Abstract
Peroxisome proliferator-activated receptor (PPAR) γ coactivator 1α (PGC-1α) is a coactivator of various nuclear receptors and other transcription factors, which is involved in the regulation of energy metabolism, thermogenesis, and other biological processes that control phenotypic characteristics of various organ systems including skeletal muscle. PGC-1α in skeletal muscle is considered to be involved in contractile protein function, mitochondrial function, metabolic regulation, intracellular signaling, and transcriptional responses. Branched-chain amino acid (BCAA) metabolism mainly occurs in skeletal muscle mitochondria, and enzymes related to BCAA metabolism are increased by exercise. Using murine skeletal muscle overexpressing PGC-1α and cultured cells, we investigated whether PGC-1α stimulates BCAA metabolism by increasing the expression of enzymes involved in BCAA metabolism. Transgenic mice overexpressing PGC-1α specifically in the skeletal muscle had increased the expression of branched-chain aminotransferase (BCAT) 2, branched-chain α-keto acid dehydrogenase (BCKDH), which catabolize BCAA. The expression of BCKDH kinase (BCKDK), which phosphorylates BCKDH and suppresses its enzymatic activity, was unchanged. The amount of BCAA in the skeletal muscle was significantly decreased in the transgenic mice compared with that in the wild-type mice. The amount of glutamic acid, a metabolite of BCAA catabolism, was increased in the transgenic mice, suggesting the activation of muscle BCAA metabolism by PGC-1α. In C2C12 cells, the overexpression of PGC-1α significantly increased the expression of BCAT2 and BCKDH but not BCKDK. Thus, PGC-1α in the skeletal muscle is considered to significantly contribute to BCAA metabolism.
Introduction
Peroxisome proliferator-activated receptor (PPAR) γ coactivator 1 α (PGC-1α) was identified as a nuclear receptor coactivator of PPARγ in brown adipose tissue and found to be upregulated in brown adipose tissue and skeletal muscle in response to cold exposure [1]. PGC-1α is now known to be involved not only in the regulation of thermogenesis but also in energy metabolism and other biological processes that are critical in controlling phenotypic characteristics of various organ systems [1]–[5]. PGC-1α coactivates a broad range of transcription factors, including PPARs, glucocorticoid receptor (GR), nuclear respiratory factors, myocyte enhancing factors, estrogen-related receptor, and forkhead box O1 [6]–[9]. PGC-1α acts through the recruitment of coactivators with histone acetyl transferase activity as well as interaction with proteins involved in transcriptional initiation and RNA processing [10].
It has recently been shown that there are several isoforms of PGC-1α mRNA [11]–[14]. We previously reported that among the PGC-1α isoforms, PGC-1α-b expression was markedly increased in response to exercise [15]. PGC-1α-b, considered to be similar in function to PGC-1α1 (originally found full-length PGC-1α [1]), structurally differs by 16 amino acids at its amino terminal [12]. We demonstrated that overexpression of PGC-1α-b in skeletal muscle but not in heart increases mitochondrial biogenesis and capillary density, contributing to improved exercise capacity [4]. Moreover, animal and cellular genetic models with altered expression of the PGC-1α gene have much evidence for the role of PGC-1α in fiber type specificity [16], [17], mitochondrial biogenesis [17]–[19], angiogenesis [20], and improved exercise performance [21].
Mammalian cells have a high capacity system for oxidative disposal of branched-chain amino acids (BCAA). In contrast to other essential amino acids, which are primarily oxidized in the liver, the most active system for the oxidation of BCAA is located in skeletal muscle cells [22]. The degradation of BCAA mainly occurs in the mitochondria via reversible transamination by branched-chain aminotransferase (BCAT) to produce the corresponding branched-chain α-keto acids (BCKA), which in turn are subjected to oxidative decarboxylation by branched-chain α-keto acid dehydrogenase (BCKDH) to produce CoA esters. The enzymes that catalyze these two reactions are common to the three BCAA (Val, Leu, and Ile). The second step enzyme, BCKDH, catalyzes an irreversible reaction that commits individual BCKA to their respective degradation pathways [23] and is considered to be the most important regulatory enzyme in the catabolism of the three BCAA [24]. BCKDH activity is regulated by BCKDH kinase (BCKDK); BCKDH phosphorylation attenuates its enzyme activity [23]. In this study, microarray analysis revealed that the BCAA catabolic pathway was coordinately activated in skeletal muscle of transgenic mice overexpressing PGC-1α. Thus, we investigated whether PGC-1α stimulates BCAA metabolism with an increase in the expression of enzymes involved in BCAA metabolism, such as BCAT, BCKDH and BCKDK, using cultured cells and murine skeletal muscle overexpressing PGC-1α.
Methods
Transgenic (Tg) mice
Tg mice overexpressing PGC-1α-b in skeletal muscle (hereafter, PGC-1α Tg mice or just Tg mice) were generated as described [12]. In brief, the human α-skeletal actin promoter was used to express PGC-1α-b in skeletal muscle (C57BL6 background). We used the B line of Tg mice in this study; Two independent lines of Tg mice showed similar phenotypes in our previous study [4]. Mice were killed by rapid neck disarticulation. A total of 32 mice were used.
