Abstract
Advances in phage therapy encourage scientific interest in interactions of phages with human and animal organisms. This has created a need for developing tools that facilitate studies of phage circulation and deposition in tissues and cells.
Here we propose a new green fluorescent protein (GFP)-based method for T4 phage molecular imaging in living systems. The method employs decoration of a phage capsid with GFP fused to the N-terminus of Hoc protein by in vivo phage display. Fluorescent phages were positively assessed as regards their applicability for detection inside living mammalian cells (by phagocytosis) and tissues (filtering and retention by lymph nodes and spleen). Molecular imaging provides innovative techniques that have brought substantial progress in life sciences. We propose it as a useful tool for studies of phage biology.
Keywords: T4 phage, molecular imaging, green fluorescence protein, phagocytosis, phage circulation
Introduction
Advances in phage therapy1-3 encourage scientific interest in interactions of phages with human and animal organisms. Phages are postulated to be an important alternative to the insufficient antibacterial drugs arsenal. This has prompted questions on detailed recognition of phage interactions with mammalian cells and tissues. Both investigations of phage biology and applications of bacteriophages in medicine have induced a need for developing tools that facilitate studies of phage circulation and deposition in tissues and cells.
Molecular imaging is potentially a highly useful tool for the investigation of phages in living systems. Molecular imaging comprises a wide spectrum of techniques that have brought substantial progress in life sciences. It utilizes various biological particles engaged in production or conversion of light. Among these, the green fluorescent protein (GFP) is a popular fluorescent marker, which was discovered in studies of jellyfish Aequorea victoria. Due to the ability of autocatalytic formation of the chromophore, GFP and GFP-like proteins have enabled novel approaches in live cell imaging, including whole body imaging and dynamic tracking.4 Thus, furnishing a phage with GFP will initiate creative methods of molecular imaging in bacteriophage studies.
Decorating a phage capsid with foreign peptides or proteins can be done by phage display technique, which has become a standard in biotechnology. Phage display relies on fusing a foreign protein to a selected phage structural protein. Fusion may result from introduction of a foreign gene to the phage genome.5 However, it can also be done by incorporation of fusion proteins expressed in bacteria: in vitro as purified compounds6 or in vivo by propagation of a phage in bacteria that express a desired fusion which is further incorporated into new phage capsids.7-9 The second method might be favorable in biological studies, since it does not require generation of genetically modified organisms (GMOs) by introducing foreign elements to phage genomes. A large bank of T4 mutants have already been constructed and exploited to facilitate mutating lytic phages; among that mutants there are ones without decorating (non-essential) capsid proteins: gpHoc or gpSoc. Deletions of that proteins enable incorporation of recombinant proteins which are fusions of gpHoc or gpSoc with foreign elements, e.g., active oligopeptides, proteins, etc.9-14
T4 phage has been shown to be a good display platform, since it has been modified successfully with extraneous proteins several times. Active anti-lysozyme IgG, domains of the HIV1-CD4 receptor, multicomponent anthrax toxin, and a peptide from Neisseria meningitidis (PorA) were fused to decorative capsid proteins Soc or Hoc and as a result displayed on the T4 capsid surface. These foreign elements retained their activity and/or immunogenicity required for anticipated biological applications.9-14
Bacteriophages decorated with GFP by modification of the phage genome have been previously proposed for detection of E. coli in the environment, i.e., in sewage water as well for the assessment of E. coli viability in the aquatic environment.15,16 However, they have never been applied in studies of phage circulation or pharmacokinetics in mammals. Although many other biological objects have been successfully visualized in cells and tissues by GFP-labeling (e.g., proteins, bacteria, cancer cells), the utility of this method for phage detection in mammals has never been determined. Here we propose a new GFP-based method for T4 phage molecular imaging in living systems. The method employs decoration of a phage capsid with GFP fused to the N-terminus of Hoc protein by in vivo phage display. Fluorescent phages were assessed as regards their applicability for detection inside living mammalian cells (by phagocytosis) and tissues (filtering and retention by spleen and lymph nodes).
