotypes create diversity in epitope specificity and T-cell repertoire but this can make selection of vaccine peptides difficult.11,12 Methods to identify peptide(s), containing ‘promiscuous’ or ‘universal’ epitope(s) that cover diverse HLA haplotypes are greatly needed.
Recent advances in in silico prediction tools have eased the process of immunodominant epitope identification. To identify the immunodominant epitopes of PsaA, we used in silico MHC affinity measurement methods that use both affinity data from the Immune Epitope Database and Analysis Resource (IEDB),13 peptide data from the SYFPEITHI14 database as well as RANKPEP,15 SVMHC16 and MHCPred tools17 to predict PsaA peptides that bind HLA-DP,-DQ and-DR alleles. Further, to correlate the predicted peptide–MHC binding affinities with in vivo immunogenicity, PsaA peptide-specific Th cell proliferation and cytokine responses were studied in mice that had been nasally challenged with S. pneumoniae strain EF3030 (capsular type 19F). Together, these in silico and in vivo studies revealed the immunodominant PsaA Th cell epitopes that might serve as potential vaccine candidates.
Materials and methods
Animals
Eight-to twelve-week-old female F1 (B6 × BALB/c) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and used in this study. Experimental mice contain MHC class II haplotype and corresponding T-cell receptor diversity that approaches those seen in man.18,19 All mice were housed in the Specific Pathogen-free Facility (SPF) of the Center for Laboratory Animal Resources (CLAR) at Morehouse School of Medicine (Atlanta, GA). Routine antibody screening for animal pathogens and routine histological analysis of organs and tissues were performed to insure that mice were pathogen free. All procedures involving mice were approved by the Institutional Animal Care and Use Committees. Moreover, the guidelines proposed by the committee for the Care of Laboratory Animal Resources Commission of Life Sciences – National Research Council were followed to minimize animal pain and distress.
S. pneumoniae strain EF3030 growth and challenge
Streptococcus pneumoniae capsular strain EF3030 used in this study is among the human isolates of capsular group 19 that were found to be relatively non-invasive in mice.20 Pneumococci were grown in Todd–Hewitt broth (Sigma-Aldrich, St Louis, MO) and stored frozen in aliquots at − 80° in 20% glycerol in sterile lactated Ringer's solution (Abbott Labs, North Chicago, IL). The EF3030 strain expresses a polysaccharide capsule when grown in Todd–Hewitt broth and has been used in previous studies.9,21 To establish nasal carriage, pneumococci were introduced into groups of mice by nasal administration. The animals were anaesthetized with ketamine (100 mg/ml) and xylazine (20 mg/ml), mixed at a 4 : 1 (volume/volume) ratio, respectively. The anaesthesia mixture was injected intramuscularly into the right hamstring muscle at a dose of 100 mg of ketamine per kg of body weight. After anaesthesia was established, the mice were inoculated with approximately 107 colony-forming units of S. pneumoniae strain EF3030 in 15 μl lactated Ringer's solution. Both experimental and control groups contained 10 mice each.
Pneumococcal antigens
Twenty-eight pneumococcal overlapping synthetic peptides, spanning the entire length of S. pneumoniae strain D39/R6 PsaA protein sequence (NCBI Accession #P0A4G3), starting with the first 15 residues at the N-terminus, was synthesized by the multipin synthesis method by Chiron Mimotopes Peptide Systems, San Diego, CA. Synthesized peptides overlapped by four amino acids (Table 1) and were acetylated at the N-terminus and ended with a C-terminal. Purity of these peptides was approximately 95%. The peptides were dissolved in a mixture (volume/volume) of 75% DMSO (Sigma-Aldrich) and 25% water, to a concentration of 70 mm, divided into small aliquots and stored frozen at −80°.
Table 1.
Overlapping pneumococcal surface adhesin A (PsaA) peptides
 |
Tissue collection and cell isolation
Mice were euthanized by CO2 inhalation for collection of spleen and cervical lymph nodes (CLNs) for single cell isolation of lymphocytes 28 days after S. pneumoniae strain EF3030 challenge. Spleen and CLNs were collected separately and single cell suspensions were prepared by aseptically removing tissues and passage through a sterile wire screen. Unpooled CD4+ T cells were further separated by autoMACS™ separator (Miltenyi Biotec, Auburn, CA) using negative selection. The remaining lymphocytes including non-CD4+ T cells, after treatment with mitomycin C (Sigma-Aldrich), were used as accessory feeder cells for PsaA peptide-specific stimulation assays.
Cytokine quantitation by Luminex™ analysis
Purified CD4+ T cells were cultured with mitomycin C-treated feeder cells at a ratio of 5 : 1. Feeder cells were treated with mitomycin C at a final concentration of 25 μg/ml for 30 min. Following treatment, the cells were washed three times with complete RPMI-1640 medium to remove traces of mitomycin C. CD4+ T cells, at a density of 5 × 106 cells/ml, were incubated with 106 mitomycin C-treated feeder cells/ml in complete medium containing 1 μm PsaA peptide at 37° with 5% CO2. For the assessment of cytokine production, 100 μl of culture supernatants from 96-well flat bottom plates (Corning, Tewsbury, MA) were harvested 3 days after ex vivo PsaA peptide stimulation to determine the levels of interleukin-2 (IL-2), IL-4, IL-5, IL-10, IL-17 and interferon-γ (IFN-γ) secreted by CD4+ T cells. Phorbol-12-myristate-13-acetate (1 μg/ml) was used as a positive control, whereas ovalbumin (1 μg/ml) and medium only were used as negative control to reduce the background reading. Cytokine levels in culture supernatants were determined by the Beadlyte™ mouse multi-cytokine detection (Bio-Rad, Hercules, CA). Briefly, the filter-bottom ELISA plates were rinsed with 100 μl Bio-plex assay buffer and liquid was removed using a Millipore™ Multiscreen Separation Vacuum Manifold System (Millipore, Billerica, MA) set at 5 mm Hg. Analyte beads in assay buffer were added to the wells followed by 50 μl of serum or standard solution. The plates were incubated for 30 min at room temperature with continuous shaking (at setting #3) using a Lab-Line™ Instrument Titer Plate Shaker (Melrose Park, IL). The filter bottom plates were washed, as before, and centrifuged at 300 g for 30 seconds. Subsequently, 50 μl of anti-mouse IL-2, IL-4, IL-5, IL-10, IL-17 and IFN-γ antibody-biotin reporter solution was added to each well, after which the plates were incubated with continuous shaking for 30 min followed by centrifugation and washing. Next, 50 μl streptavidin-phycoerythrin solution was added and plates were incubated with continuous shaking for 10 min at room temperature. Then, 125 μl Bio-plex assay buffer was added, and Beadlyte™ readings were measured using a Luminex™ System and calculated using Bio-plex™ software (Bio-Rad). The cytokine Beadlyte™ assays were capable of detecting > 5 pg/ml for each analyte. All experiments were performed in triplicate.
Cell proliferation
Lymphocyte proliferation was measured using 5-Bromo-2′-deoxy uridine (BrdU) absorption and detection (Roche Diagnostics, Indianapolis, IN). In brief, purified CD4+ T cells and mitomycin C-treated feeder cells were cultured at a density of 5 × 106 and 106 cells/ml, respectively, in complete medium containing 1 μm of each PsaA peptide at 37° in 5% CO2. After 2 days of ex vivo antigen stimulation, cells were transferred to polystyrene 96-well plates (Corning). BrdU labelling solution (10 μm final concentration) was added to each well and plates were incubated for 18 hr at 37° with 5% CO2. The cells were then fixed and incubated with 100 μl nuclease in each well for 30 min at 37° with 5% CO2. Following nuclease treatment, cells were washed with complete RPMI-1640 mediuum and incubated with anti-BrdU antibody–peroxidase conjugate solution for 30 min at 37° with 5% CO2. BrdU incorporation was developed with 2,2′-azino-bis 3-ethylbenzthiazoline-6-sulphonic acid (ABTS) solution and optical density (OD) was read at 450 nm. Antigen-specific CD4+ T-cell proliferation was obtained by measuring BrdU incorporation, according to the manufacturer's instructions (Roche Diagnostics). BrdU absorption or optical density at 450 nm (OD450) was detected using a scanning multi-well SpectraMax 250 spectrophotometer (Molecular Devices, Sunnyvale, CA). The proliferation index (PI) was calculated as follows: PI = OD450 in peptide-stimulated wells/OD450 in unstimulated wells × 100. The results were expressed as mean ± standard error of mean (SEM) of the response of three replicates. Statistical significance was assessed by Student's t-test.
