Abstract
Aspergillus fumigatus is an opportunistic pathogen that is responsible for a life-threatening fungal infection known as invasive aspergillosis. Current therapies for the treatment of this disease continue to be associated with a poor outcome, so there is a need for more information about aspects of the fungus–host interaction that could offer novel targets for drug intervention. One attractive possibility is the unfolded protein response (UPR), an intracellular signaling network that helps the fungus meet the demand for secretion in the host environment. The major function of the UPR is to mitigate ER stress by maintaining an equilibrium between the load of client proteins entering the endoplasmic reticulum (ER) and the protein folding capacity of the organelle. However, recent findings suggest that A. fumigatus, as well as several other pathogenic fungi, also rely upon this pathway for virulence. In this review, we provide an update on the A. fumigatus UPR, discuss emerging evidence that the UPR is situated at the nexus of a number of physiological functions that are vital for the virulence of this fungus, and suggest exciting possibilities for future therapeutic targeting of this pathway for the treatment of aspergillosis.
Keywords: Aspergillus fumigatus, ER stress, ERAD, UPR, aspergillosis, unfolded protein response, virulence
Aspergillus and Aspergillosis
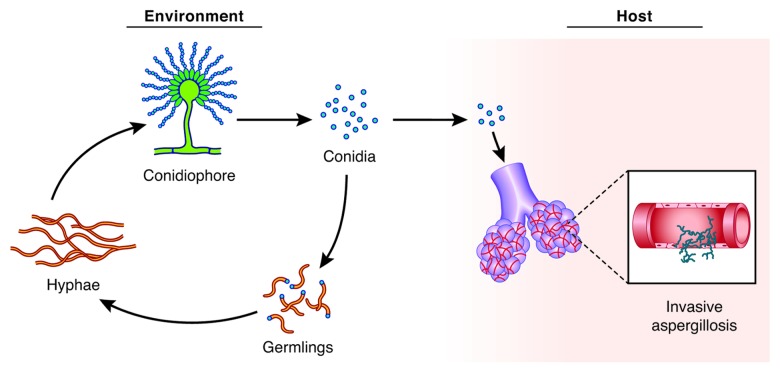
Aspergillosis refers to a spectrum of diseases caused by filamentous fungi within the genus Aspergillus, a group of environmental molds that are found ubiquitously in nature, particularly in decaying organic debris.1 Over the past few decades, Aspergillus species have emerged as important pathogens of both animals and humans, and are the most common molds associated with invasive fungal disease.2,3 Although hundreds of species have been identified within this genus,4 A. fumigatus accounts for the vast majority of Aspergillus infections.2,3 The organism propagates itself by the airborne distribution of asexual spores (conidia), which are unavoidably inhaled by most individuals on a daily basis. The outcome of this initial fungus-host interaction depends largely on the immunological status of the host; healthy individuals readily clear the inhaled conidia, but immunocompromised patients can have impaired conidial clearance mechanisms that permit germination in the lung.1 When the conidia germinate into hyphae they secrete abundant hydrolytic enzymes,5 which erode the pulmonary epithelial barrier and allow the fungus to penetrate the vasculature and disseminate to other tissues (Fig. 1).
Figure 1. Lifecycle and pathogenesis of invasive aspergillosis. The asexual lifecycle of A. fumigatus in the environment is shown on the left. Conidia germinate into hyphae when they encounter moist conditions and a nutrient source. In response to environmental cues, the hyphae initiate a developmental program to generate a spore-forming structure, the conidiophore, upon which chains of conidia are added and released into the atmosphere. Upon inhalation into the lung, the conidia may germinate into hyphae if the immune system is compromised. The hyphae secrete large quantities of hydrolytic enzymes, which allows them to enter the vasculature and cause disseminated disease (invasive aspergillosis). Current evidence indicates that the host environment is a source of ER stress to this fungus, requiring UPR activation.
Despite the best treatment with current antifungal drugs, invasive aspergillosis continues to have a very poor prognosis, resulting in the highest per person hospitalization costs among the systemic mycoses.6,7 Individuals who are at greatest risk for this infection include those with hematologic malignancies, solid organ transplants, bone marrow transplants, and advanced AIDS. Indeed, invasive aspergillosis has become one of the most feared opportunistic infections in transplant units worldwide, with high rates of morbidity and mortality that generate over $600 million in annual hospitalization costs in the United States.2,7 In addition, the poor response to current antifungal therapy is further compounded by issues of drug toxicity, as well as emerging antifungal resistance.8 The incidence of invasive aspergillosis continues to increase in proportion to the rise in the immunosuppressed population, but this has not been matched by an increase in the number of effective antifungal agents to treat the infection, emphasizing the need for the identification of new antifungal targets for therapeutic intervention. In this review, we summarize evidence that the unfolded protein response (UPR) may be a point of vulnerability in A. fumigatus that could be exploited for the design of novel antifungal therapies.