Results and Discussion
Previous studies of phage display on T4-like phages have shown that effective presentation of large foreign proteins can be done successfully by their fusion to the N-terminal part of decorative protein Hoc.8,10 Thus, to ensure good exposition of GFP on the capsid, N-terminal fusion of GFP to Hoc was exploited in this study. In order to obtain functional E. coli clones expressing GFP-Hoc fusion, a new expression vector based on pDEST17 containing the gfp-hoc sequence was created. Expression was verified in E. coli B834 strain; production of expected fusion was confirmed by SDS-page of the complete protein profile of induced expression bacteria (Fig. 1). Fluorescence of the produced GFP-Hoc fusion was further assessed by fluorescence measurement of induced expression bacteria; fluorescence of GFP-Hoc fusion expressing bacteria was significantly higher in comparison to the control (P = 0.0253) (Fig. 2).
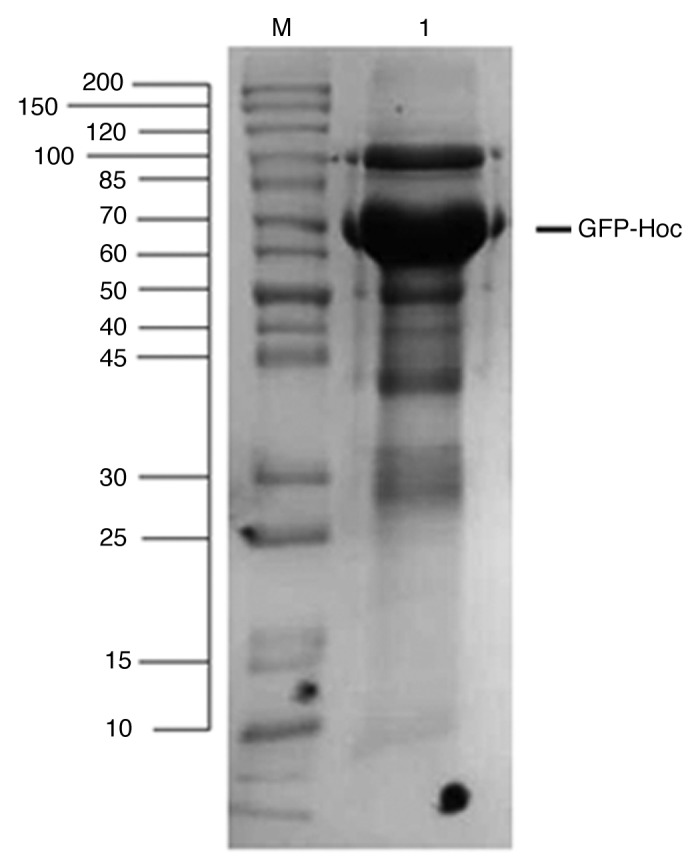
Figure 1. Expression of recombinant GFP-Hoc fusion in E. coli. Expression plasmids containing GFP-hoc fusion were tested for their effectiveness in production of GFP-Hoc proteins as showed by SDS-PAGE. M, marker; 1, induced GFP-Hoc expressing bacteria.
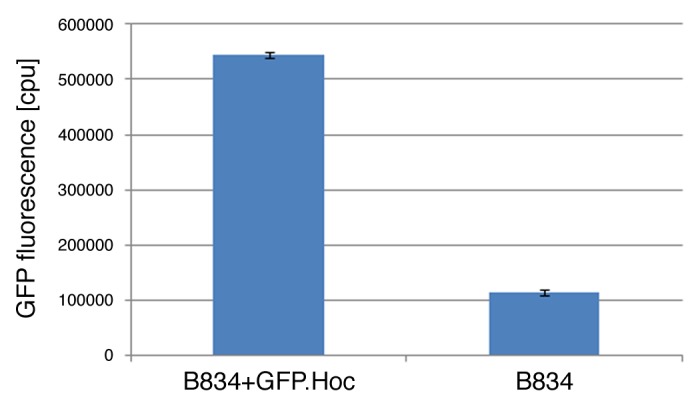
Figure 2. Fluorescence of GFP-Hoc fusion in E. coli. Effective fluorescence of the GFP-Hoc fusion was assessed by comparison of fluorescence in E. coli induced for GFP-Hoc production and control E. coli (expressing protein Hoc without GFP).