To quantify non-specific cytokine and proliferation responses of PsaA peptides we calculated the cut-off value (dash lines) using mean value ± SEM of control responses (naive cells from control group mice).
MHC class II epitope prediction using external tools
IEDB (http://www.immuneepitope.org/), SYFPEITHI (http://www.syfpeithi.de/), SVMHC (http://www.bs.informatik.unituebingen.de/SVMHC/), RANKPEP (http://bio.dfci.harvard.edu/RANKPEP/), and MHCPred (http://www.jenner.ac.uk/MHCPred) external software(s) were used to predict peptide binding affinities to mouse I-A and I-E as well as to HLA-DR,-DP and-DQ. In brief, for average relative binding evaluation, 10-fold cross validation IEDB results were used to estimate overall performance. Because the binding of peptides to MHC class II molecules is not dependent on exact size, the derivation of MHC class II average relative binding matrices followed an iterative procedure. For the first iterative step, a matrix was generated from a set of nine-residue core sequences randomly obtained from each peptide sequence in the training set. For subsequent cycles, nine-residue core sequences were used to generate a matrix. The overall binding affinity of a peptide was predicted using the highest scoring nine-residue core sequence. For SYFPEITHI prediction, we matched each testing peptide with three glycine residues at both ends before evaluation. This was recommended by the creators of SYFPEITHI to ensure that all potential binders were correctly presented to the peptide-binding prediction algorithm. For all other methods, the original tested peptides were submitted directly for prediction. To assign a single MHC binding affinity prediction for peptides longer than nine amino acids in the context of predictive tools, we took the highest affinity prediction of all possible 9-mers within the longer peptide as the initial result. For each MHC class II molecule whose binding can be predicted by three or more algorithms, the top three methods were selected that gave the best performance. For each method, peptides were tested and ranked by their scores with higher ranks for better binders. For each tested peptide, three rankings from different methods were taken and the median rank was taken as the consensus score. Peptides were classified into binders (IC50 < 500 nm) and non-binders (IC50 ≥ 500 nm), as practical cut-offs. Three-dimensional structure analysis was performed using the YASARA program.
Statistics
Data are expressed as the mean ± SEM and compared using a two-tailed Student's t-test or an unpaired Mann–Whitney U-test. The results were analysed using Microsoft Excel and were considered statistically significant if P values were < 0·01. When cytokine levels were below the detection limit, they were recorded as one-half of the lower detection limit for statistical analysis.
Results
Peptide selection, binding analysis and overview of PsaA predicted secondary structure
Individual but overlapping synthetic PsaA peptides, 15 amino acids in length, were used in this study (Table 1). The complete amino acid sequence of full-length PsaA was used to predict the protein structure as well as β-turn (t) using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/)22 and COUDES (http://bioserv.rpbs.jussieu.fr/Coudes/index.html)23 methods. Coiled–coiled (C) as well as helical (H) structures were noted throughout the PsaA protein (Fig. 1). Small coiled-helix (PsaA68–81) and (PsaA231–268) domains showed no β-turns or potential asparagine (N) endopeptidase sites. An array of complex secondary structures and numerous potential N endopeptidase sites were also observed in PsaA. The latter sites are typically found in bacterial cell wall-associated domains and are known to effect antigen processing and to decrease MHC presentation.24
Figure 1.
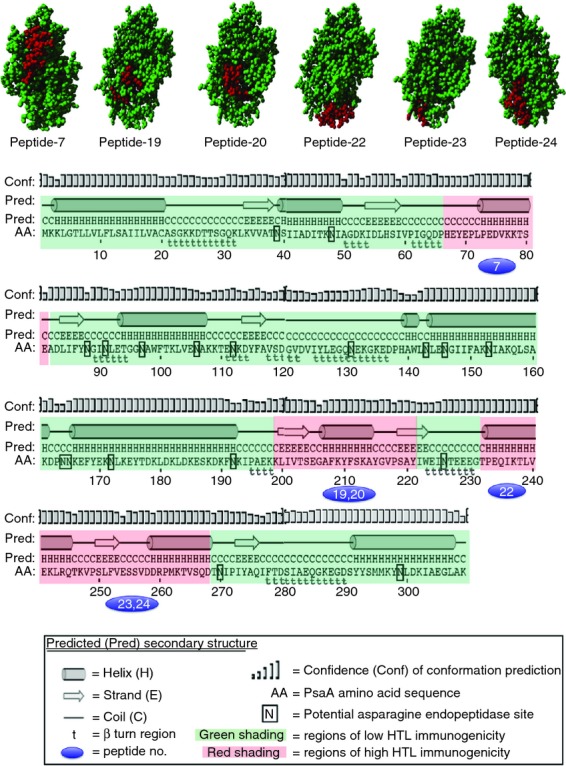
Modular pneumococcal surface adhesin A (PsaA) amino acid sequence showing regions of predicted immunogenicity and secondary structure. The three-dimensional structure shows the immunogenic peptide (red colour) regions in PsaA. The PsaA amino acid (AA) sequence was used to predict helical (H), coiled (C), strand (E), turns (t), and asparagine endopeptidase sites (N). Regions shaded with green show regions of low immunogenicity.
Overlapping PsaA peptides were used to determine MHC II binding affinities in silico. These data span a total of 16 human and four mouse MHC class II types. Each of these PsaA peptides was scanned using IEDB, MHCPred, RANKPEP, SVMHC and SYFPEITHI MHC class II epitope databases. Five different databases were used to predict the binding affinity of PsaA peptides to cover the maximum human HLA alleles. The hypothetical binding affinities of PsaA peptide with MHC alleles from different databases were similar. The selected PsaA peptides were compared with archived peptide datasets previously used to measure peptide–MHC class II affinities. Peptides were classified into binders (IC50 < 500 nm) and non-binders (IC50 ≥ 500 nm) based on in silico-derived binding affinities. This analysis revealed that nearly all PsaA peptides could potentially bind a variety of mouse and human MHC class II molecules (Table 2).
Table 2.
Overview of pneumococcal surface adhesin A (PsaA) peptide predicted binding affinities to MHC class II alleles
 |
Proliferative responses of S. pneumoniae-specific systemic and mucosal CD4+ T cells following PsaA peptide stimulation
To better determine whether the predicted PsaA peptide-MHC class II binding affinities corresponded with Th cell proliferation, PsaA peptide-specific CD4+ T-cell responses were characterized 28 days after S. pneumoniae strain EF3030 or mock challenge. PsaA peptide-specific proliferative responses by spleen-or CLN-derived CD4+ T cells from S. pneumoniae strain EF3030-challenged mice were found to be significantly higher than those by naive CD4+ T cells from mock challenged mice (Fig. 2). CLN-and spleen-derived Th cells from S. pneumoniae strain EF3030-challenged mice significantly proliferated in response to PsaA peptides 7, 19, 20, 22, 23 and 24 compared with naive controls. CLN CD4+ T cells PsaA peptide-specific proliferation responses were moderately higher than similar cells isolated from the spleen of S. pneumoniae strain EF3030-challenged mice.
Figure 2.