Ethics Statement
Mice were cared for in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals and our institutional guidelines. All animal experiments were performed with the approval of the National Institute of Health and Nutrition Ethics Committee on Animal Research (approval ID: No 908, 1008, and 1111) and Institutional Animal Care and Use Committee of University of Shizuoka.
cDNA microarray analysis
RNA was isolated from skeletal muscle (gastrocnemius) of Tg mice (age, 12 weeks) and age-matched WT control mice. Samples from WT and Tg mice (N = 5) were pooled and used. Each sample was labeled with a cyanine 3-CTP using the Low Input Quick Amp Labeling Kit (Agilent Technologies, Inc., Santa Clara, CA) and hybridized to the Agilent whole mouse genome microarray (4×44K), which contains 41,534 genes including expressed sequence tags. Signal detection and data analysis were performed according to the manufacturer's instructions [25].
Functional annotation analysis in genes up-regulated by PGC-1α overexpression
We conducted pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database resource with DAVID v6.7 [26], which is a web application providing a comprehensive set of functional annotation tools to understand the biological meaning of a large list of genes. A list of gene symbols that showed increased expression in skeletal muscle of PGC-1α Tg mice was submitted, and a significant overrepresentation of the KEGG pathway was detected.
Bioinformatics analysis of transcription factors enriched in the BCAA metabolic pathway genes up-regulated in PGC-1α Tg mice
We employed ChIP Enrichment analysis (ChEA) software [27] to explore the transcription factors involved in the regulation of genes whose expression was induced by PGC-1α overexpression classified as BCAA metabolic pathway by DAVID v6.7 [26]. ChEA is a tool that computes over-representation of transcription factor targets from the database of ChIP-seq and ChIP-chip experiments [27]. The database as of Dec 23, 2013 contains 471,284 extracted entries, from 228 publications, describing the binding of 203 transcription factors to 47,119 target genes (http://amp.pham.mssm.edu/lib/chea.jsp).
Quantitative real-time RT-PCR analysis
Total RNA was prepared usnig TRIzol (Life Technologies Japan, Tokyo, Japan). cDNA was synthesized from 1 μg of total RNA using the QuantiTect Rev. Transcription Kit (QIAGEN K.K, Tokyo, Japan). Gene expression levels were measured as described [25]. The following primers were used:
BCAT2 Fw, 5′-CGGACCCTTCATTCGTCAGA -3′; BCAT2 Rv, 5′-CCATAGTTCCCCCCCAACTT-3′; BCKDHa Fw, 5′-CCAGGGTTGGTGGGATGAG-3′; BCKDHa Rv, 5′-GGCTTCCATGACCTTCTTTCG-3′; BCKDK Fw, 5′-GATCCGAATGCTGGCTACTCA-3′; BCKDK Rv, 5′-GCCAACAAAATCAGGCTTGTC-3′; PGC-1α Fw, 5′-CGGAAATCATATCCAACCAG-3′; PGC-1α Rv, 5′-TGAGGACCGCTAGCAAGTTTG-3′; 36B4 Fw, 5′-GGCCCTGCACTCTCGCTTTC-3′; 36B4 Rv, 5′-TGCCAGGACGCGCTTGT -3′;
Stable cell lines
PlatE cells were cultured in 90-mm dishes and transfected at 70% confluence using Lipofectamine 2000 (Life Technologies Japan, Tokyo, Japan) according to the manufacturer's instructions using 2 μg pMX-derived expression plasmid [28] containing PGC-1α cDNA or vector alone. Virus-containing supernatants were harvested 48 h after transfection and added to dishes of C2C12 cells, which were selected using 5 μg/ml puromycin to eliminate uninfected cells. After drug selection, virally infected stable cells were cultured to confluence in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal calf serum, and the medium was changed every 2 days. On 3 day after confluence, cells were used for RNA preparation.
Western blotting analysis
Frozen skeletal muscle (gastrocnemius) was homogenized in RIPA Lysis Buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing 0.2 mM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (1/100 volume) (Sigma Aldrich Japan, K.K. Tokyo, Japan). The supernatant was separated by centrifugation at 20,400 g for 15 min at 4°C. Protein from the supernatant (20 μg) was applied onto an SDS-PAGE gel. A commercially available precast ready-made gel (10% acrylamide, e-PAGEL, ATTO Co. Tokyo, Japan) was used. The following primary antibodies were used for Western blotting: anti-PGC-1α (Rabbit polyclonal IgG) against the carboxyl terminus 777–797 (Millipore, Billerica, MA), and anti-BCKDH (rabbit polyclonal, [29]).
Amino acid analysis
Skeletal muscle (gastrocnemius) and blood from PGC-1α Tg mice (in the feeding condition) were used for amino acid analysis. In addition, C2C12 cells overexpressing PGC-1α were examined for the amino acid content. Skeletal muscle and C2C12 cells were extracted with methanol/chloroform/water (5/5/2 in volume), and centrifuged. The supernatant was dried with nitrogen gas and dissolved in water. Amino acid levels were measured by HPLC assays (SRL, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed using Student's t test. Data were expressed as the mean ± SE. P value <0.05 was considered statistically significant. Standard P-values (Fisher's exact test) and Benjamini P-values were evaluated for functional annotation analysis. The Fisher exact test with the Bonferroni's correction P-values were evaluated for transcription factor search analysis.