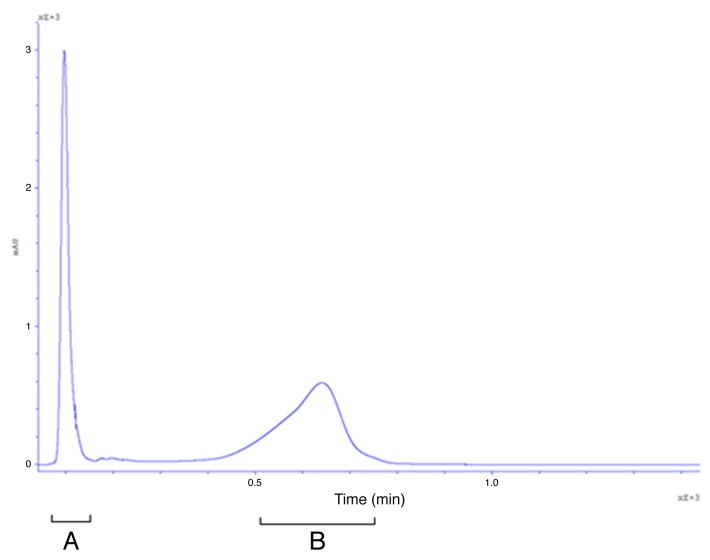
E. coli B834 strain effectively expressing fluorescent GFP-Hoc fusion was used for propagation of HAP1 phage (T4 phage without protein Hoc, display platform).17 The phage display was thus completed in vivo, in bacterial cells infected by the phage after induction for recombinant protein expression. Then, phages were separated from bacterial debris, also from GFP-Hoc fusions not incorporated into the phage capsid by size exclusion chromatography (Fig. 3), and dialyzed against PBS. The final phage titer of purified GFP-labeled phages was 1 × 1011 pfu per ml. Fluorescence of this preparation was significantly higher (P = 0.0495) than that of the control, which was an identically purified but non-modified phage (Fig. 4).
Figure 3. Separation of modified bacteriophages by fast protein liquid chromatography. (A) Fraction containing bacteriophages. (B) Fraction containing non-incorporated GFP-Hoc fusions and other proteins.
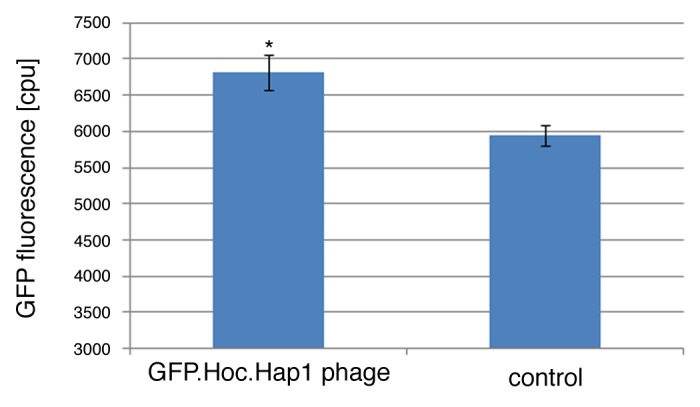
Figure 4. Comparison of fluorescence in GFP-modified and control phage. Effective fluorescence of the purified GFP.Hoc.HAP1 phages was compared with the fluorescence of HAP1 phage (without GFP, purified identically by FPLC).
According to previous reports, macrophages are not crucial for the clearance of phages from blood, however they have been clearly shown as capable of phage phagocytosis.18,19 In order to determine how GFP-labeled bacteriophages can be visualized in mammalian cells, fluorescent bacteriophages (GFP.Hoc.HAP1 phages) were incubated with murine macrophages (J774A.1). After the incubation, labeled phages were visualized inside the cells by fluorescent microscopy. Since 10 min of incubation did not result in visible uptake of the phages, while 20 and 40 min revealed an increasing fluorescent signal in the cells (Fig. 5), the amount of uptake was dependant on incubation time. This is in line with studies of the uptake of phages by rabbit macrophages performed by Aronow et al.20 who used electron microscopy. These authors showed an increasing uptake of a phage by macrophages from approximately 7 min to 30 min. Intracellular destruction of ingested phages occurred some 2 h after their uptake.