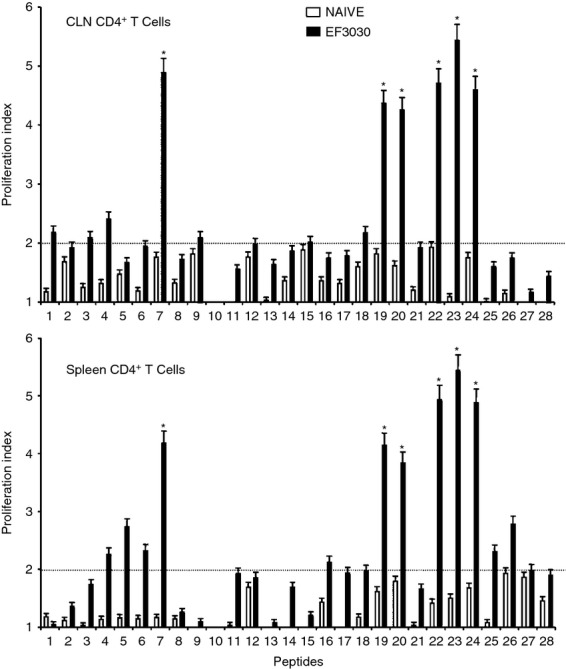
Proliferation responses of pneumococcal surface adhesin A (PsaA) peptide-specific systemic and mucosal CD4+ T cells during pneumococcal carriage. Cervical lymph node (CLN) and splenic lymphocytes were isolated, 28 days after intranasal challenge, from both Streptococcus pneumoniae strain EF3030 challenged (▮), and naive (□) F1 (B6 × BALB/c) group mice. CD4+ T cells were incubated with 1 μm of PsaA peptide (15-amino acid peptides that overlapped every 11 residues) plus mitomycin C-treated naive syngeneic feeder cells, for 3 days, at a ratio of 5 : 1 × 106 cells. Proliferation was measured by 5-bromo-2′-deoxy uridine incorporation, which was measured by ELISA. The data presented are the mean optical density at 450 nm (OD450). Experimental groups consisted of 10 mice. The results were expressed as mean ± standard error of mean (SEM) of the response from three replicates. (*) represents the peptides showing significantly higher proliferation response. Dotted line (–) represents the cut-off value above which the values were considered significant.
Cytokine production by S. pneumoniae-specific helper T cells following PsaA peptide stimulation
In general, pneumococcal infection resulted in significantly higher Th cell cytokine secretion by ex vivo PsaA peptide-stimulated CD4+ T cells from the spleen as well as from CLNs of S. pneumoniae strain EF3030-challenged mice, compared with mock-challenged mice. In contrast to proliferation responses, spleen-derived CD4+ T cells from S. pneumoniae strain EF3030-challenged mice showed higher levels of IL-2 secretion following PsaA peptide ex vivo stimulation than CLNs derived from CD4+ T cells. Interferon-γ and IL-2 secreting Th cells from CLNs and spleen of pneumococcus-challenged mice significantly responded to PsaA peptides 7,19, 20, 22, 23 and 24 (Figs 3 and 4).
Figure 3.
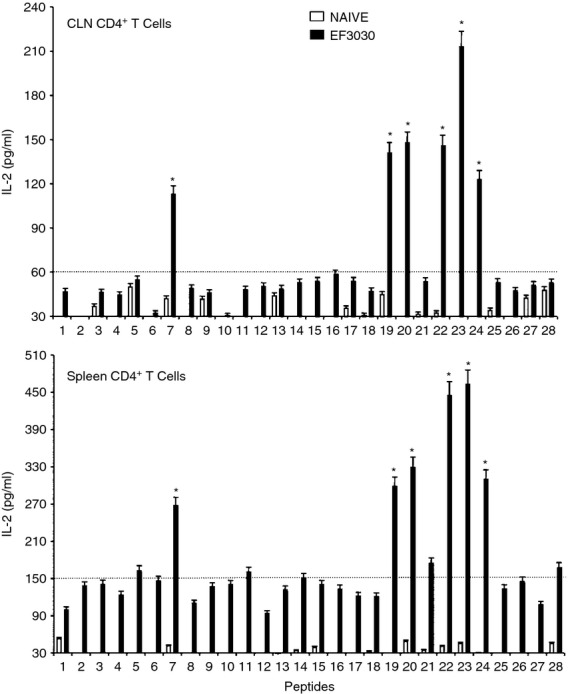
Pneumococcal surface adhesin A (PsaA) peptide-specific interleukin-2 (IL-2) secretion by CD4+ T cells following pneumococcal challenge. Groups of 10 F1 (B6 × BALB/c) mice were intranasally challenged with 107 colony-fomring units of Streptococcus pneumoniae strain EF3030 in a 15 μl volume of Ringer's solution. Cervical lymph node (CLN) and splenic lymphocytes were isolated, 28 days after intranasal challenge, from both S. pneumoniae strain EF3030 challenged (▮), and naive (□) group mice. CD4+ T cells were incubated with 1 μm of PsaA peptide (15 amino acid peptides that overlapped every 11 residues) plus mitomycin C-treated naive syngeneic feeder cells, for 3 days, at a ratio of 5 : 1 × 106 cells. The results were expressed as mean ± standard error of mean (SEM) of the response from three replicates. The IL-2 production of cultured supernatants was determined by Luminex capable of detecting >2 pg/ml of IL-2. (*) represents the peptides showing significantly higher cytokine response. Dotted line represents the cut-off value above which the values were considered significant.
Figure 4.
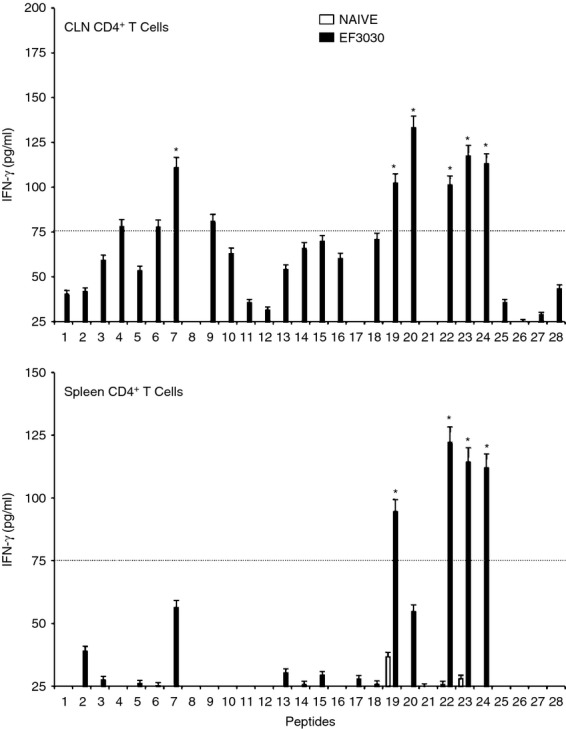
Pneumococcal surface adhesin A (PsaA) peptide-specific interferon-γ (IFN-γ) secretion by CD4+ T cells following pneumococcal challenge. Groups of 10 F1 (B6 × BALB/c) mice were intranasally challenged with 107 colony-forming units of Streptococcus pneumoniae strain EF3030 in a 15 μl volume of Ringer's solution. Cervical lymph node (CLN) and splenic lymphocytes were isolated, 28 days after intranasal challenge, from both S. pneumoniae strain EF3030 challenged (▮), and naive (□) group mice. CD4+ T cells were incubated with 1 μm of PsaA peptide (15 amino acid peptides that overlapped every 11 residues) plus mitomycin C-treated naive syngeneic feeder cells, for 3 days, at a ratio of 5 : 1 × 106 cells. The results were expressed as mean ± standard error of mean (SEM) of the response from three replicates. IFN-γ production of cultured supernatants was determined by Luminex capable of detecting > 2 pg/ml of IFN-γ. (*) represents the peptides showing significantly higher cytokine response. Dotted line represents the cut-off value above which the values were considered significant.
CD4+ T cells isolated from the spleen and CLNs of challenged mice also secreted Th2 cytokines after ex vivo stimulation of PsaA peptides. While the cells isolated from mock-challenged mice did not significantly respond to PsaA peptides, CD4+ T cells from S. pneumoniae strain EF3030-challenged mice showed significant IL-4 and IL-5 secretion in response to PsaA peptide ex vivo stimulation (Figs 5 and 6). In particular, there were comparatively higher responses to PsaA peptides 7, 19, 20, 22, 23 and 24 by both splenic and CLN Th cells. CLN and splenic CD4+ T cells from S. pneumoniae strain EF3030-challenged mice also significantly secreted IL-10 after PsaA peptide re-stimulation, with comparatively higher responses to PsaA peptides 7, 19, 20, 22, 23 and 24 (Fig. 7).
Figure 5.