Results And Discussion
Increased BCAA metabolism in skeletal muscle of PGC-1α Tg mice
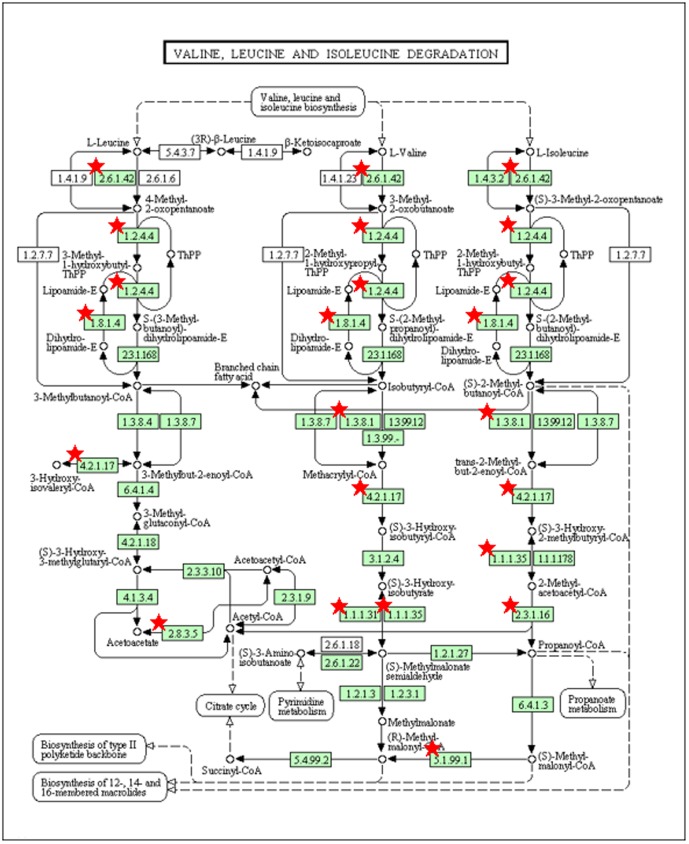
To characterize the phenotype of the skeletal muscle of Tg mice, we performed microarray analysis of gene expression. Microarray analysis revealed that the expression of many genes was changed in Tg mice as compared with that in WT mice. Among these, 315 genes (Table S1) were up-regulated (more than 2.5-hold) and used to conduct pathway analysis, which detected 7 categories (Table 1), including oxidative phosphorylation, TCA cycle, and fatty acid metabolism, which were related to mitochondrial function. These results were consistent with previous reports that PGC-1α increases the mitochondrial number and enhances their function [3], [5]. We observed pathway categories of Parkinson's disease, Huntington's disease and Alzheimer's disease (Table 1), and individual genes increased in the categories were all related to mitochondrial functions (data not shown). In addition, we found that BCAA metabolic pathway (i.e., Val, Leu, Ile degradation, Table 1). As shown in Figure 1, gene expression of many enzymes involved in BCAA catabolism was increased, suggesting that PGC-1α stimulates BCAA metabolism in skeletal muscle. Thus, we examined whether PGC-1α stimulates BCAA metabolism.
Table 1. Pathway analysis.
| Categories of pathway analysis | P-Value | Benjamini |
| Oxidative phosphorylation | 1.10E-13 | 1.00E-11 |
| Parkinson's disease | 1.70E-13 | 8.10E-12 |
| Citrate cycle (TCA cycle) | 3.10E-12 | 9.70E-11 |
| Huntington's disease | 9.10E-12 | 2.10E-10 |
| Alzheimer's disease | 5.30E-10 | 9.90E-09 |
| Valine, leucine and isoleucine degradation | 7.10E-09 | 1.10E-07 |
| Fatty acid metabolism | 9.10E-08 | 1.20E-06 |
Compared with WT mice, 315 genes were found to be up-regulated in PGC-1α Tg mice by microarray and classified into KEGG pathway analysis as described in Methods.
Figure 1. Pathway map of Val, Leu, and Ile degradation.
Individual gene name of the KEGG pathway of Val, Leu, and Ile degradation extracted by pathway analysis is shown as a metabolic map. Red asterisks indicate increased gene expression by microarray of Tg mice. Gene names corresponding to enzyme numbers with red asterisks are as follows: 2.8.3.5, 3-oxoacid CoA transferase 1; 2.3.1.16, acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase); 1.3.8.1, acyl-Coenzyme A dehydrogenase, short chain; 2.6.1.42, branched chain aminotransferase 2, mitochondrial; 1.2.4.4, branched chain ketoacid dehydrogenase E1, alpha polypeptide; 1.8.1.4, dihydrolipoamide dehydrogenase; 1.1.1.35, hydroxyacyl-Coenzyme A dehydrogenase; 4.2.1.17, hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit; 5.1.99.1, methylmalonyl CoA epimerase; 1.1.1.31, 3-hydroxyisobutyrate dehydrogenase.