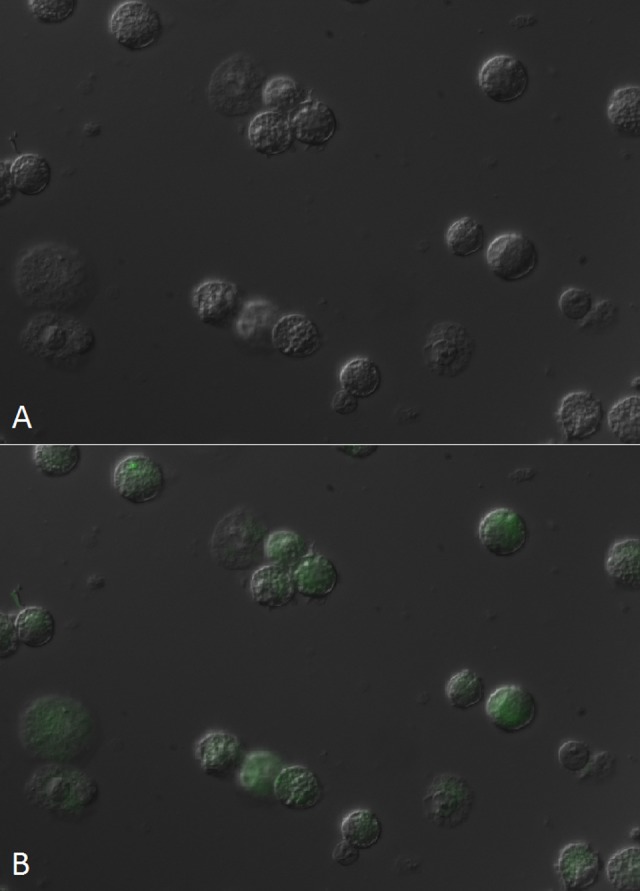
Figure 5. Visualization of GFP-modified bacteriophages in phagocytic cells J774A.1 (murine macrophages). Fluorescence of murine macrophage cell line was visualized in fluorescent microscopy. (A) control, phagocytes before incubation with GFP.Hoc.HAP1 phage, (B) phagocytes after 40 min incubation with GFP.Hoc.HAP1 phage.
In order to determine if GFP-labeled bacteriophages can also be visualized in murine tissues, fluorescent phages were injected i.v. into the lateral tail vein. After 1.5 h, fluorescence of particular organs was detected in imaging hood, revealing a visible signal of GFP.Hoc.HAP1 phages in the spleen (Fig. 6B) and in the lymph node (Fig. 6D). In control mice injected with non-modified phages, only background signal from the tissues was visible (Fig. 6A and C), however much lower than that of the GFP labeled phage.
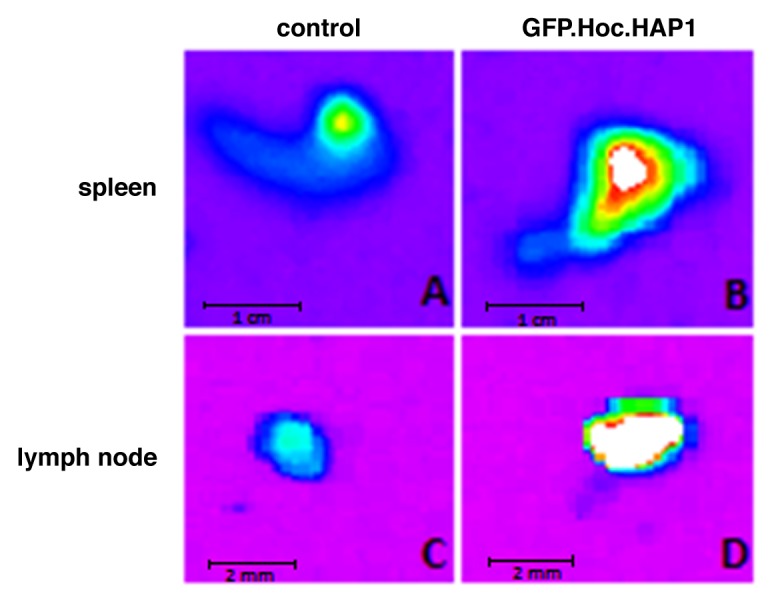
Figure 6. Visualization of GFP-modified bacteriophages in murine tissues. GFP.Hoc.HAP1 phages and non-modified HAP1 phages (control) were injected into murine lateral tail vein (1011 pfu/mouse). Fluorescence of spleen and lymph nodes was visualized in an imaging hood. (A) spleen of mice injected with non-modified HAP1 phage, (B) spleen of mice injected with GFP.Hoc.HAP1 phage, (C) lymph node of mice injected with non-modified HAP1 phage, (D) lymph node of mice injected with GFP.Hoc.HAP1 phage.