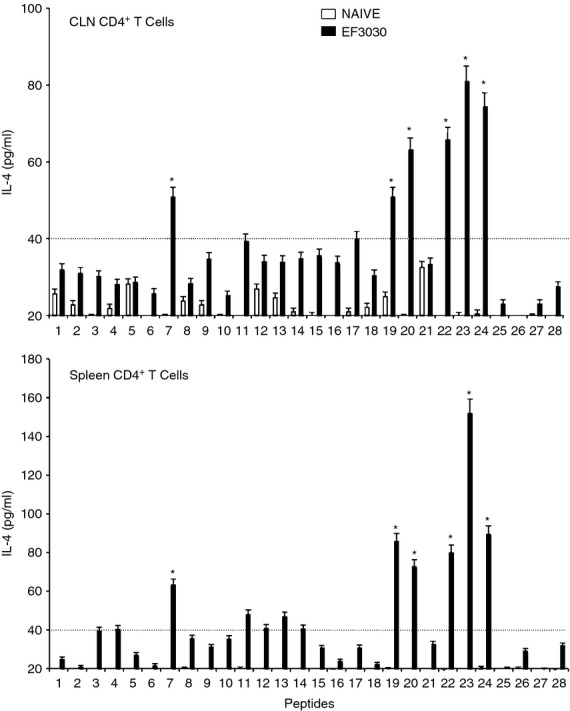
Pneumococcal surface adhesin A (PsaA) peptide-specific interleukin-4 (IL-4) secretion by CD4+ T cells following pneumococcal challenge. Groups of 10 F1 (B6 × BALB/c) mice were intranasally challenged with 107 colony-forming units of Streptococcus pneumoniae strain EF3030 in a 15 μl volume of Ringer's solution. Cervical lymph node (CLN) and splenic lymphocytes were isolated, 28 days after intranasal challenge, from both S. pneumoniae strain EF3030 challenged (▮), and naive (□) group mice. CD4+ T cells were incubated with 1 μm of PsaA peptide (15 amino acid peptides that overlapped every 11 residues) plus mitomycin C-treated naive syngeneic feeder cells, for 3 days, at a ratio of 5 : 1 × 106 cells. The results were expressed as mean ± standard error of mean (SEM) of the response from three replicates. IL-4 production of cultured supernatants was determined by Luminex capable of detecting > 2 pg/ml of IL-4. (*) represents the peptides showing significantly higher cytokine response. Dotted line represents the cut-off value above which the values were considered significant.
Figure 6.
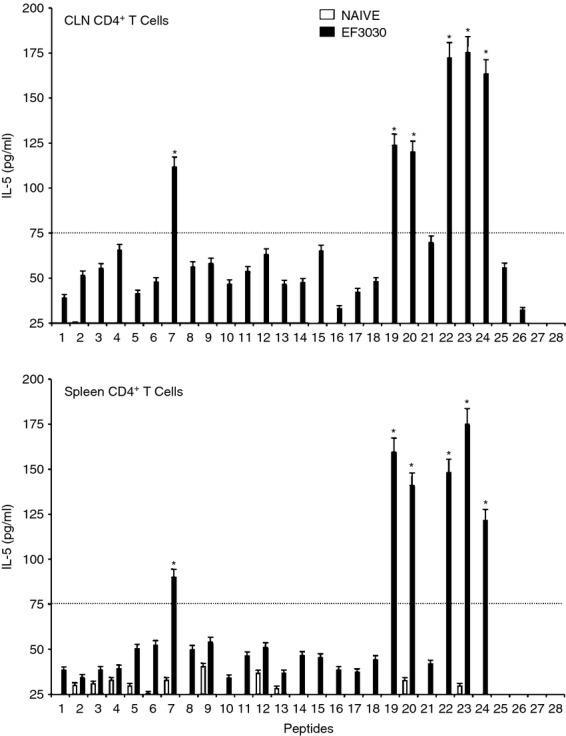
Pneumococcal surface adhesin A (PsaA) peptide-specific interleukin-5 (IL-5) secretion by CD4+ T cells following pneumococcal challenge. Groups of 10 F1 (B6 × BALB/c) mice were intranasally challenged with 107 colony-forming units of Streptococcus pneumoniae strain EF3030 in a 15 μl volume of Ringer's solution. Cervical lymph node (CLN) and splenic lymphocytes were isolated, 28 days after intranasal challenge, from both S. pneumoniae strain EF3030 challenged (▮), and naive (□) group mice. CD4+ T cells were incubated with 1 μm of PsaA peptide (15 amino acid peptides that overlapped every 11 residues) plus mitomycin C-treated naive syngeneic feeder cells, for 3 days, at a ratio of 5 : 1 × 106 cells. The results were expressed as mean ± standard error of mean (SEM) of the response from three replicates. IL-5 production of cultured supernatants was determined by Luminex capable of detecting > 2 pg/ml of IL-5. (*) represents the peptides showing significantly higher cytokine response. Dotted line represents the cut-off value above which the values were considered significant.
Figure 7.
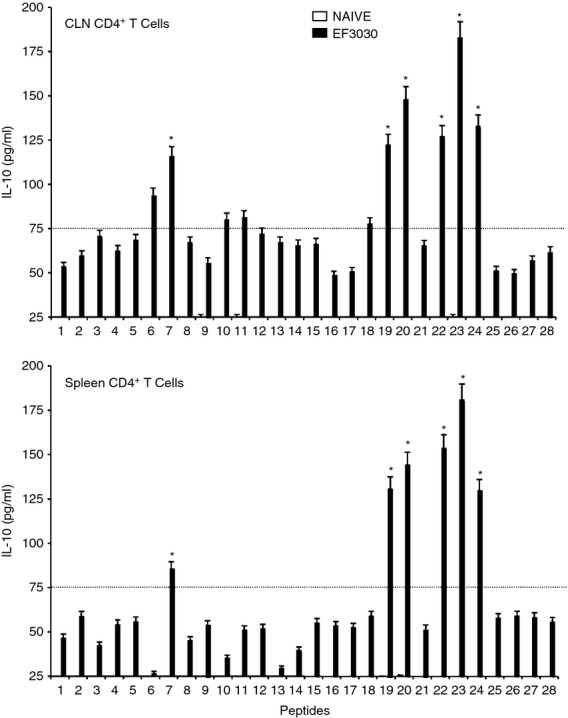
Pneumococcal surface adhesin A (PsaA) peptide-specific interleukin-10 (IL-10) secretion by CD4+ T cells following pneumococcal challenge. Groups of 10 F1 (B6 × BALB/c) mice were intranasally challenged with 107 colony-forming units of Streptococcus pneumoniae strain EF3030 in a 15 μl volume of Ringer's solution. Cervical lymph node (CLN) and splenic lymphocytes were isolated, 28 days after intranasal challenge, from both S. pneumoniae strain EF3030 challenged (▮), and naive (□) group mice. CD4+ T cells were incubated with 1 μm of PsaA peptide (15 amino acid peptides that overlapped every 11 residues) plus mitomycin C-treated naive syngeneic feeder cells, for 3 days, at a ratio of 5 : 1 × 106 cells. The results were expressed as mean ± standard error of mean (SEM) of the response from three replicates. IL-10 production of cultured supernatants was determined by Luminex capable of detecting > 2 pg/ml of IL-10. (*) represents the peptides showing significantly higher cytokine response. Dotted line represents the cut-off value above which the values were considered significant.
Th cells from S. pneumoniae strain EF3030-and mock-challenged mice were also evaluated for their ability to secrete IL-17 in response to PsaA peptides. Both CLN and splenic CD4+ T cells from S. pneumoniae strain EF3030-challenged mice significantly secreted IL-17 following PsaA peptide re-stimulation, compared with mock-challenged mice (Fig. 8). Compared with the other cytokine secretion profiles studied, IL-17 secreted by pneumococcus-challenged mice was most indicative of antigen specific Th cell responsiveness. As seen with Th cell cytokine and proliferative responses, there was comparatively more IL-17 secretion by bacteria-challenged Th cells re-stimulated ex vivo with peptides 7, 19, 20, 22, 23 and 24.
Figure 8.