Levels of BCAA and its catabolic enzymes in skeletal muscle of PGC-1α Tg mice
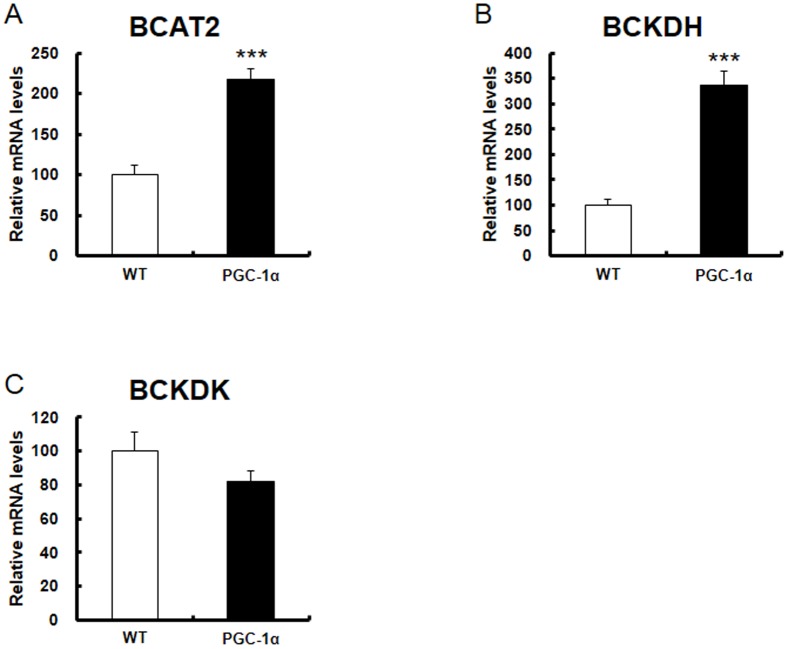
We examined the gene expression of BCAA metabolic enzymes (BCAT2, BCKDH, BCKDK). RNA was obtained from the skeletal muscle of Tg and WT mice. The expression of BCAT2 (2.0-fold) and BCKDH (3.5-fold) was significantly increased in Tg mice compared with that in WT mice (Fig. 2A, B). Meanwhile, the expression of BCKDK was decreased (Fig. 2C). Subsequently, we examined protein expression of BCAA metabolic enzymes by Western blot analysis. PGC-1α protein (100 kDa) increased 4-fold in Tg mice compared with WT mice (Fig. 3A). In this experiment, we also observed increased 45 kDa and 25 kDa bands, whose physiological significance in currently unclear. Using a BCKDH antibody, we observed the strongest band at 55 kDa (arrowhead), which corresponded to the E2 subunit, and was slightly (1.3-fold) increased in Tg mice. The faint band in WT mice at approximately 45 kDa (arrowhead, long exposure), which probably represents E1α subunits [29], increased in Tg mice significantly (1.5-fold). The band at approximately 35 kDa (arrowhead), which probably represents E1β subunits [29], increased markedly (11-fold). Thus, we observed an increased protein level of BCKDH (Fig. 3B), which is consistent with the increased mRNA level. Next we examined BCAA levels from skeletal muscle in Tg mice and WT mice. Val and Leu levels were significantly decreased in Tg mice compared with that in WT mice (Fig. 4A). Ile was detected in WT mice but observed only at trace levels in Tg mice (Fig. 4A). The level of Glu, a metabolite of BCAA catabolism, was increased, in contrast to the decreased BCAA level. Levels of other amino acids are shown in Table 2. We also measured BCAA levels in blood (plasma). BCAA levels tended to be decreased in blood as observed in skeletal muscle (although not significant, P = 0.067 for Val, P = 0.072 for Leu and P = 0.063 for Ile; Fig. 4B and Table 3). These findings indicate that changes in the expression of BCAA metabolic enzymes are functional, and accompanied by enzyme activation.
Figure 2. Gene expression of BCAA metabolic enzyme in skeletal muscle of PGC-1α Tg mice.
Expression of A) BCAT2, B) BCKDH, and C) BCKDK genes in WT (control; open columns, N = 9) and PGC-1α Tg (filled columns, N = 7) mice by quantitative real-time RT-PCR. RNA was obtained from mice with feeding condition. These samples were as used in [4]. In the sample, PGC-1α expression was 30 fold higher in Tg mice than in WT mice (Fig. 1 of [4]). The relative values are shown (the control is set as 100). ***P<0.001.
Figure 3. Protein expression of PGC-1α and BCAA metabolic enzymes in skeletal muscle of PGC-1α -Tg mice.