These results show that molecular imaging can be utilized for investigation of bacteriophages in living systems. Visualization can be done both in living mammalian cells and in tissues/organs, including quantitation of the results. This offers a good method for studies of time-dependent effects (like presented phagocytosis of bacteriophages) or multiple comparison of phage circulation aspects e.g., in tissues. T4-like phages can be effectively furnished with the popular biofluorescent marker GFP by phage display technique, engaging simple fusion of GFP to one of the decorative capsid proteins. Since this technique does not employ complex molecular or chemical modifications of the phage, we propose it as a useful tool for further studies of phage biology.
In previous studies of Mullaney et al.,21 GFP was targeted into T4 heads as a probe of the internal environment. That approach offered an idea for the method of phage visualization (in vitro). Internal localization of a fluorescent marker makes Hoc protein available for other fusions, including fusions with targeting molecules or active compounds, which demonstrates a potential for developing visualization methods in the field of bacteriophage biology and applications.
Visualization of bacteriophages in living systems can be a potent tool for progress in bacteriophage therapy, in the time of the insufficient antibacterial drugs arsenal. Easy observation and quantification of phage deposition in particular tissues and organs may induce progress in determination of phage dosage, timing regiments and other aspects of phage pharmacokinetics, which are critical for efficacy and safety of phage therapy. Thus, this novel technique may contribute to the development of new alternatives in antibacterial treatment.
Materials and Methods
Construction of gfp-hoc fusion in the expression vector
For gene cloning, a combination of Gateway recombination cloning technology (Invitrogen, Life Technologies Corporation) and a standard restriction/ligation approach was applied. Three steps allowed construction of expression plasmids: i) construction of an entry clone in a non-expression vector, pDONR™221 (a pUC origin and universal M13 sequencing sites, kanamycin resistance) (Invitrogen, Life Technologies Corporation, http://tools.invitrogen.com/content/sfs/vectors/pdonr221_pdonrzeo_map.pdf) by introduction of the hoc gene and restriction sites at the 5′-terminus of the gene, then ii) introduction of the gfp gene into an entry clone using of restriction sites, and iii) transfer of the coding fragment (gfp-hoc fusion) to a destination expression vector, pDEST17 (Invitrogen, Life Technologies Corporation, http://tools.invitrogen.com/content/sfs/vectors/pdest15_map.pdf) for production of the recombinant proteins. Recombinant engineering (i and iii) was done according to the Gateway technology manual. Both the hoc gene and the gfp gene were cloned into the vector as PCR products. Recombination sites (hoc) or restriction sites (gfp) that allowed incorporation of the products into the vector were introduced with PCR primers (hoc forward primer: GGCAAAGTTT GTACAAAAAA GCAGGCTCCC GGGAAAGAA TTCATGACTT TTACAGTTGA TATAACTC, hoc reverse primer: GGGGACCACT TTGTACAAGA AAGCTGGGTC CTATGGATAG GTATAGATGA TACC gfp forward primer: ACCCGGGAAA ACCTGTATTT TCAGGGCAGC AGCAGCATGA GTAAAGGAGA AGAACTTTTC, gfp reverse primer: AAAGAATTCG CTTGGGGCGC TTGGGGCTTT GTATAGTTCA TCCATGCCAT GTGTAATCC). The high solubility motif APSAPS was introduced between GFP and Hoc by a relevant sequence in the gfp reverse primer (marked in bold). Polymerase chain reaction (PCR) was performed on a template of T4 total DNA (hoc) or pcDNA-DEST53 (Invitrogen, Life Technologies Corporation) (gfp). PCR products of the hoc gene were introduced into the pDONR221 using BP Clonase™ II Enzyme Mix (Invitrogen, Life Technologies Corporation) according to the manufacturer’s instructions. Constructions were verified by automated Sanger sequencing with standard M13 primers using a 3730 DNA Analyzer, Applied Biosystems, Hitachi, DNA Sequencing KitBig Dye™ Terminator Cycle Sequencing version 1.1 (Oligo, Institute of Biochemistry and Biophysics, Warsaw, Poland). The apropriate clones were used for restriction/ligation with gfp PCR products; restriction enzymes: EcoRI and SmaI (FastDigest, Fermentas) and ligase (LigaFast, Promega) were used. Constructions were verified by automated Sanger sequencing with standard M13 primers using a 3730 DNA Analyzer, Applied Biosystems, Hitachi, DNA Sequencing KitBig Dye™ Terminator Cycle Sequencing version 1.1 (Oligo, Institute of Biochemistry and Biophysics, Warsaw, Poland). Proper fusions of the genes (gfp-hoc) were transferred by recombination to the expression vector pDEST17 (Invitrogen, Life Technologies Corporation) in the LR reaction, according to the manufacturer’s instructions.