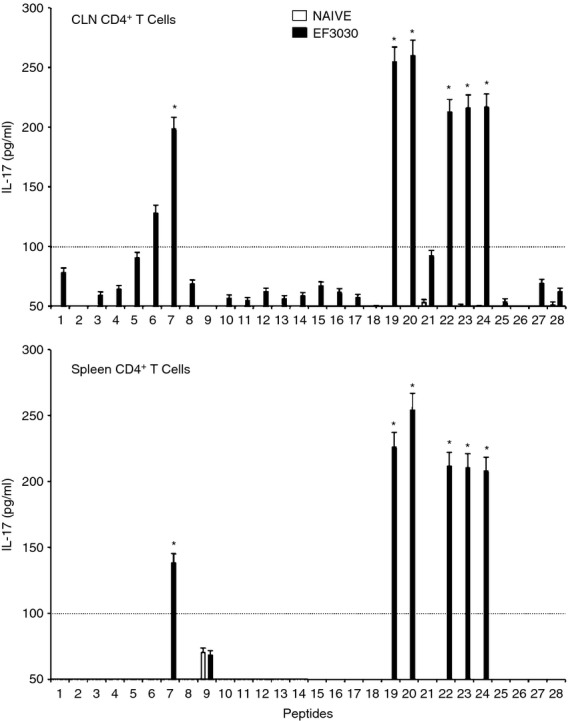
Pneumococcal surface adhesin A (PsaA) peptide-specific interleukin-17 (IL-17) secretion by CD4+ T cells following pneumococcal challenge. Groups of 10 F1 (B6 × BALB/c) mice were intranasally challenged with 107 colony-forming units of Streptococcus pneumoniae strain EF3030 in a 15 μl volume of Ringer's solution. Cervical lymph node (CLN) and splenic lymphocytes were isolated, 28 days after intranasal challenge, from both S. pneumoniae strain EF3030 challenged (▮), and naive (□) group mice. CD4+ T cells were incubated with 1 μm of PsaA peptide (15 amino acid peptides that overlapped every 11 residues) plus mitomycin C-treated naive syngeneic feeder cells, for 3 days, at a ratio of 5 : 1 × 106 cells. The results were expressed as mean ± standard error of mean (SEM) of the response from three replicates. IL-17 production of cultured supernatants was determined by Luminex capable of detecting > 2 pg/ml of IL-17. (*) represents the peptides showing significantly higher cytokine response. Dotted line represents the cut-off value above which the values were considered significant.
In summary, CD4+ T cells from S. pneumoniae strain EF3030-challenged mice consistently mounted significant yet selective proliferation, and IFN-γ, IL-2, IL-4, IL-5 and IL-10 secretion, largely in response to PsaA peptides 7, 19, 20, 22, 23 and 24. These elevated cytokine secretion patterns by ex vivo re-stimulated Th cells did not directly correlate with predictive MHC II binding affinities, but were associated with the absence of asparagine endopeptidase sites. The peptide-binding affinity analysis by in silico methods suggested that peptides 1, 2, 7, 14, 15, 18, 19, 20, 22, 23 and 24 would be among the strongest MHC class II binders and presumably the best inducers of Th cell cytokine and proliferation responses. However, peptides 7, 19, 20, 22, 23 and 24 were shown via ex vivo analysis to be the greatest stimulators of Th cell responses. This could be best explained by the absence of endopeptidase sites in peptides 7, 19, 20, 22, 23 and 24, which prevents them from being degraded, resulting in more efficient presentation by MHC class II molecules.
Predicted PsaA peptide–MHC class II allele binding affinities and correlation with proliferation and cytokine responses
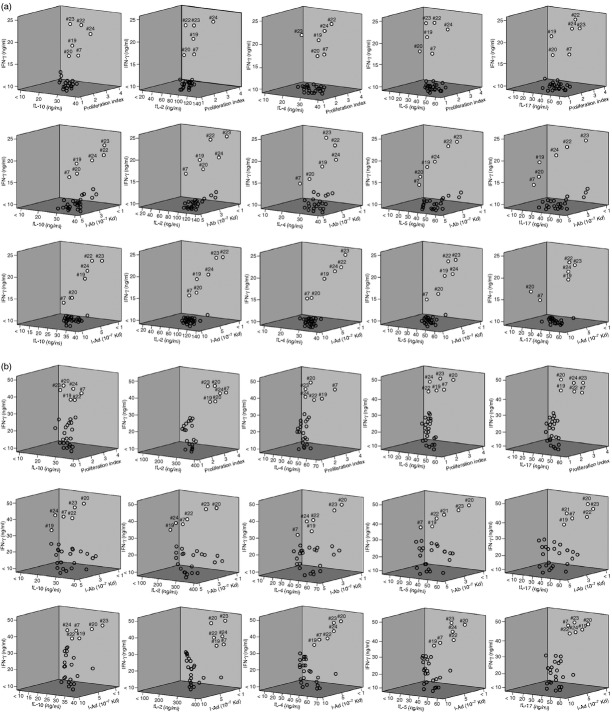
The PsaA peptides 7, 19, 20, 22, 23 and 24 mounted significant Th cell responses and displayed strong predictive binding affinities to several HLA-DR,-DQ and-DP as well as I-Ab and I-Ed haplotypes. This is best illustrated by viewing a three-dimensional plot of the proliferation index as well as IL-2, IFN-γ, IL-4, IL-5, IL-10 and IL-17 responses compared with MHC allele-binding affinities. Three-dimensional plots of IFN-γ and IL-10 secretion relative to proliferation of I-A/I-E to predict peptide-binding by CD4+ T cells, explain IFN-γ, IL-10 and proliferation responses of PsaA peptide-specific CD4+ T cells isolated from F1 (B6 × BALB/c) mice, 28 days after S. pneumoniae challenge and predicted I-A or I-E binding affinities. The y-axis and x-axis indicate the concentrations (ng/ml) of IFN-γ and IL-10, respectively, secreted by PsaA peptide-stimulated CD4+ T cells. The z-axis represents the predicted I-A or I-E binding affinities (Kd). PsaA peptides 7, 19, 20, 22, 23 and 24 appear as white circles, which have higher binding affinity of responses while remaining peptides are open circles (Fig. 9a and b).
Figure 9.
(a) Three-dimensional plots of T helper type 1 (Th1)/Th2 cytokine secretion relative to proliferation or I-A and I-E predicted peptide-binding by cervical lymph node (CLN)-derived CD4+ T cells. The panels summarize interleukin-2 (IL-2), interferon-γ (IFN-γ), IL-4, IL-5, IL-10 and IL-17, proliferation responses, and predicted I-A binding affinities of pneumococcal surface adhesin A (PsaA) peptide-specific CD4+ T cells isolated from cervical lymph nodes of F1 (B6 × BALB/c) mice, 28 days after Streptococcus pneumoniae strain EF3030 challenge. The y-axis and x-axis indicate the concentration (ng/ml) of cytokine, secreted by PsaA peptide-stimulated CD4+ T cells. The z-axis represents the predicted I-A or I-E binding affinities (Kd). PsaA peptides 7, 19, 20, 22, 23 and 24 appear as white circles, while remaining peptides are open circles. (b) Three-dimensional plots of Th1/Th2 cytokine secretion relative to proliferation or I-A and I-E predicted peptide-binding by spleen-derived CD4+ T cells. The panels summarize IFN-γ, IL-10, IL-2, IL-4, IL-5, IL-17, proliferation responses, and predicted I-A-binding affinities of PsaA peptide-specific CD4+ T cells isolated from spleen of F1 (B6 × BALB/c) mice, 28 days after S. pneumoniae strain EF3030-challenge. The y-axis and x-axis indicate the concentration (ng/ml) of IFN-γ and IL-10, IL-2, IL-4, IL-5 and IL-17, respectively, secreted by PsaA peptide-stimulated CD4+ T cells. The z-axis represents the predicted I-A binding affinities (Kd). PsaA peptides 7, 19, 20, 22, 23 and 24 appear as white circles, while remaining peptides are open circles.
The Th cells that recognized and responded to these peptides expressed significantly more T helper cytokines and had greater proliferative responses than those able to respond to other PsaA peptides. These peptides were also predicted to bind with high affinity to both mouse and human MHC II. PsaA peptides 19, 20, 21 and 22 were predicted to bind I-Ab/I-Ad, I-Ab/I-Eb, I-Ab and I-Ab/I-Ad, respectively, with IC50 < 500 nm. From these, PsaA peptide 20 was predicted to have marginal binding affinities to I-Ab and I-Eb with IC50 = 485 and 493 nm, respectively. This also corresponded with relatively high IL-10 responsiveness. Splenic CD4+ T cells stimulated with peptide 20 or 23 showed comparatively higher IL-10 secretion. Similar to PsaA peptide 20, peptide 23 was predicted to have moderate I-Ab and I-Eb binding affinity, i.e. IC50 = 452 and 412 nm, respectively. It is noteworthy that several PsaA peptides that were predicted to tightly bind I-A and/or I-E alleles did not always correspond with elevated cytokine secretion (e.g. peptides 1, 2, 4, 14 etc.) and always had asparagine endopeptidase sites.