Filamentous Fungi are Secretion Factories
Filamentous fungi such as A. fumigatus possess complex membrane trafficking systems that are specialized for high-capacity secretion.9-11 In fact, their ability to serve as “secretion factories” has been widely exploited by the biotechnology industry for the production of proteins of economic importance.12 Secretion is also important to A. fumigatus in the host. For example, the transition from the environment to the host is associated with the upregulation of mRNAs encoding secreted hydrolytic enzymes, as well as membrane transporters that are dedicated to nutrient uptake.13 A. fumigatus proteases can be readily detected in host tissue during infection, and there is strong genetic evidence that the fungus relies on the enzymatic breakdown of proteins and phospholipids to support growth in the host.14-18 The secretory pathway provides a mechanism to deliver these proteins into, and across, the plasma membrane, allowing the organism to degrade host tissues into component molecules that are suitable for transport into the cytoplasm. All proteins that enter the secretory pathway are initially processed and folded in the endoplasmic reticulum (ER). However, when the level of nascent proteins entering the ER lumen exceeds the organelle’s capacity to process that load, unfolded proteins can accumulate. For this reason, dynamic changes in the demand for secretory activity during infection places the fungal ER at risk for the accumulation of unfolded proteins. ER folding capacity may also be perturbed by adverse host conditions such as mammalian body temperature, oxidative stress, hypoxia, and nutrient limitation.19 Since unfolded proteins are toxic to cell physiology, their presence triggers a series of integrated adaptive responses, collectively known as the UPR.
The Fungal UPR: Lessons from Yeast
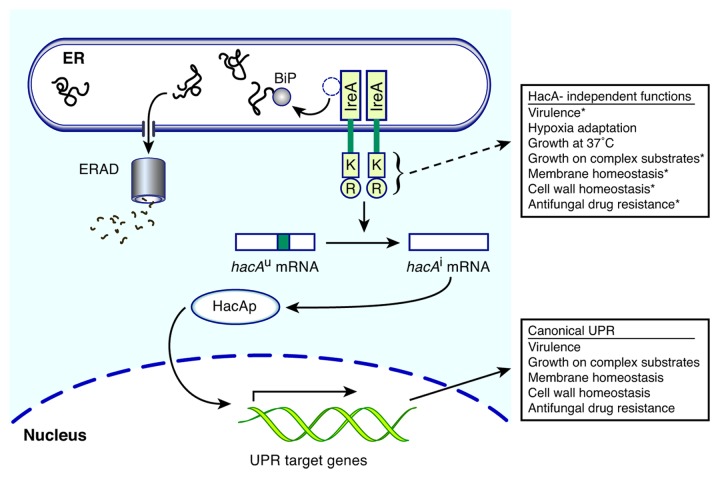
The UPR is a stress response pathway that is charged with maintaining the fidelity of the protein folding activities of the ER.20 In metazoans, a trio of coordinated signaling pathways comprise the UPR, each of which is controlled by a stress sensor embedded in the ER membrane: IRE1, ATF6, and PERK.21 However, only the Ire1 sensor has been reported in fungi.22 The archetypal pathway of fungal UPR signaling was elucidated in the model yeast Saccharomyces cerevisiae (Fig. 2). Ire1 is a single-pass ER membrane protein with a lumenal stress sensing domain and a cytosolic effector domain that contains a protein kinase domain linked to an endoribonuclease (RNase).21 When unfolded protein levels are low, Ire1 is bound to the ER-resident Hsp70 chaperone BiP, which helps to maintain Ire1 in an inactive state. The presence of high levels of unfolded proteins triggers dissociation of BiP from Ire1, allowing for Ire1 oligomerization and trans-autophosphorylation. These events alter the conformation of Ire1, which activates the RNase domain. The substrate of the RNase activity is a cytoplasmic mRNA known as HAC1 in S. cerevisiae (hacA in A. fumigatus). The removal of an unconventional intron from the HAC1u mRNA (u for uninduced), creates a frame-shift in the induced form of the mRNA (HAC1i) that specifies the translation of the bZIP transcription factor, Hac1p. As the master transcriptional regulator of the UPR, Hac1p restores ER homeostasis by moving to the nucleus and orchestrating transcriptional changes to rebalance protein folding homeostasis in the ER. Despite UPR activation, some proteins may ultimately fail to achieve their proper conformation. Since aberrant polypeptides are deleterious to the cell, they are disposed of through ER-associated degradation (ERAD), a pathway that works in conjunction with the UPR to relieve the burden of unfolded proteins on the ER (Fig. 2). ERAD recognizes unfolded proteins and escorts them through a channel in the ER membrane into the cytosol, where they are targeted for proteasomal degradation.23
Figure 2. The A. fumigatus UPR is a regulatory hub for virulence. When unfolded proteins accumulate in the ER lumen, the chaperone BiP dissociates from the ER stress sensor IreA to assist with folding, which allows oligomerization of IreA and activation of its cytosolic kinase (K) and RNase (R) domains. In the canonical UPR pathway, the IreA RNase splices the cytoplasmic mRNA hacA, which alters the reading frame to allow translation of the bZIP transcription factor HacA. HacA then migrates to the nucleus and orchestrates transcriptional changes to strengthen ER folding capacity. Proteins that are terminally misfolded are retrotranslocated to the cytoplasm through a protein complex in the ER membrane and targeted for degradation by the proteasome (ERAD). These adaptive responses directly support ER homeostasis, but also indirectly support biological processes that promote the virulence and antifungal drug resistance of A. fumigatus. Current evidence suggests that IreA also directs a HacA-independent pathway of unknown mechanism that cooperates with the canonical UPR to support virulence and drug susceptibility (dashed arrow). *These traits are more defective in the ΔireA mutant than the ΔhacA mutant, suggesting that they are supported by HacA-independent functions of IreA as well as the canonical UPR.