Expression clones were tested in E. coli B834(DE3) F− ompT hsdSB(rB− mB−) gal dcm met (DE3) (EMD, Europe); the strains were checked for their ability to express the recombinant proteins gfp-hoc as follows: they were cultured with intensive aeration in LB high salts (10 g/l of NaCl) culture medium (Sigma-Aldrich, Europe or AppliChem, Europe) with appropriate selection antibiotics (ampicillin) at 37 °C and induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) (0.2 mM) in the exponential growth phase as determined by OD600 measurements (usually at OD600 = 0.8). Further expression was conducted overnight at 25 °C. Bacteria were harvested by centrifugation (6000 rpm, 5 min) and analyzed by SDS-page and fluorescence measurement in a multilabel plate reader (EnSpire, Perkin Elmer).
Phage display
As a display platform, HAP1 phage from the IIET Microorganisms Collection was used; HAP1 is a mutant of T4 phage (American Type Culture Collection, USA) with a nonsense mutation in the hoc gene with no functional Hoc.22
Bacterial cells were transformed with an expression vector containing gfp-hoc fusion. Transformed bacteria were grown at 37 °C in Luria broth (LB) with ampicillin as a selection antibiotic until optical density (OD600) 0.7 was reached. Next they were transferred to fresh LB medium containing 0.2 mM IPTG; IPTG was an expression inductor. After induction of protein expression in bacteria, 106 pfu HAP1 (in 1 ml volume) was added. This infected culture was incubated at 37 °C for 8 h. Next the culture was incubated for 3 d at 4 °C. Lysates were filtered by 0.22 µm sterile syringe filters. Phage titer was determined by Routine Test Dilution (RTD).23 Phage modified with GFP by phage display was denominated: “GFP.Hoc.HAP1 phage”.
Purification of the GFP-modified phages (Fast protein liquid chromatography, FPLC)
GFP.Hoc.HAP1 phages were purified by size-exclusion chromatography as described by Boratyński et al.24; briefly, the suspension was chromatographed on a sepharose 4B column (2.5 × 95 cm, eluent 0.063 M phosphate buffer, pH 7.2, flow 0.3 ml min−1). Bacteriophages were eluted in the highest molecular weight fraction, as confirmed by fraction titration. Phages were filtered by 0.22 µm sterile syringe filters. Phage titer was determined by RTD.23
Additionally, fluorescence of subsequent fractions was assessed in an imaging hood: In-Vivo MS FX PRO (Carestream Molecular Imaging). The GFP-displaying phage (GFP.Hoc.HAP1) fraction, other fractions resulting from FPLC, and a control phage (HAP1) were dropped into the detection area and analyzed (examined volume: 10 µl, excitation: 470 nm, detection: 535 nm). intensity of fluorescence was normalized as numerical data by Carestream Imaging software.
Phage imaging in living cells and tissues
J774A.1 cells (cell line, murine macrophages, ATCC collection) were incubated at 37 °C with GFP.Hoc.HAP1 phage for 10, 20, or 40 min (106 pfu per 106 cells), washed with PBS three times, and analyzed. Detection of the signal was performed in a fluorescent Fully Automated Inverted Research Microscope for Biomedical Research: Leica DMI6000B.