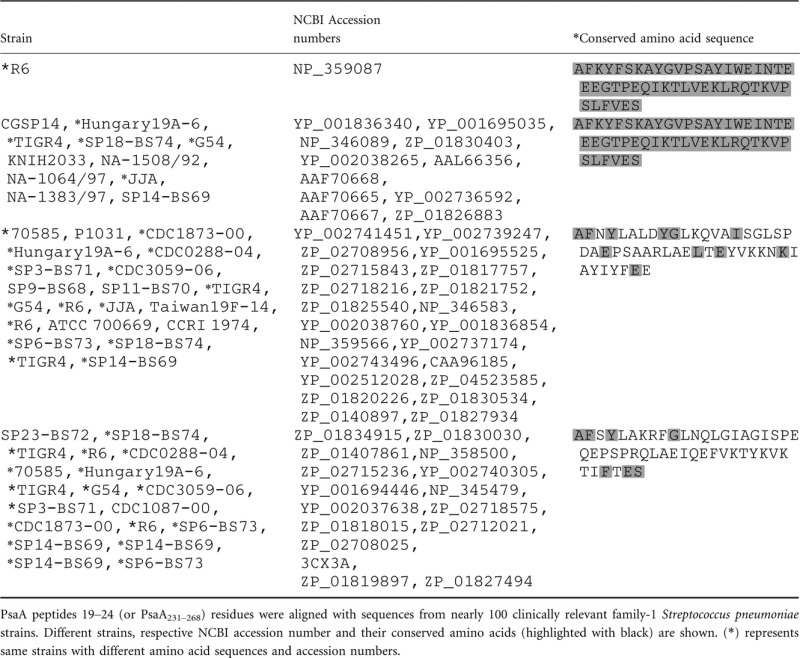
The amino acid sequence comprising PsaA peptides 19–24 (or PsaA231–268) was aligned with sequences from nearly 100 clinically relevant Family 1, S. pneumoniae strains. PsaA231–268 was found to be highly conserved among several S. pneumoniae strains, with 10% of strains having sequence similarity of more than 90% (Table 3).
Table 3.
Alignment of pneumococcal surface adhesin A231–268 (PsaA231–268) amino acid sequences from family 1 pneumococcal strains
 |
Discussion
The immune system is remarkably robust in mounting specific responses to a multitude of foreign antigens and therefore plays a vital role in survival. Th cells are crucial for generating efficient humoral and cellular immune responses following recognition of foreign antigen in the context of MHC class II. Enormous polymorphisms of MHC genes lead to differences in antigen presentation to immune effector cells deployed against pathogens. While peptide vaccines potentially circumvent the problem of using heat-killed or attenuated pathogens, or whole antigen, as vaccines, this approach is impeded by this exhaustive MHC repertoire.25 Hence, the identification of optimal or common Th cell epitope(s) is imperative in mounting a broad protective immune response against different strains of a pathogen. The identification and characterization of ‘promiscuous’ or ‘universal’ peptides that can bind to multiple HLA (β chain) alleles would overcome many of these problems. Recently, Yu et al.26 have used the HLA-transgenic mice to map Th cell epitopes; however, the limited number of HLA transgenic mice does not represent all populations in man. Hence, the present study is the first of many to map clinically relevant Th cell pneumococcal epitopes. We have used in silico methods for predicting class II-restricted peptides and evaluated immunogenicity by ex vivo peptide re-stimulation of experimental pneumococcal challenge.
The precise role of peptide MHC class II interactions that determine protective pneumococcal immunity are not known. A number of studies have shown a cause and effect relationship between human MHC genes and resistance to infection18,27 as well as autoimmune diseases.19 Murine I-A is highly homologous to human HLA-DQ28 and restricts antigen-specific Th cells in mice, whereas I-E is homologous to HLA-DR29 and has been reported to control non-responsiveness to antigens through antigen-specific suppressor cells.27 Further studies will be required to determine whether I-E or I-A as well as DQ or DR molecules might be involved in pneumococcal antigen non-responsiveness or cytokine secretion in mouse or human, respectively. To this end, many of the PsaA peptides were predicted to bind I-A, whereas relatively few were predicted to bind I-E.
PsaA is a highly conserved, immunogenic adhesin protein that induces both B-cell and T-cell responses, and is therefore considered as a promising vaccine candidate for combating pneumococcal infection. Streptococcus pneumoniae strain EF3030, a novel human isolate of capsular group 19 pneumococci, which has a greater propensity to cause nasal or pulmonary infections than sepsis when given intranasally, was used to accomplish the objectives of this study. Further, F1 (B6 × BALB/c) mice have reduced susceptibility to S. pneumoniae strain EF3030 and express functional I-Ab, I-Ad, I-Eb and I-Ed. These mice also contain MHC class II haplotypes and corresponding T-cell receptor diversity that more closely represents diverse human HLA alleles. We used this approach previously to identify immunogenic Th cell epitopes in PspA.30
Interleukin-2, produced by CD4+ Th1 type cells, promotes T-cell proliferation. Our experimental data show that epitopes from certain PsaA peptides mounted comparatively high IL-2 and proliferation Th cell recall responses in mice challenged with S. pneumoniae strain EF3030. Another Th1 cytokine, IFN-γ, is also required for protective pneumococcal immunity.31 Interferon-γ blockade accelerated the death of animals during pneumococcal infection,32 whereas treatment of mice with IFN-γ enhanced the survival of mice.33 However, too much IFN-γ in the lung has been shown to inhibit alveolar macrophage-mediated microbial clearance by down-regulating the expression of MARCO, which subsequently enhances the susceptibility to secondary pneumococcal infection.34 The CLN-and spleen-derived Th cells from S. pneumoniae strain EF3030-challenged mice secreted significant amounts of IFN-γ following ex vivo PsaA peptide stimulation.
Interleukin-10 is an anti-inflammatory cytokine that serves as a counterbalance for potentially harmful inflammatory effects of pro-inflammatory cytokines. This is particularly important in severe pneumococcal infections, in which the delicate balance between these two classes of cytokines plays a crucial role in outcome of the disease.35 In the absence of IL-10, a marked increase in pro-inflammatory cytokines is reported during pneumococcal infection;36 its absence hampers effective clearance of infection, and reduces survival of pneumococcal infection.37 Here, we assessed IL-10 secretion and found that certain PsaA peptides stimulated pneumococcal strain EF3030-primed CD4+ T cells to secrete IL-10. Interestingly, Th cells from CLN mounted IL-10 responses to more peptides, than similar cells isolated from the spleen. Perhaps this contributes to establishing pneumococcal carriage by supporting selective pneumococcal clearance by CLN ≫ spleen antigen-presenting cells after stimulation with CD4+ T-cell-derived IL-10, whereas IFN-γ-secreting Th cells might support spleen ≫ CLN macrophages activation and/or internalization of S. pneumoniae.
Interleukin-10 may also help the host by increasing the expression of MARCO. During a secondary bacterial infection, IFN-γ inhibits the MARCO expression, thereby reducing the macrophage activity required for pneumococcal clearance.34 We have previously shown that inhibition of CCL5, a potent chemoattractant for a variety of immune cells including monocytes, macrophages and dendritic cells, resulted in lower IFN-γ-secreting CD4+ T cells and significantly more PsaA-specific IL-10-producing CD4+ T cells, which corresponded with the transition from pneumococcal carriage to lethal pneumonia.38 Hence, the precise role of IL-10 in providing pneumococcal immunity remains unclear and needs further investigations, but studies suggest that excessive IL-10 responses play a deleterious role in pneumococcal immunity. However, moderate levels of IL-10 are required for optimal adaptive (humoral) immune responses to pneumococcal infection and reduced mucosal hyperplasia.