The A. fumigatus UPR follows this basic paradigm, with a few notable differences. The proximal ER stress sensor is IreA, which is comprised of a similar domain organization to that of yeast Ire1.24 However, the unconventional intron in the downstream target mRNA, hacAu, is only 20 nucleotides in length,25 which contrasts the 252 nucleotides in the corresponding intron of S. cerevisiae HAC1 mRNA. The long intron in S. cerevisiae HAC1 has been shown to block translation of the unspliced mRNA by forming a stem-loop structure that interacts with the 5′UTR, thereby stalling ribosomes.26 The removal of this intron by Ire1-mediated splicing releases the translation block, allowing the spliced mRNA to be translated. The hacAu intron in A. fumigatus is similar in length to what has been described in mammals, Caenorhabditis elegans, Candida albicans, and other filamentous fungi, but it is considered unlikely to be involved in a translational block of the 5′UTR because of its small size.27-30 In fact, current evidence indicates that the unspliced mRNA in mammals is actually translated, and a hydrophobic segment in the nascent protein helps to target the mRNA to the ER membrane, thereby facilitating splicing.31 Since the hacAu mRNA of A. fumigatus associates with polysomes (K.K., unpublished), it will be interesting to determine whether it is translated into a protein product that also helps to tether the mRNA to the ER membrane.
Another distinction between the yeast and A. fumigatus UPR is the size of the reading frame shift caused by removal of the unconventional intron in the HAC1/hacA mRNAs. In A. fumigatus, the hacAu mRNA is predicted to encode a protein of 433 amino acids. Removal of the unconventional intron alters the reading frame such that 220 amino acids at the c-terminus of the HacAu protein are replaced with a unique c-terminal domain comprised of 129 amino acids. In yeast, only 10 amino acids at the c-terminus of the unspliced open reading frame are replaced with 18 unique amino acids following splicing. Although the functional significance of these variations is currently unknown, it is likely to reflect differences in the way each species has optimized the UPR to suit specialized needs.
Components of the UPR have now been studied in a number of fungal pathogens, including Aspergillus niger,30,32-34 Aspergillus nidulans,29 Cryptococcus neoformans,35-37 Alternaria brassicicola,28 Candida albicans,28 and Candida glabrata.38 The basic aspects of the response are conserved; an Ire1-like sensor in the ER membrane signals to the nucleus to modify the transcriptome and strengthen ER homeostasis. However, there are intriguing differences between species, most notably in C. glabrata where the ER stress response involves Ire1 functions that appear to be independent of HAC1 mRNA splicing.38 It will be interesting to determine the extent to which other eukaryotic pathogens have customized ER stress response pathways to suit their individual requirements in the host environment.
The UPR Transcriptome in A. fumigatus
The major functional output of the yeast UPR is the transcriptional upregulation of mRNAs that support ER function.39 The extent to which A. fumigatus follows this model was ascertained by comparing the gene expression profile of WT A. fumigatus to that of a ΔhacA and a ΔireA mutant in the presence and absence of ER stress.24 HacA and IreA were shown to collectively influence over 13% of the open reading frames in the A. fumigatus genome, illustrating the remarkable scope of gene regulation mediated by this pathway. However, the UPR transcriptome was strikingly different depending on the level of ER stress: high levels of exogenously applied ER stress triggered signaling through the canonical IreA–HacA pathway, resulting in the activation of a program of gene expression that focuses almost exclusively on supporting the secretory pathway, such as protein folding, ER glycosylation, ER degradation, ER translocation, and vesicular trafficking.24 By contrast, the transcriptional response of the UPR in the absence of any added ER stress was greater in size and more diverse in function, suggesting that the UPR is constantly modifying the output of the pathway in proportion to stress level, even during normal growth. It is interesting to speculate that the challenge of delivering cell wall and membrane components to rapidly growing hyphal tips in filamentous fungi creates fluctuations in ER stress that require dynamic changes in UPR activity to maintain ER homeostasis.