Female C57/Bl6 (8 wk) mice were purchased from the Center of Experimental Medicine, Medical University of Bialystok, and kept in specific pathogen free (SPF) conditions in the Animal Breeding Centre of the Institute of Immunology and Experimental Therapy (IIET). All experiments were approved by the 1st Local Committee for Experiments with the Use of Laboratory Animals, Wroclaw, Poland (no. 64/2009).
Mice were injected intravenously (i.v.) with GFP.Hoc.HAP1 phage 2 × 1011 pfu per mouse, control mice were injected i.v. with the phage without GFP protein (HAP1) 2 × 1011 pfu per mouse. 90 min later mice were sacrificed and lymph nodes were excised. Imaging was performed in In-Vivo MS FX PRO (Carestream Molecular Imaging).
Statistical methods
ANOVA, Mann-Whitney U test, and the Statistica 6.0 software packages were applied (www.statsoft.pl).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by EU Structural Funds (Operational Programme Innovative Economy “Optimization of characteristics and preparation of bacteriophage preparations for therapeutic purposes”). PM is a recipient of the “Start” grant Foundation for Polish Science.
Glossary
Abbreviations:
- FPLC
fast protein liquid chromatography
- GFP
green fluorescent protein
- i.v.
intravenously
- IIET
Institute of Immunology and Experimental Therapy
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LB
luria broth
- M
marker
- PCR
polymerase chain reaction
- RTD
routine test dilution
- SPF
specific pathogen free
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/28364
References
- 1.Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage. 2011;1:66–85. doi: 10.4161/bact.1.2.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Górski A, Międzybrodzki R, Borysowski J, Dąbrowska K, Wierzbicki P, Ohams M, Korczak-Kowalska G, Olszowska-Zaremba N, Łusiak-Szelachowska M, Kłak M, et al. Phage as a modulator of immune responses: practical implications for phage therapy. Adv Virus Res. 2012;83:41–71. doi: 10.1016/B978-0-12-394438-2.00002-5. [DOI] [PubMed] [Google Scholar]
- 3.Kutter E, De Vos D, Gvasalia G, Alavidze Z, Gogokhia L, Kuhl S, Abedon ST. Phage therapy in clinical practice: treatment of human infections. Curr Pharm Biotechnol. 2010;11:69–86. doi: 10.2174/138920110790725401. [DOI] [PubMed] [Google Scholar]
- 4.Wiedenmann J, Oswald F, Nienhaus GU. Fluorescent proteins for live cell imaging: opportunities, limitations, and challenges. IUBMB Life. 2009;61:1029–42. doi: 10.1002/iub.256. [DOI] [PubMed] [Google Scholar]
- 5.Beghetto E, Gargano N. Lambda-display: a powerful tool for antigen discovery. Molecules. 2011;16:3089–105. doi: 10.3390/molecules16043089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Shivachandra SB, Zhang Z, Rao VB. Assembly of the small outer capsid protein, Soc, on bacteriophage T4: a novel system for high density display of multiple large anthrax toxins and foreign proteins on phage capsid. J Mol Biol. 2007;370:1006–19. doi: 10.1016/j.jmb.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oślizło A, Miernikiewicz P, Piotrowicz A, Owczarek B, Kopciuch A, Figura G, Dąbrowska K. Purification of phage display-modified bacteriophage T4 by affinity chromatography. BMC Biotechnol. 2011;11:59. doi: 10.1186/1472-6750-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceglarek I, Piotrowicz A, Lecion D, Miernikiewicz P, Owczarek B, Hodyra K, Harhala M, Górski A, Dąbrowska K. A novel approach for separating bacteriophages from other bacteriophages using affinity chromatography and phage display. Sci Rep. 2013;3:3220. doi: 10.1038/srep03220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren Z, Black LW. Phage T4 SOC and HOC display of biologically active, full-length proteins on the viral capsid. Gene. 1998;215:439–44. doi: 10.1016/S0378-1119(98)00298-4. [DOI] [PubMed] [Google Scholar]