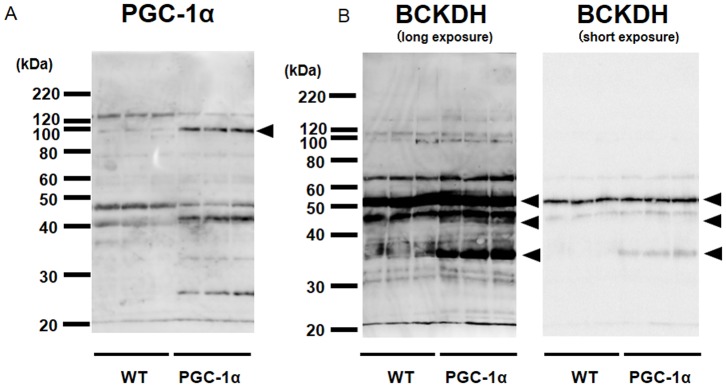
Total lysates from skeletal muscle were subjected to SDS-PAGE, followed by Western blot analysis with indicated antibodies. Typical blots are shown. Densitometric analysis was performed on the bands indicated (arrowheads). Molecular size marked was indicated on the left side of blots. Tg and WT mice were sacrificed at 12 weeks of age (N = 3 for WT and N = 3 for Tg mice). In these samples, we confirmed increased mRNA expression of BCAT2 and BCKDH but not BCKDK as observed in Figure 1 (data not shown).
Figure 4. BCAA content in skeletal muscle and blood of PGC-1α Tg mice.

Val, Leu, and Ile levels in (A) skeletal muscle and (B) blood. Open columns represent for WT (N = 4) and filled columns represent Tg (N = 4). ***P<0.001, **P<0.01. T.R., trace level.
Table 2. Amino acid content in skeletal muscle of PGC-1α Tg mice.
| Amino acid | WT | PGC-1α Tg |
| Alanine | 1768.3 | 1388.6 |
| Arginine | 218.1 | 446.4* |
| Asparagine | 61.9 | 53.1 |
| Aspartic acid | 163.4 | 257.3** |
| Cystine | TR | TR |
| Glutamic acid | 505.9 | 1237.5*** |
| Glutamine | 1341 | 1557.9 |
| Glycine | 3165.9 | 655.9** |
| Histidine | 104.3 | 101.3 |
| Isoleucine | 44.4 | TR |
| Leucine | 61.1 | 40.3*** |
| Lysine | 574 | 1074.1* |
| Methionine | 49.9 | 35.7 |
| Phenylalanine | 12.4 | TR |
| Proline | 169.1 | ND |
| Serine | 357.7 | 186.3* |
| Threonine | 242.6 | 186.3 |
| Tryptophan | ND | ND |
| Tyrosine | 77 | 75 |
| Valine | 120.8 | 68.4** |
The samples were used as in Figure 4. ***P<0.001, **P<0.01, *P<0.05. TR, trace level. ND, not detected.
Table 3. Amino acid content in blood of PGC-1α Tg mice.
| Amino acid | WT | PGC-1α Tg |
| Alanine | 381.4 | 313.9 |
| Arginine | 112.3 | 114.5 |
| Asparagine | 45.7 | 38.9 |
| Aspartic acid | 7.7 | 6.7 |
| Cystine | 8.7 | 5.1 |
| Glutamic acid | 42 | 35.2* |
| Glutamine | 631.4 | 726 |
| Glycine | 306.2 | 334 |
| Histidine | 60.1 | 58 |
| Isoleucine | 63.7 | 43.6 |
| Leucine | 115.3 | 81.3 |
| Lysine | 277.7 | 256.4 |
| Methionine | 56.1 | 53.1 |
| Phenylalanine | 60.5 | 59.7 |
| Proline | 105 | 96.1 |
| Serine | 134.8 | 125.5 |
| Threonine | 117.2 | 121.6 |
| Tryptophan | 41.8 | 39.7 |
| Tyrosine | 99.4 | 82 |
| Valine | 171.1 | 122.9 |
The samples were used as in Figure 4.
*P<0.05.
BCAA metabolism gene expression in C2C12 cells overexpressing PGC-1α
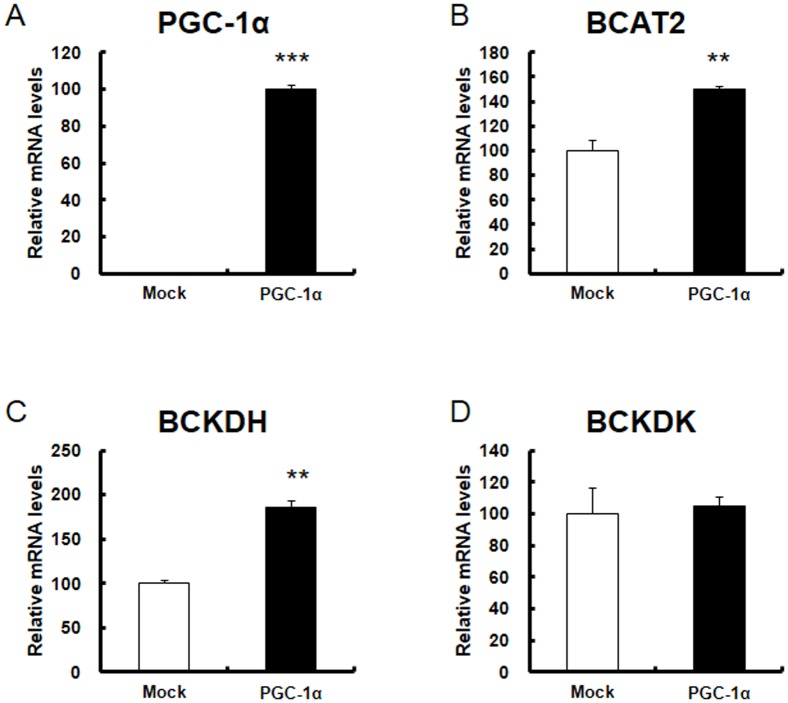
Next, to examine whether the effect of PGC-1α on increased BCAA metabolism was cell autonomous, we used C2C12 cells, which are ectopically overexpressed PGC-1α by retrovirus, and examined BCAA metabolism gene expression. Gene expression of BCAT2 (1.5-fold) and BCKDH (2-fold) was increased but that of BCKDK was not (Fig. 5), as observed in Tg mice. These data suggest that PGC-1α regulates BCAA catabolic gene expression in a cell autonomous manner. Then, we examined the amino acid levels in the cells. Ile was observed only at a trace level. Val was slightly lower in mock cells than in cells overexpressing PGC-1α (P = 0.162). Leu levels were detectable in mock cells; however, it was detected only at a trace level in cells overexpressing PGC-1α (Table 4). In summary, BCAA levels appeared to be decreased in C2C12 cells overexpressing PGC-1α, suggesting that BCAA catabolism is regulated by PGC-1α in muscle cells.