Streptococcus pneumoniae has been reported to induce typically Th1 but not Th2 responses.35 Mice primed to mount Th2 cell responses followed by pneumococcal infection showed an increase in the number of pneumococci compared with naive or Th1-primed groups.39 Interleukin-4 is a Th2-cell-derived cytokine that plays a central role in directing the development of the Th2 phenotype responses in lung. Indeed, elevated IL-4 responses have been associated with an increased risk in pneumococcal infection.40 Similarly, IL-5 is a Th2 cytokine that triggers B-cell activation and differentiation into plasma cells. In the present study we demonstrated that PsaA231–268-specific Th cells from S. pneumoniae strain EF3030-challenged mice secreted significant amounts of IL-4 (spleen ≫ CLN) and IL-5 (spleen ≤ CLN) largely in response to PsaA peptides 19, 20, 21 and 22. However, the uncertain role of IL-4 and IL-5 in pneumococcal cellular immunity makes correlations of these cytokines with protective immunity difficult.
Interleukin-17A-secreting CD4+ T cells (Th17) mediate resistance to mucosal colonization by multiple pathogens41 but their role with respect to pneumococcal immunity has not been studied in detail. However, recent studies have reported that IL-17A supports antibody responses to pneumococcal capsular polysaccharides42 and enhances the pneumococcal killing by human neutrophils.43 A fusion protein of PsaA with the pneumolysin non-toxic derivative PdT coupled with cell wall polysaccharide was reported to induce higher serum antibody titres and greater priming for IL-17A responses than an equimolar mixture of the three antigens.44 Moffitt et al.45 identified the antigens that provide near complete protection from pneumococcal colonization through a Th17-mediated mechanism. These findings suggest that a vaccine candidate that can promote IL-17A responses and elicit protective antibodies may be more effective in protection against pneumococcal mucosal colonization and invasive disease. Recently, Li et al. demonstrated the role of Th17 in diversifying selection and variation of S. pneumoniae antigens. Th17 cell-based immunity creates little selective pressure for antigenic variation and may limit the immune escape of antigenic variants and result in broader and longer protection. We found that selected PsaA peptides (7, 19, 20, 22, 23 and 24) induce pneumococcal strain EF3030-primed CD4+ T cells to secrete IL-17, which may provide longer immunity against divergent strains of S. pneumoniae. Also, higher induction of IL-17 producers by PsaA231–268 peptides could contribute to the conservation of this region, which otherwise would be under immune selection.46 Additional studies on the role of Th cell-derived IL-17 would greatly contribute to the field and will be required to understand how secretion of this cytokine correlates with pneumococcal immunity.
PsaA plays a pivotal role in pneumococcal adherence and colonization, which is a primary step in S. pneumoniae infection. Therefore, from the pathogen's perspective, it would be critical to maintain PsaA function, while reducing detection of T-cell immunodominant epitopes. Several potential, N endopeptidase sites along with β-turn secondary structures would optimally expose these sites for cleavage, which in certain cases would prevent binding to several MHC class II alleles and corresponding induction of cellular immunity.
The secondary structure of proteins consists of regular elements such as α-helices and β-sheets, and irregular elements such as β-bulges, random coils and tight turns. Tight turns are the connecting elements that link one segment of secondary structure to the next and are classified as δ-, γ-, β-, α-and π-turns according to the number of residues involved. β-turns have important biological tasks.29 We predicted the β-turns in PsaA using a highly accurate secondary structure prediction software, PSIPRED, which showed that the β-turns were abundant in PsaA but interestingly none were found in immunodominant Th cell epitope regions. Analysis also revealed that PsaA has only α-helical (no β-sheet) secondary structure content and is predominantly a coiled-coil structure. PsaA peptides with β-turn or continuous strand predicted secondary structures were not considered immunodominant.
In addition to protein secondary structure, several proteases such as N endopeptidases are implicated in antigen and the MHC class II-bound invariant chain processing.47 The level and activity of N endopeptidases can directly control the proteolysis and presentation of T-cell epitopes48 and is required for both antigen and invariant chain (Ii) processing.49 Hence, N endopeptidase can have both positive and negative effects on the outcome of antigen processing.24,50 The proteolytic separation of MHC class II-bound epitopes has been found to be a rate-limiting step in the presentation of T-cell epitopes.51 We observed that Th cells from pneumococcus-challenged mice were incapable of mounting significant cytokine or proliferative responses to peptides containing N-endopeptidase sites. Notably, peptides 7, 19, 20, 22, 23 and 24 sequences are devoid of β-turns and asparagine endopeptidase sites (Fig. 1).
This study supports the use of in silico and in vivo methods to validate T-cell responsiveness to peptide-based vaccines. Here we show that PsaA cellular immunity is mediated by the availability of T-cell epitopes, driven in part by MHC binding affinity as well as lack of β-turns and N endopeptidase sites. It is important to mention that while select peptides provoked a potent Th cell response, additional studies will be required to determine whether these epitopes confer protection against S. pneumoniae infection or carriage. Moreover, studies involving bacterial challenges with pneumococcal strains with PsaA peptides less conserved than strain EF3030 will be important to illustrate the broad use of immunogenic PsaA peptides as vaccines. This study is limited to the recall responses and analysing whether such responses of other peptides are capable of inducing the response of naive T cells will be the focus of future studies.
Acknowledgments
This study was supported by funds from the National Institute of Health Grants AI057808, MD007602, and RR03034. The contents of this manuscript benefitted from many fruitful conversations with colleagues at Morehouse School of Medicine and the University of Alabama at Birmingham.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Hausdorff WP, Bryant J, Paradiso PR, Siber GR. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin Infect Dis. 2000;30:100–21. doi: 10.1086/313608. [DOI] [PubMed] [Google Scholar]
- 2.Berkley JA, Lowe BS, Mwangi I, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005;352:39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 3.Klugman KP. Pneumococcal resistance to antibiotics. Clin Microbiol Rev. 1990;3:171–96. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajam G, Anderton JM, Carlone GM, Sampson JS, Ades EW. Pneumococcal surface adhesin A (PsaA): a review. Crit Rev Microbiol. 2008;34:131–42. doi: 10.1080/10408410802275352. [DOI] [PubMed] [Google Scholar]
- 5.Sampson JS, Furlow Z, Whitney AM, Williams D, Facklam R, Carlone GM. Limited diversity of Streptococcus pneumoniae psaA among pneumococcal vaccine serotypes. Infect Immun. 1997;65:1967–71. doi: 10.1128/iai.65.5.1967-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison KE, Lake D, Crook J, Carlone GM, Ades E, Facklam R, Sampson JS. Confirmation of psaA in all 90 serotypes of Streptococcus pneumoniae by PCR and potential of this assay for identification and diagnosis. J Clin Microbiol. 2000;38:434–7. doi: 10.1128/jcm.38.1.434-437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dintilhac A, Alloing G, Granadel C, Claverys JP. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–39. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 8.McAllister LJ, Tseng H-J, Ogunniyi AD, Jennings MP, McEwan AG, Paton JC. Molecular analysis of the psa permease complex of Streptococcus pneumoniae. Mol Microbiol. 2004;53:889–901. doi: 10.1111/j.1365-2958.2004.04164.x. [DOI] [PubMed] [Google Scholar]
- 9.Briles DE, Ades E, Paton JC, et al. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun. 2000;68:796–800. doi: 10.1128/iai.68.2.796-800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palaniappan R, Singh S, Singh UP, et al. Differential PsaA-, PspA-, PspC-, and PdB-specific immune responses in a mouse model of pneumococcal carriage. Infect Immun. 2005;73:1006–13. doi: 10.1128/IAI.73.2.1006-1013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammer J, Sturniolo T, Sinigaglia F. HLA class II peptide binding specificity and autoimmunity. Adv Immunol. 1997;66:67–100. doi: 10.1016/s0065-2776(08)60596-9. [DOI] [PubMed] [Google Scholar]