- 10.Ren ZJ, Lewis GK, Wingfield PT, Locke EG, Steven AC, Black LW. Phage display of intact domains at high copy number: a system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 1996;5:1833–43. doi: 10.1002/pro.5560050909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang J, Abu-Shilbayeh L, Rao VB. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect Immun. 1997;65:4770–7. doi: 10.1128/iai.65.11.4770-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathaliyawala T, Rao M, Maclean DM, Birx DL, Alving CR, Rao VB. Assembly of HIV antigens on bacteriophage T4: a novel in vitro approach to construct multicomponent HIV vaccines. J Virol. 2006;80:7688–98. doi: 10.1128/JVI.00235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shivachandra SB, Rao M, Janosi L, Sathaliyawala T, Matyas GR, Alving CR, Leppla SH, Rao VB. In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc-capsid interactions: a strategy for efficient display of large full-length proteins. Virology. 2006;345:190–8. doi: 10.1016/j.virol.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 14.Shivachandra SB, Li Q, Peachman KK, Matyas GR, Leppla SH, Alving CR, Rao M, Rao VB. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine. 2007;25:1225–35. doi: 10.1016/j.vaccine.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Namura M, Hijikata T, Miyanaga K, Tanji Y. Detection of Escherichia coli with fluorescent labeled phages that have a broad host range to E. coli in sewage water. Biotechnol Prog. 2008;24:481–6. doi: 10.1021/bp070326c. [DOI] [PubMed] [Google Scholar]
- 16.Awais R, Fukudomi H, Miyanaga K, Unno H, Tanji Y. A recombinant bacteriophage-based assay for the discriminative detection of culturable and viable but nonculturable Escherichia coli O157:H7. Biotechnol Prog. 2006;22:853–9. doi: 10.1021/bp060020q. [DOI] [PubMed] [Google Scholar]
- 17.Dąbrowska K, Zembala M, Boratynski J, Switala-Jelen K, Wietrzyk J, Opolski A, Szczaurska K, Kujawa M, Godlewska J, Gorski A. Hoc protein regulates the biological effects of T4 phage in mammals. Arch Microbiol. 2007;187:489–98. doi: 10.1007/s00203-007-0216-y. [DOI] [PubMed] [Google Scholar]
- 18.Górski A, Międzybrodzki R, Borysowski J, Dąbrowska K, Wierzbicki P, Ohams M, Korczak-Kowalska G, Olszowska-Zaremba N, Łusiak-Szelachowska M, Kłak M, et al. Phage as a modulator of immune responses: practical implications for phage therapy. Adv Virus Res. 2012;83:41–71. doi: 10.1016/B978-0-12-394438-2.00002-5. [DOI] [PubMed] [Google Scholar]
- 19.Uchiyama J, Maeda Y, Takemura I, Chess-Williams R, Wakiguchi H, Matsuzaki S. Blood kinetics of four intraperitoneally administered therapeutic candidate bacteriophages in healthy and neutropenic mice. Microbiol Immunol. 2009;53:301–4. doi: 10.1111/j.1348-0421.2009.00125.x. [DOI] [PubMed] [Google Scholar]
- 20.Aronow R, Danon D, Shahar A, Aronson M. Electron microscopy of in vitro endocytosis of T2 phage by cells from rabbit peritoneal exudate. J Exp Med. 1964;120:943–54. doi: 10.1084/jem.120.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullaney JM, Thompson RB, Gryczynski Z, Black LW. Green fluorescent protein as a probe of rotational mobility within bacteriophage T4. J Virol Methods. 2000;88:35–40. doi: 10.1016/S0166-0934(00)00166-X. [DOI] [PubMed] [Google Scholar]
- 22.Dąbrowska K, Zembala M, Boratynski J, Switala-Jelen K, Wietrzyk J, Opolski A, Szczaurska K, Kujawa M, Godlewska J, Gorski A. Hoc protein regulates the biological effects of T4 phage in mammals. Arch Microbiol. 2007;187:489–98. doi: 10.1007/s00203-007-0216-y. [DOI] [PubMed] [Google Scholar]
- 23.Collier LH. Chlamydia. Topley and Wilson’s Principles of Bacteriology, Virology and Immunity. 8th edition. Published Edward Arnold 1990; 629-646 [Google Scholar]
- 24.Boratyński J, Syper D, Weber-Dabrowska B, Łusiak-Szelachowska M, Poźniak G, Górski A. Preparation of endotoxin-free bacteriophages. Cell Mol Biol Lett. 2004;9:253–9. [PubMed] [Google Scholar]