A surprising finding from this analysis of the A. fumigatus UPR was that a large fraction of differentially expressed genes in the absence of added ER stress were dependent upon IreA, but not HacA, for expression. This suggests that IreA controls dual signaling circuits that are both HacA-dependent and HacA-independent in this organism. The mechanism by which IreA affects gene expression independently of HacA is currently unknown, but is an area under active investigation.24 Extensive Hac1-independent functions for Ire1 have not been reported in yeast, suggesting that the increased complexity of the secretory pathway in filamentous fungi relative to that of yeast may have driven the need for expanded functions for the IreA sensor, similar to what has been described for the ortholog in higher eukaryotes.40,41
The UPR: A Regulatory Hub for Virulence Traits
Since A. fumigatus is a potent opportunistic pathogen, understanding the stress response pathways that the fungus exploits during the fungus-host interaction is an important goal. UPR mutants of A. fumigatus are either attenuated for virulence (ΔhacA) or avirulent (ΔireA), indicating a major role for this pathway in the pathogenicity of this fungus.24,25 Surprisingly however, the ΔireA mutant had more severe phenotypic defects in virulence-related traits than the ΔhacA mutant, suggesting that IreA function is not limited to hacA mRNA processing and may link to other stress response pathways in a HacA-independent manner (Fig. 2). Phenotypic analysis of the ΔhacA and ΔireA mutants suggested that the mechanism by which the UPR supports the virulence of A. fumigatus is multifactorial, involving the following adaptive traits.
The UPR supports host-temperature adaptation
A. fumigatus, like most human pathogens, grows well at 37 °C. In fact, it is one of the most thermally adaptable fungal species, capable of growth at a wide range of temperatures.42 The ΔhacA mutant of A. fumigatus grew normally at temperatures up to 37 °C, but was unable to grow at 45 °C.25 By contrast, the ΔireA mutant had a 50% reduction in growth rate at 37 °C and was unable to grow at 42 °C, indicating that IreA has HacA-independent functions that are important for adaptation to mammalian body temperature.25,43 The mechanism for this thermosensitivity is likely to be due to a loss of cell integrity since both mutants exhibited hyphal tip lysis at non-permissive growth temperatures.24,25 It is remarkable that the corresponding mutants in S. cerevisiae do not exhibit temperature-sensitive phenotypes, revealing a fundamental difference between A. fumigatus and yeast with respect to their reliance on the UPR for mechanisms that support growth at elevated temperatures.25,39
The UPR supports membrane and cell wall homeostasis
The plasma membrane and cell wall constitute the major interface between the fungus and host environment, and are currently the major targets of antifungal drugs in clinical use for the treatment of invasive aspergillosis.44 The polysaccharide composition of the cell wall is abnormal in the ΔhacA and ΔireA mutants, which may contribute to the increased fragility of their cell walls at higher temperatures.24,25 In addition, both UPR mutants showed decreased expression of mRNAs encoding enzymes in the ergosterol biosynthetic pathway, which correlated with reduced total ergosterol levels. Ergosterol is the major sterol in fungal membranes and, like mammalian cholesterol, helps to maintain membrane homeostasis by optimizing fluidity.45 A reduction in membrane ergosterol is likely to increase membrane fluidity, which could also contribute to the heightened temperature sensitivity of these mutants.
The antifungal drugs used for the treatment of invasive aspergillosis target membrane ergosterol (polyenes and azoles) or cell wall β-glucan synthesis (echinocandins). Mutants of A. fumigatus that are deficient in UPR function showed increased susceptibility to all 3 of these antifungal drug classes, most likely due to the loss of membrane and cell wall homeostasis created by the absence of a functional UPR.24,25 This effect was particularly striking for caspofungin, where the normally fungistatic activity of this drug against A. fumigatus became fungicidal in the absence of either hacA or ireA.
The UPR supports nutritional versatility
The ΔhacA and ΔireA mutants grew well on reduced carbon and nitrogen substrates, but were growth impaired on polymeric nutrient sources such as explants of mouse lung tissue. The inability to grow well on a complex nutrient source correlated with reduced secretory capacity, suggesting that the attenuated virulence of these strains is partly due to an impaired secretory system that is unable to extract sufficient nutrients from host tissues.24,25 A. fumigatus also faces iron starvation during infection because the host immune system sequesters this essential nutrient as a microbial defense mechanism.46 However, the fungus has a number of iron uptake mechanisms at its disposal, the deletion of which has been shown to abrogate virulence.47 UPR deletion in A. fumigatus was associated with defects in the expression of mRNAs encoding components of siderophore-mediated iron acquisition and reductive iron assimilation. This suggested that UPR mutants would be defective in iron homeostasis, which was confirmed by showing impaired growth under iron-limiting conditions.24 Taken together, the impaired ability of A. fumigatus UPR mutants to grow on either polymeric extracellular substrates or iron-limited media suggests that the UPR is important to the nutritional versatility that is required of this fungus in the host environment.