Figure 5. Gene expression of BCAA metabolic enzymes in cultured C2C12 cells overexpressing PGC-1α.
Total RNA was isolated from the cells and analyzed by quantitative real-time RT-PCR with primers for A) PGC-1α, B) BCAT2, C) BCKDHa, and D) BCKDK. Open columns represent mock cells (N = 3), and filled columns represent PGC-1α-overexpressed cells (N = 3). Each value represents mean ± SE (N = 3). The relative values are shown (the control is set as 100). For PGC-1α expression, the value was set as 100 in the PGC-1α overexpressed cells. ***P<0.001, **P<0.01.
Table 4. Amino acid content in C2C12 cells overexpressing of PGC-1α.
| Amino acid | Mock | PGC-1α |
| Alanine | 35.7 | 62.7* |
| Arginine | ND | ND |
| Asparagine | ND | ND |
| Aspartic acid | 44.3 | 55.3 |
| Cystine | TR | TR |
| Glutamic acid | 116.3 | 87.1 |
| Glutamine | 24.4 | 20.6 |
| Glycine | 67.1 | 61.3 |
| Histidine | ND | ND |
| Isoleucine | TR | TR |
| Leucine | 9 | TR |
| Lysine | 14.6 | 15.9 |
| Methionine | ND | ND |
| Phenylalanine | ND | ND |
| Proline | ND | ND |
| Serine | 9 | 9.5 |
| Threonine | 27.4 | 35.2 |
| Tryptophan | ND | ND |
| Tyrosine | TR | TR |
| Valine | 10.2 | 8.7 |
The samples were used as in Figure 5.
*P<0.05. TR, trace level. ND, not detected.
Changed level of other amino acids caused by PGC-1α overexpression
In addition to BCAA, the levels of other amino acids were also changed. For example, in skeletal muscle of Tg mice, Glu levels were significantly increased (Table 2). During BCAA degradation by BCAT, α-keto glutarate is catabolized to Glu. Thus, increasing Glu levels are consistent with the stimulation of BCAA degradation in muscle. Alternatively, in cells overexpressing PGC-1α, Glu was not increased, but Ala was increased (Table 4). The reason for this may be that BCAT catalyzes amino base transfer from BCAA to pyruvate, thereby producing Ala. However, in Tg mice, the expression of genes involved in glycolysis is markedly decreased [4], resulting in inadequate pyruvate, a product of the glycolysis pathway, for Ala production, or Ala may be moved and used in other tissues in animals.
How did PGC-1α increase expression of BCAA metabolism enzyme?
PGC-1α Tg mice have muscle, which has a much higher oxidative capacity [4]. BCAT2 and BCKDH are mitochondrial enzymes [30]. Therefore, the increased expression of BCAT2 and BCKDH could be due to an increased number of mitochondria in the muscles of the Tg mice. Alternatively, PGC-1α may activate BCAA metabolism via the coactivation of glucocorticoid receptor (GR) and PPARα. PGC-1α is a transcriptional coactivator of nuclear receptor and other transcriptional factors, some of which have been reported to activate the transcription of the BCAT2 gene. For example, the expression of BCAT2 was decreased in KLF15-KO mice [31], and the rat BCAT2 promoter was activated by KLF15 and GR [32]. Moreover, PPARα activated the BCKDH complex in the liver [22]. In addition, bioinformatics analysis revealed that several nuclear receptors, including PPAR and estrogen receptor-related receptor (ERR), are significantly frequently recruited to the BCAA metabolic pathway genes up-regulated in skeletal muscle of PGC-1α Tg mice (Table 5). These data suggest that PGC-1α activates BCAA metabolism through multiple nuclear receptors.