- 12.Sturniolo T, Bono E, Ding J, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–61. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 13.Sette A, Fleri W, Peters B, Sathiamurthy M, Bui H-H, Wilson S. A roadmap for the immunomics of category A-C pathogens. Immunity. 2005;22:155–61. doi: 10.1016/j.immuni.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–9. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 15.Reche PA, Glutting J-P, Zhang H, Reinherz EL. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56:405–19. doi: 10.1007/s00251-004-0709-7. [DOI] [PubMed] [Google Scholar]
- 16.Dönnes P, Kohlbacher O. SVMHC: a server for prediction of MHC-binding peptides. Nucleic Acids Res. 2006;34:W194–7. doi: 10.1093/nar/gkl284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parry CS. Flanking p10 contribution and sequence bias in matrix based epitope prediction: revisiting the assumption of independent binding pockets. BMC Struct Biol. 2008;8:44. doi: 10.1186/1472-6807-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridgway WM, Fathman CG. MHC structure and autoimmune T cell repertoire development. Curr Opin Immunol. 1999;11:638–42. doi: 10.1016/s0952-7915(99)00030-8. [DOI] [PubMed] [Google Scholar]
- 19.Gregersen PK. HLA class II polymorphism: implications for genetic susceptibility to autoimmune disease. Lab Invest. 1989;61:5–19. [PubMed] [Google Scholar]
- 20.Briles DE, Crain MJ, Gray BM, Forman C, Yother J. Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect Immun. 1992;60:111–6. doi: 10.1128/iai.60.1.111-116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aaberge IS, Eng J, Lermark G, Løvik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–52. doi: 10.1016/s0882-4010(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 22.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–5. doi: 10.1093/bioinformatics/16.4.404. (Oxford, England) [DOI] [PubMed] [Google Scholar]
- 23.Fuchs PFJ, Alix AJP. High accuracy prediction of β-turns and their types using propensities and multiple alignments. Proteins. 2005;59:828–39. doi: 10.1002/prot.20461. [DOI] [PubMed] [Google Scholar]
- 24.Manoury B, Hewitt EW, Morrice N, Dando PM, Barrett AJ, Watts C. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396:695–9. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- 25.Reche PA, Reinherz EL. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol. 2003;331:623–41. doi: 10.1016/s0022-2836(03)00750-2. [DOI] [PubMed] [Google Scholar]
- 26.Yu J-J, Goluguri T, Guentzel MN, Chambers JP, Murthy AK, Klose KE, Forsthuber TG, Arulanandam BP. Francisella tularensis T-cell antigen identification using humanized HLA-DR4 transgenic mice. Clin Vaccine Immunol. 2010;17:215–22. doi: 10.1128/CVI.00361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ottenhoff TH, Walford C, Nishimura Y, Reddy NB, Sasazuki T. HLA-DQ molecules and the control of Mycobacterium leprae-specific T cell nonresponsiveness in lepromatous leprosy patients. Eur J Immunol. 1990;20:2347–50. doi: 10.1002/eji.1830201027. [DOI] [PubMed] [Google Scholar]
- 28.Bono MR, Strominger JL. Direct evidence of homology between human DC-1 antigen and murine I-A molecules. Nature. 1982;299:836–40. doi: 10.1038/299836a0. [DOI] [PubMed] [Google Scholar]
- 29.Hurley CK, Nunez G, Winchester R, Finn OJ, Levy R, Capra JD. The human HLA-DR antigens are encoded by multiple β-chain loci. J Immunol. 1982;129:2103–8. (Baltimore, Md: 1950) [PubMed] [Google Scholar]
- 30.Singh R, Singh S, Sharma PK, Singh UP, Briles DE, Hollingshead SK, Lillard JW. Helper T cell epitope-mapping reveals MHC-peptide binding affinities that correlate with T helper cell responses to pneumococcal surface protein A. PLoS ONE. 2010;5:e9432. doi: 10.1371/journal.pone.0009432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marqués JM, Rial A, Muñoz N, et al. Protection against Streptococcus pneumoniae serotype 1 acute infection shows a signature of Th17-and IFN-γ-mediated immunity. Immunobiology. 2012;217:420–9. doi: 10.1016/j.imbio.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 32.Rubins JB, Pomeroy C. Role of γ interferon in the pathogenesis of bacteremic pneumococcal pneumonia. Infect Immun. 1997;65:2975–7. doi: 10.1128/iai.65.7.2975-2977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weigent DA, Huff TL, Peterson JW, Stanton GJ, Baron S. Role of interferon in streptococcal infection in the mouse. Microb Pathog. 1986;1:399–407. doi: 10.1016/0882-4010(86)90071-9. [DOI] [PubMed] [Google Scholar]
- 34.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-γ during recovery from influenza infection. Nat Med. 2008;14:558–64. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 35.Khan AQ, Shen Y, Wu Z-Q, Wynn TA, Snapper CM. Endogenous pro-and anti-inflammatory cytokines differentially regulate an in vivo humoral response to Streptococcus pneumoniae. Infect Immun. 2002;70:749–61. doi: 10.1128/iai.70.2.749-761.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuchiya K, Komori M, Zheng QY, Ferrieri P, Lin J. Interleukin-10 is an essential modulator of mucoid metaplasia in a mouse otitis media model. Ann Otol Rhinol Laryngol. 2008;117:630–6. doi: 10.1177/000348940811700814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Poll T, Marchant A, Keogh CV, Goldman M, Lowry SF. Interleukin-10 impairs host defense in murine pneumococcal pneumonia. J Infect Dis. 1996;174:994–1000. doi: 10.1093/infdis/174.5.994. [DOI] [PubMed] [Google Scholar]
- 38.Palaniappan R, Singh S, Singh UP, et al. CCL5 modulates pneumococcal immunity and carriage. J Immunol. 2006;176:2346–56. doi: 10.4049/jimmunol.176.4.2346. (Baltimore, Md: 1950) [DOI] [PubMed] [Google Scholar]
- 39.Yu X, Sperling A, Blair C, Thompson K, Naclerio R. Antigen stimulation of TH2 cells augments acute bacterial sinusitis in mice. J Allergy Clin Immunol. 2004;114:328–34. doi: 10.1016/j.jaci.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 40.Kang CI, Rouse MS, Patel R, Kita H, Juhn YJ. Allergic airway inflammation and susceptibility to pneumococcal pneumonia in a murine model with real-time in vivo evaluation. Clin Exp Immunol. 2009;156:552–61. doi: 10.1111/j.1365-2249.2009.03925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 2009;126:177–85. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malley R, Srivastava A, Lipsitch M, Thompson CM, Watkins C, Tzianabos A, Anderson PW. Antibody-independent, interleukin-17A-mediated, cross-serotype immunity to pneumococci in mice immunized intranasally with the cell wall polysaccharide. Infect Immun. 2006;74:2187–95. doi: 10.1128/IAI.74.4.2187-2195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y-J, Gross J, Bogaert D, et al. Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog. 2008;4:e1000159. doi: 10.1371/journal.ppat.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y-J, Forte S, Thompson CM, Anderson PW, Malley R. Protection against Pneumococcal colonization and fatal pneumonia by a trivalent conjugate of a fusion protein with the cell wall polysaccharide. Infect Immun. 2009;77:2076–83. doi: 10.1128/IAI.01554-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moffitt KL, Gierahn TM, Lu Y-J, Gouveia P, Alderson M, Flechtner JB, Higgins DE, Malley R. TH17-based vaccine design for prevention of Streptococcus pneumoniae colonization. Cell Host Microbe. 2011;9:158–65. doi: 10.1016/j.chom.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Gierahn T, Thompson MC, et al. Distinct effects on diversifying selection by two mechanisms of immunity against Streptococcus pneumoniae. PLoS Pathog. 2012;8:e1002989. doi: 10.1371/journal.ppat.1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villadangos JA, Riese RJ, Peters C, Chapman HA, Ploegh HL. Degradation of mouse invariant chain: roles of cathepsins S and D and the influence of major histocompatibility complex polymorphism. J Exp Med. 1997;186:549–60. doi: 10.1084/jem.186.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manoury B, Mazzeo D, Fugger L, et al. Destructive processing by asparagine endopeptidase limits presentation of a dominant T cell epitope in MBP. Nat Immunol. 2002;3:169–74. doi: 10.1038/ni754. [DOI] [PubMed] [Google Scholar]
- 49.Amigorena S, Webster P, Drake J, Newcomb J, Cresswell P, Mellman I. Invariant chain cleavage and peptide loading in major histocompatibility complex class II vesicles. J Exp Med. 1995;181:1729–41. doi: 10.1084/jem.181.5.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antoniou AN, Blackwood SL, Mazzeo D, Watts C. Control of antigen presentation by a single protease cleavage site. Immunity. 2000;12:391–8. doi: 10.1016/s1074-7613(00)80191-0. [DOI] [PubMed] [Google Scholar]
- 51.Castellino F, Zappacosta F, Coligan JE, Germain RN. Large protein fragments as substrates for endocytic antigen capture by MHC class II molecules. J Immunol. 1998;161:4048–57. (Baltimore, Md: 1950) [PubMed] [Google Scholar]