The UPR supports hypoxia adaptation
In nature, A. fumigatus resides in compost, an environment in which oxygen levels can range from atmospheric to hypoxic.48 In order to thrive in this niche, A. fumigatus has evolved adaptive mechanisms to support growth under a broad range of oxygen concentrations. The ΔireA mutant was growth impaired under hypoxia, but the ΔhacA mutant grew normally, suggesting that IreA has HacA-independent functions that are important when oxygen is limiting.24,48 A. fumigatus also encounters areas of limited oxygen during infection, particularly in deep tissues or microenvironments that are damaged by fungal growth and host inflammation. Since the ability to adapt to hypoxia is a well-established virulence determinant for A. fumigatus,49 the hypoxic growth defect in the ΔireA mutant is likely to contribute to the inability of this strain to establish a productive infection.24 The exact mechanism by which IreA promotes hypoxia adaptation is not yet clear. However, since the ΔireA mutant has reduced levels of mRNAs encoding proteins involved in oxidative phosphorylation,24 it is conceivable that the hypoxia sensitivity of this strain is due, at least in part, to a defect in mitochondrial respiration.
The UPR supports virulence in conjunction with ERAD
The requirement for hacA and ireA during infection implies that the host environment triggers physiological changes in the fungus that lead to the accumulation of unfolded proteins. Misfolded proteins are ultimately disposed of by ER-associated degradation (ERAD), a process that transports abnormal proteins across the ER membrane back into the cytosol, where they are ubiquitinated and degraded by the proteasome.23 The derA gene of A. fumigatus encodes the homolog of S. cerevisiae Der1p (Degraded in the ER), a protein that is part of an ER-membrane complex that targets unfolded ER proteins for ERAD (Fig. 2).23 Deletion of DerA had no effect on A. fumigatus in the presence of ER stress. However, the ΔderA mutant showed increased UPR activity, suggesting that the UPR can compensate for the loss of ERAD function. This was confirmed by disrupting the UPR in the ΔderA mutant; a ΔderA/ΔhacA mutant showed a more severe reduction in growth rate and antifungal drug resistance than either of the single gene deletion mutants. Moreover, the ΔderA/ΔhacA mutant was avirulent, which contrasted the wt virulence of ΔderA and the partial virulence of ΔhacA.50 Similar findings were observed by deleting the hrdA gene, encoding a ubiquitin ligase that is part of the same ERAD complex.51 Together, these findings established that ERAD cooperates with the UPR to support functions of the secretory pathway that are necessary for virulence. However, the UPR has the dominant role with respect to pathogenicity.
Therapeutic Implications
Ire1 has dual enzymatic domains, which makes it a prime target for small molecule intervention.52,53 In fact, strategies to disrupt human Ire1 pharmacologically are already in the development pipeline for the treatment of secretory tumors such as multiple myeloma.54-56 In this case, the selectivity of the drug for the cancer is based on the fact that the target is a secretory cell and therefore relies heavily upon the UPR for survival. The high secretory capacity of A. fumigatus, and the importance of the UPR to virulence,24 suggest that an analogous IreA-targeting approach could be an effective antifungal strategy. Pharmacologic disruption of IreA function would be predicted to inhibit the canonical UPR, as well as the HacA-independent functions of this pathway, which would have the following beneficial effects:
1) Decrease overall secretory efficiency
Loss of IreA function would impair secretory capacity and limit the acquisition of nutrients from host tissues. The impaired release of hydrolytic enzymes would also improve outcome by minimizing damage to host tissue.
2) Induce fungal death
Loss of IreA function would overwhelm ER folding capacity, resulting in toxic levels of unfolded proteins that would be expected to have fungistatic and/or fungicidal effects on the organism.
3) Enhance the activity of current antifungal drugs
IreA contributes to the repair of the fungal membrane and cell wall caused by azole, polyene, and echinocandin antifungal drugs.24 This suggests that combining an IreA inhibitor with one or more current antifungals could increase efficacy, reduce toxicity, and prevent the emergence of resistance better than monotherapy regimens. In this regard, it is worth noting that a recent yeast synthetic lethal screen showed that Ire1 can compensate for surprisingly diverse biochemical dysfunctions.57 In fact, it was so effective that numerous genes that were previously identified as “nonessential” would actually be essential if not for Ire1’s intervention. This raises the possibility that pharmacologic inhibition of IreA could expand the realm of drug susceptibility beyond the standard antifungal drug classes, which has the potential for use against other pathogenic molds, such as Aspergillus terreus, Fusarium spp., Scedosporium spp., and zygomycetes, that show intrinsic resistance to current antifungal compounds.