Table 5. Bioinformatics analysis of transcription factors enriched in the BCAA metabolic pathway genes up-regulated in PGC-1α Tg mice.
| Transcription factor | P-Value | Target gene |
| KLF4 | 2.80E-07 | ACAA2;ACADS;BCAT2;BCKDHA;HADHA;MCEE;OXCT1 |
| PPARG | 3.68E-07 | ACAA2;ACADS;BCAT2;HADHB |
| EKLF | 5.14E-06 | ACADS;BCAT2;HADH;HIBADH;OXCT1 |
| ESRRB | 1.04E-05 | ACADS;DLD;HIBADH;MCEE;OXCT1 |
| PPARD | 1.16E-05 | HADHA;HADHB;OXCT1 |
| ZFP42 | 1.22E-05 | ACADS;BCAT2;BCKDHA;HADHA;HADHB |
| WT1 | 4.22E-04 | BCAT2;HADHB;HIBADH;OXCT1 |
| NR0B1 | 4.49E-04 | ACADS;BCAT2;HADHA;HADHB |
| TET1 | 6.17E-04 | ACAA2;ACADS;HADHA;OXCT1 |
| GATA4 | 9.09E-04 | ACAA2;BCAT2;HADHA;HADHB |
List of transcription factors, which are statistically identified as ones that can be recruited to the BCAA metabolic genes, up-regulated in PGC-1α Tg mice. Target genes were previously found in ChIP assay for interacting with indicated transcription factors in the literature [27]. Abbreviations of the transcription factors are as follows, KLF4, Krueppel-like factor 4; PPARG, Constitutive coactivator of peroxisome proliferator-activated receptor gamma (Constitutive coactivator of PPAR-gamma) (Constitutive coactivator of PPARG); EKLF, Krueppel-like factor 1 (Erythroid krueppel-like transcription factor); ESRRB, Steroid hormone receptor ERR2 (Estrogen receptor-like 2) (Estrogen-related receptor beta) (ERR-beta); PPARD, Peroxisome proliferator-activated receptor delta (PPAR-delta); ZFP42, Zinc finger protein 42; WT1, Wilms tumor protein; NR0B1, Nuclear receptor subfamily 0 group B member 1 (Nuclear receptor DAX-1); TET1, Methylcytosine dioxygenase TET1 (EC 1.14.11.n2) (CXXC-type zinc finger protein 6) (Ten-eleven translocation 1 gene protein homolog); GATA4, Transcription factor GATA-4 (GATA-binding factor 4).
Conclusion
In this study, we investigated whether PGC-1α stimulates BCAA metabolism by increasing the expression of involved enzymes, such as BCAT and BCKDH, using cultured cells and murine skeletal muscle overexpressing PGC-1α. Our data suggest that BCAA degradation is mediated by increased expression of PGC-1α, and the physiological relevance, such as condition of induced expression in PGC-1α i.e., exercise, remain to be analyzed by further experiments.
Supporting Information
List of genes up-regulated in Tg mice by microarray. RNA obtained from WT and Tg mice was pooled and used for microarray analysis as described in Methods. Hybridized signals outside the linear range were excluded, and values were normalized by the 75th percentile calculation. Genes with a calculated value less than 100 in Tg mice were deleted. Genes up-regulated more than 2.5 fold in Tg mice were listed. Pooled array data were confirmed by performing quantitative real-time RT-PCR with representative probes and similar increased was observed in all samples, indicating that microarray data represent the expression change in each group (data not shown).
(PDF)
Acknowledgments
Microarray data analysis was, in part, performed at the Medical Research Support Center, Graduate School of Medicine, Kyoto University.
Funding Statement
This study was supported in part by Fuji Foundation for Protein Research, Sapporo Bioscience Foundation, The Kieikai Research Foundation, The Uehara Memorial Foundation, Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Puigserver P, Wu Z, Park CW, Graves R, Wright M, et al. (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839. [DOI] [PubMed] [Google Scholar]
- 2. Liang H, Balas B, Tantiwong P, Dube J, Goodpaster BH, et al. (2009) Whole body overexpression of PGC-1alpha has opposite effects on hepatic and muscle insulin sensitivity. Am J Physiol Endocrinol Metab 296: E945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Puigserver P, Spiegelman BM (2003) Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev 24: 78–90. [DOI] [PubMed] [Google Scholar]
- 4. Tadaishi M, Miura S, Kai Y, Kano Y, Oishi Y, et al. (2011) Skeletal muscle-specific expression of PGC-1alpha-b, an exercise-responsive isoform, increases exercise capacity and peak oxygen uptake. PLoS One 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wareski P, Vaarmann A, Choubey V, Safiulina D, Liiv J, et al. (2009) PGC-1alpha and PGC-1beta regulate mitochondrial density in neurons. J Biol Chem 284: 21379–21385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, et al. (2003) PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci U S A 100: 12378–12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelly DP, Scarpulla RC (2004) Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev 18: 357–368. [DOI] [PubMed] [Google Scholar]
- 8. Lin J, Handschin C, Spiegelman BM (2005) Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1: 361–370. [DOI] [PubMed] [Google Scholar]
- 9. Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P (2008) Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett 582: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG (2003) Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell 12: 1137–1149. [DOI] [PubMed] [Google Scholar]