An important challenge for future IreA-targeting therapy is developing selectivity for the fungal protein. In this regard, atomic-level structural analysis of IreA would be an important next step to help guide medicinal chemistry efforts. However, even if an IreA-targeting drug were to partially inhibit human Ire1, current evidence indicates that redundancy between the three mammalian ER stress response pathways could protect human tissues from a loss of Ire1 function. For example, although a mouse Ire1 knockout is a developmental lethal, this is due to an extraembryonic defect in the placenta. A conditional knockout of Ire1 in the embryo is viable, showing only a mild phenotype of reduced insulin and immunoglobulin levels.58,59 Furthermore, Ire1-targeting strategies for the treatment of human cancer are already in the development pipeline, with successful outcomes reported in animal models.54-56 These compounds do not discriminate between normal and cancer cells at the biochemical level of target inhibition. Rather, it is the increased reliance of the tumor cell on the UPR for survival that confers selectivity. One final consideration is the inevitable emergence of resistance to pharmacologic inhibition of any antifungal target. Simultaneous targeting of the two enzymatic domains of IreA may offer one useful strategy to minimize the evolution of resistance. However, continued analysis of the HacA-dependent and HacA-independent functions of IreA is warranted to determine whether additional downstream targets could be identified to augment the effects of IreA inhibition, as well as to prevent the emergence of resistance. For example, compounds that target both IreA and ERAD (Fig. 2) could be the basis for a therapeutically useful combination to exacerbate ER stress (ERAD inhibition) and prevent subsequent adaptive responses (IreA inhibition). A similar strategy has already shown promise for targeting secretory tumor cells that rely extensively on the UPR and ERAD for survival.60
In summary, the data outlined in this review highlight the pivotal role that the UPR plays as a regulator of both virulence and antimicrobial drug susceptibility in A. fumigatus. These findings raise the exciting possibility that supplementation of conventional antifungal drug therapy with novel UPR-targeting agents could improve therapeutic outcome in aspergillosis, as well as other infections caused by eukaryotic pathogens that exploit the UPR for the support of highly developed secretory systems.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Supported by NIH grant R01AI072297 and a Cystic Fibrosis Foundation grant to D.S.A.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/26571
References
- 1.Segal BH. Aspergillosis. N Engl J Med. 2009;360:1870–84. doi: 10.1056/NEJMra0808853. [DOI] [PubMed] [Google Scholar]
- 2.Shoham S, Marr KA. Invasive fungal infections in solid organ transplant recipients. Future Microbiol. 2012;7:639–55. doi: 10.2217/fmb.12.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. The PATH (Prospective Antifungal Therapy) Alliance® registry and invasive fungal infections: update 2012. Diagn Microbiol Infect Dis. 2012;73:293–300. doi: 10.1016/j.diagmicrobio.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Hawksworth DL. Naming Aspergillus species: progress towards one name for each species. Med Mycol. 2011;49(Suppl 1):S70–6. doi: 10.3109/13693786.2010.504753. [DOI] [PubMed] [Google Scholar]
- 5.Farnell E, Rousseau K, Thornton DJ, Bowyer P, Herrick SE. Expression and secretion of Aspergillus fumigatus proteases are regulated in response to different protein substrates. Fungal Biol. 2012;116:1003–12. doi: 10.1016/j.funbio.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim A, Nicolau DP, Kuti JL. Hospital costs and outcomes among intravenous antifungal therapies for patients with invasive aspergillosis in the United States. Mycoses. 2011;54:e301–12. doi: 10.1111/j.1439-0507.2010.01903.x. [DOI] [PubMed] [Google Scholar]
- 7.Wilson LS, Reyes CM, Stolpman M, Speckman J, Allen K, Beney J. The direct cost and incidence of systemic fungal infections. Value Health. 2002;5:26–34. doi: 10.1046/j.1524-4733.2002.51108.x. [DOI] [PubMed] [Google Scholar]
- 8.van der Linden JW, Camps SM, Kampinga GA, Arends JP, Debets-Ossenkopp YJ, Haas PJ, Rijnders BJ, Kuijper EJ, van Tiel FH, Varga J, et al. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin Infect Dis. 2013;57:513–20. doi: 10.1093/cid/cit320. [DOI] [PubMed] [Google Scholar]