- 11. Chinsomboon J, Ruas J, Gupta RK, Thom R, Shoag J, et al. (2009) The transcriptional coactivator PGC-1alpha mediates exercise-induced angiogenesis in skeletal muscle. Proc Natl Acad Sci U S A 106: 21401–21406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miura S, Kai Y, Kamei Y, Ezaki O (2008) Isoform-specific increases in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to beta2-adrenergic receptor activation and exercise. Endocrinology 149: 4527–4533. [DOI] [PubMed] [Google Scholar]
- 13. Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, et al. (2012) A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshioka T, Inagaki K, Noguchi T, Sakai M, Ogawa W, et al. (2009) Identification and characterization of an alternative promoter of the human PGC-1alpha gene. Biochem Biophys Res Commun 381: 537–543. [DOI] [PubMed] [Google Scholar]
- 15. Tadaishi M, Miura S, Kai Y, Kawasaki E, Koshinaka K, et al. (2011) Effect of exercise intensity and AICAR on isoform-specific expressions of murine skeletal muscle PGC-1alpha mRNA: a role of beta2-adrenergic receptor activation. Am J Physiol Endocrinol Metab 300: E341–349. [DOI] [PubMed] [Google Scholar]
- 16. Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, et al. (2002) Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418: 797–801. [DOI] [PubMed] [Google Scholar]
- 17. Miura S, Kai Y, Ono M, Ezaki O (2003) Overexpression of peroxisome proliferator-activated receptor gamma coactivator-1alpha down-regulates GLUT4 mRNA in skeletal muscles. J Biol Chem 278: 31385–31390. [DOI] [PubMed] [Google Scholar]
- 18. Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, et al. (2007) Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem 282: 30014–30021. [DOI] [PubMed] [Google Scholar]
- 19. Miura S, Tomitsuka E, Kamei Y, Yamazaki T, Kai Y, et al. (2006) Overexpression of peroxisome proliferator-activated receptor gamma co-activator-1alpha leads to muscle atrophy with depletion of ATP. Am J Pathol 169: 1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, et al. (2008) HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature 451: 1008–1012. [DOI] [PubMed] [Google Scholar]
- 21. Calvo JA, Daniels TG, Wang X, Paul A, Lin J, et al. (2008) Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol 104: 1304–1312. [DOI] [PubMed] [Google Scholar]
- 22. Shimomura Y, Murakami T, Nakai N, Nagasaki M, Harris RA (2004) Exercise promotes BCAA catabolism: effects of BCAA supplementation on skeletal muscle during exercise. J Nutr 134: 1583S–1587S. [DOI] [PubMed] [Google Scholar]
- 23. Shimomura Y, Obayashi M, Murakami T, Harris RA (2001) Regulation of branched-chain amino acid catabolism: nutritional and hormonal regulation of activity and expression of the branched-chain alpha-keto acid dehydrogenase kinase. Curr Opin Clin Nutr Metab Care 4: 419–423. [DOI] [PubMed] [Google Scholar]
- 24. Shimomura Y, Honda T, Shiraki M, Murakami T, Sato J, et al. (2006) Branched-chain amino acid catabolism in exercise and liver disease. J Nutr 136: 250S–253S. [DOI] [PubMed] [Google Scholar]
- 25. Takahashi M, Kamei Y, Ehara T, Yuan X, Suganami T, et al. (2013) Analysis of DNA methylation change induced by Dnmt3b in mouse hepatocytes. Biochem Biophys Res Commun 434: 873–878. [DOI] [PubMed] [Google Scholar]
- 26. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 27. Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, et al. (2010) ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26: 2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Misawa K, Nosaka T, Morita S, Kaneko A, Nakahata T, et al. (2000) A method to identify cDNAs based on localization of green fluorescent protein fusion products. Proc Natl Acad Sci U S A 97: 3062–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kobayashi R, Shimomura Y, Otsuka M, Popov KM, Harris RA (2000) Experimental hyperthyroidism causes inactivation of the branched-chain alpha-ketoacid dehydrogenase complex in rat liver. Arch Biochem Biophys 375: 55–61. [DOI] [PubMed] [Google Scholar]
- 30. Shimomura Y, Fujii H, Suzuki M, Murakami T, Fujitsuka N, et al. (1995) Branched-chain alpha-keto acid dehydrogenase complex in rat skeletal muscle: regulation of the activity and gene expression by nutrition and physical exercise. J Nutr 125: 1762S–1765S. [DOI] [PubMed] [Google Scholar]
- 31. Jeyaraj D, Scheer FA, Ripperger JA, Haldar SM, Lu Y, et al. (2012) Klf15 orchestrates circadian nitrogen homeostasis. Cell Metab 15: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shimizu N, Yoshikawa N, Ito N, Maruyama T, Suzuki Y, et al. (2011) Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab 13: 170–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes up-regulated in Tg mice by microarray. RNA obtained from WT and Tg mice was pooled and used for microarray analysis as described in Methods. Hybridized signals outside the linear range were excluded, and values were normalized by the 75th percentile calculation. Genes with a calculated value less than 100 in Tg mice were deleted. Genes up-regulated more than 2.5 fold in Tg mice were listed. Pooled array data were confirmed by performing quantitative real-time RT-PCR with representative probes and similar increased was observed in all samples, indicating that microarray data represent the expression change in each group (data not shown).
(PDF)