- 9.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–15. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 10.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–61. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 11.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–6. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 12.Su X, Schmitz G, Zhang M, Mackie RI, Cann IK. Heterologous gene expression in filamentous fungi. Adv Appl Microbiol. 2012;81:1–61. doi: 10.1016/B978-0-12-394382-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 13.McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, Loss O, Cairns T, Goldman G, Armstrong-James D, et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008;4:e1000154. doi: 10.1371/journal.ppat.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markaryan A, Morozova I, Yu H, Kolattukudy PE. Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect Immun. 1994;62:2149–57. doi: 10.1128/iai.62.6.2149-2157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moutaouakil M, Monod M, Prévost MC, Bouchara JP, Paris S, Latgé JP. Identification of the 33-kDa alkaline protease of Aspergillus fumigatus in vitro and in vivo. J Med Microbiol. 1993;39:393–9. doi: 10.1099/00222615-39-5-393. [DOI] [PubMed] [Google Scholar]
- 16.Reichard U, Büttner S, Eiffert H, Staib F, Rüchel R. Purification and characterisation of an extracellular serine proteinase from Aspergillus fumigatus and its detection in tissue. J Med Microbiol. 1990;33:243–51. doi: 10.1099/00222615-33-4-243. [DOI] [PubMed] [Google Scholar]
- 17.Ibrahim-Granet O, Dubourdeau M, Latgé JP, Ave P, Huerre M, Brakhage AA, Brock M. Methylcitrate synthase from Aspergillus fumigatus is essential for manifestation of invasive aspergillosis. Cell Microbiol. 2008;10:134–48. doi: 10.1111/j.1462-5822.2007.01025.x. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Gao M, Han X, Tao S, Zheng D, Cheng Y, Yu R, Han G, Schmidt M, Han L. Disruption of the phospholipase D gene attenuates the virulence of Aspergillus fumigatus. Infect Immun. 2012;80:429–40. doi: 10.1128/IAI.05830-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann T, Sasse C, Schedler A, Hasenberg M, Gunzer M, Krappmann S. Shaping the fungal adaptome--stress responses of Aspergillus fumigatus. Int J Med Microbiol. 2011;301:408–16. doi: 10.1016/j.ijmm.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Moore KA, Hollien J. The unfolded protein response in secretory cell function. Annu Rev Genet. 2012;46:165–83. doi: 10.1146/annurev-genet-110711-155644. [DOI] [PubMed] [Google Scholar]
- 21.Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol. 2012;28:251–77. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- 22.Kohno K. Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. J Biochem. 2010;147:27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- 23.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–7. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng X, Krishnan K, Richie DL, Aimanianda V, Hartl L, Grahl N, Powers-Fletcher MV, Zhang M, Fuller KK, Nierman WC, et al. HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus. PLoS Pathog. 2011;7:e1002330. doi: 10.1371/journal.ppat.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, Miley MD, White S, McCarthy JW, Latgé JP, Feldmesser M, et al. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 2009;5:e1000258. doi: 10.1371/journal.ppat.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rüegsegger U, Leber JH, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–14. doi: 10.1016/S0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 27.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–6. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 28.Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, Yin Z, Brown AJ, Archer DB. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet Biol. 2008;45:1235–47. doi: 10.1016/j.fgb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Saloheimo M, Valkonen M, Penttilä M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol Microbiol. 2003;47:1149–61. doi: 10.1046/j.1365-2958.2003.03363.x. [DOI] [PubMed] [Google Scholar]
- 30.Mulder HJ, Saloheimo M, Penttilä M, Madrid SM. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol Genet Genomics. 2004;271:130–40. doi: 10.1007/s00438-003-0965-5. [DOI] [PubMed] [Google Scholar]
- 31.Yanagitani K, Imagawa Y, Iwawaki T, Hosoda A, Saito M, Kimata Y, Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 32.Mulder HJ, Nikolaev I. HacA-dependent transcriptional switch releases hacA mRNA from a translational block upon endoplasmic reticulum stress. Eukaryot Cell. 2009;8:665–75. doi: 10.1128/EC.00131-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillemette T, van Peij N, Goosen T, Lanthaler K, Robson GD, van den Hondel CA, Stam H, Archer DB. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics. 2007;8:158. doi: 10.1186/1471-2164-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulder HJ, Nikolaev I, Madrid SM. HACA, the transcriptional activator of the unfolded protein response (UPR) in Aspergillus niger, binds to partly palindromic UPR elements of the consensus sequence 5′-CAN(G/A)NTGT/GCCT-3′. Fungal Genet Biol. 2006;43:560–72. doi: 10.1016/j.fgb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Jung KW, Kang HA, Bahn YS. Essential roles of the Kar2/BiP molecular chaperone downstream of the UPR pathway in Cryptococcus neoformans. PLoS One. 2013;8:e58956. doi: 10.1371/journal.pone.0058956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheon SA, Jung KW, Chen YL, Heitman J, Bahn YS, Kang HA. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog. 2011;7:e1002177. doi: 10.1371/journal.ppat.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Havel VE, Wool NK, Ayad D, Downey KM, Wilson CF, Larsen P, Djordjevic JT, Panepinto JC. Ccr4 promotes resolution of the endoplasmic reticulum stress response during host temperature adaptation in Cryptococcus neoformans. Eukaryot Cell. 2011;10:895–901. doi: 10.1128/EC.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyazaki T, Nakayama H, Nagayoshi Y, Kakeya H, Kohno S. Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata. PLoS Pathog. 2013;9:e1003160. doi: 10.1371/journal.ppat.1003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–58. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 40.Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–31. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhabhra R, Askew DS. Thermotolerance and virulence of Aspergillus fumigatus: role of the fungal nucleolus. Med Mycol. 2005;43(Suppl 1):S87–93. doi: 10.1080/13693780400029486. [DOI] [PubMed] [Google Scholar]
- 43.Richie DL, Feng X, Krishnan K, Askew DS. Secretion stress and antifungal resistance: an Achilles’ heel of Aspergillus fumigatus? Med Mycol. 2011;49(Suppl 1):S101–6. doi: 10.3109/13693786.2010.497504. [DOI] [PubMed] [Google Scholar]
- 44.Tada R, Latgé JP, Aimanianda V. Undressing the fungal cell wall/cell membrane--the antifungal drug targets. Curr Pharm Des. 2013;19:3738–47. doi: 10.2174/1381612811319200012. [DOI] [PubMed] [Google Scholar]
- 45.Sturley SL. Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim Biophys Acta. 2000;1529:155–63. doi: 10.1016/S1388-1981(00)00145-1. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg ED. The role of iron in protozoan and fungal infectious diseases. J Eukaryot Microbiol. 1999;46:231–8. doi: 10.1111/j.1550-7408.1999.tb05119.x. [DOI] [PubMed] [Google Scholar]
- 47.Schrettl M, Haas H. Iron homeostasis--Achilles’ heel of Aspergillus fumigatus? Curr Opin Microbiol. 2011;14:400–5. doi: 10.1016/j.mib.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grahl N, Shepardson KM, Chung D, Cramer RA. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot Cell. 2012;11:560–70. doi: 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA., Jr. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richie DL, Feng X, Hartl L, Aimanianda V, Krishnan K, Powers-Fletcher MV, Watson DS, Galande AK, White SM, Willett T, et al. The virulence of the opportunistic fungal pathogen Aspergillus fumigatus requires cooperation between the endoplasmic reticulum-associated degradation pathway (ERAD) and the unfolded protein response (UPR) Virulence. 2011;2:12–21. doi: 10.4161/viru.2.1.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnan K, Feng X, Powers-Fletcher MV, Bick G, Richie DL, Woollett LA, Askew DS. Effects of a defective endoplasmic reticulum-associated degradation pathway on the stress response, virulence, and antifungal drug susceptibility of the mold pathogen Aspergillus fumigatus. Eukaryot Cell. 2013;12:512–9. doi: 10.1128/EC.00319-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kraskiewicz H, FitzGerald U. InterfERing with endoplasmic reticulum stress. Trends Pharmacol Sci. 2012;33:53–63. doi: 10.1016/j.tips.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Wang L, Perera BG, Hari SB, Bhhatarai B, Backes BJ, Seeliger MA, Schürer SC, Oakes SA, Papa FR, Maly DJ. Divergent allosteric control of the IRE1α endoribonuclease using kinase inhibitors. Nat Chem Biol. 2012;8:982–9. doi: 10.1038/nchembio.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volkmann K, Lucas JL, Vuga D, Wang X, Brumm D, Stiles C, Kriebel D, Der-Sarkissian A, Krishnan K, Schweitzer C, et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286:12743–55. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F, Niwa M, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–4. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ri M, Tashiro E, Oikawa D, Shinjo S, Tokuda M, Yokouchi Y, Narita T, Masaki A, Ito A, Ding J, et al. Identification of Toyocamycin, an agent cytotoxic for multiple myeloma cells, as a potent inhibitor of ER stress-induced XBP1 mRNA splicing. Blood Cancer J. 2012;2:e79. doi: 10.1038/bcj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thibault G, Ismail N, Ng DT. The unfolded protein response supports cellular robustness as a broad-spectrum compensatory pathway. Proc Natl Acad Sci U S A. 2011;108:20597–602. doi: 10.1073/pnas.1117184109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwawaki T, Akai R, Kohno K. IRE1α disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS One. 2010;5:e13052. doi: 10.1371/journal.pone.0013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci U S A. 2009;106:16657–62. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, Hu Y, Fabre C, Minami J, Ohguchi H, et al. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–81. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]