At more than one million copies per genome, Alu elements represent the most abundant primate-specific repetitive element, constituting more than 10% of the human genome.1 These small interspersed elements (SINEs) can be transcribed by RNA polymerase III; alternatively, they can be embedded in transcripts synthesized by RNA polymerase II, where they have the potential to regulate host mRNA expression by a variety of mechanisms.1,2 Compared with other repetitive elements, Alu elements are uniquely characterized by their relatively large length, which can be up to 300 nucleotides (i.e., ~2× longer than mouse B elements) and their high (70%) degree of homology.3 These features generally allow a pair of inverted-repeat Alu elements (IRAlus) that reside within the same transcript to base-pair, forming extensive yet imperfect stretches of double-stranded RNA (dsRNA). Such structures can be bound at multiple sites by the dsRNA-binding proteins (dsRBPs) called adenosine deaminases acting on RNA (ADARs), which leave the molecular footprint of A-to-I conversions on both RNA strands.1 In fact, >90% of ADARs-mediated A-to-I editing in human cells occurs at IRAlus. When IRAlus reside within 3′-untranslated regions (3′ UTRs), they act as cis-regulatory elements that can modulate the metabolism of 3′ UTR IRAlus-containing mRNAs (3′ UTR IRAlus mRNAs) in multiple ways. In the nucleus, in addition or alternatively to binding ADARs, certain 3′ UTR IRAlus can bind the paraspeckle component p54nrb, which inhibits the nuclear export of the mRNA in which they reside, sequestering it within nuclear paraspeckles.1,4 In the cytoplasm, some 3′ UTR IRAlus have been shown to inhibit the translation of their host mRNA, enhancing its accumulation in stress granules through unidentified mechanisms.5
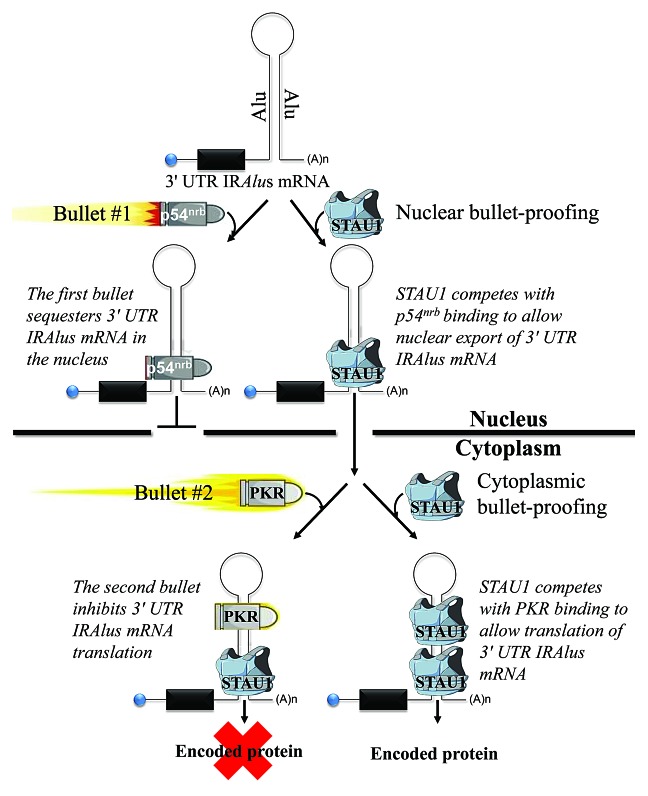
The exceptionally long dsRNA structures of IRAlus and the lack of strict sequence specificity that typifies the binding sites of those dsRBPs that have been studied spurred us to determine if competition among different dsRBPs for binding to 3′ UTR IRAlus affects the A-to-I editing, nuclear-to-cytoplasmic distribution, and/or cytoplasmic translation of 3′ UTR IRAlus mRNAs. We had previously shown that the dsRBP human Staufen1 (STAU1) binds to intermolecular Alu-element duplexes, formed between long noncoding RNAs (lncRNAs) and mRNAs, or between 2 different mRNAs, and that binding triggers STAU1-mediated mRNA decay (SMD).2,6 In the current study, we found that STAU1 also binds to intramolecular 3′ UTR IRAlus to post-transcriptionally enhance 3′ UTR IRAlus mRNA expression. Expression is enhanced via a 2-pronged mechanism (Fig. 1). This mechanism enables 3′ UTR IRAlus mRNAs to avert nuclear retention (the first “bullet”) and, subsequently, inhibition of cytoplasmic translation (the second “bullet”).7
Figure 1. dsRNA-binding protein warfare: STAU1 binds one class of mRNA 3′ UTR IRAlus, reducing the impact of 2 “bullets” and thereby enhancing 3′ UTR IRAlus gene expression. STAU1 is shown as a bulletproof vest. Bullet #1 is p54nrb, which inhibits 3′ UTR IRAlus mRNA nuclear export. Bullet #2 is PKR, which inhibits 3′ UTR IRAlus mRNA translation and, less so, global-cell translation.
We used cell fractionation, fluorescent in situ hybridization (FISH), and analyses of RNA−protein complexes to demonstrate that nuclear STAU1 binding to those 3′ UTR IRAlus studied promotes the nuclear export of 3′ UTR IRAlus mRNAs by precluding binding of the dsRBP p54nrb, thereby preventing nuclear retention.7 Prior to our study, removal of 3′ UTR IRAlus through alternative RNA 3′-end cleavage and polyadenylation was the only reported mechanism to promote the nuclear export of mRNAs that derive from 3′ UTR IRAlus genes.1,4 STAU1 did not significantly change the level of ADAR1-mediated A-to-I editing in those 3′ UTR IRAlus studied.7
We also found that STAU1 binding to those 3′ UTR IRAlus studied promotes the translation of 3′ UTR IRAlus mRNAs by precluding the binding of yet another dsRBP, protein kinase R (PKR).7 PKR is a key player in the innate immune response. PKR dimerization and subsequent autophosphorylation on dsRNAs result in the PKR-mediated phosphorylation of eukaryotic translation initiation factor 2α (eIF2α), which inhibits cellular translation.8 Notably, the ratio of STAU1-to-PKR binding to cytoplasmic IRAlus mRNAs reaches beyond those mRNAs actually bound by either or both dsRNA-binding proteins to affect global-cell translation, albeit to a lesser extent.7 This was observed upon downregulating p54nrb abundance, which elevated the cytoplasmic level of 3′ UTR IRAlus mRNAs and induced a PKR-mediated shutdown of global-cell translation.7 Consistent with this view, elevated levels of cytoplasmic 3′ UTR IRAlus mRNAs that were due to the presence of exogenous STAU1 expression did not trigger a PKR-mediated shutdown of global-cell translation.7 Thus, 3′ UTR IRAlus can function not only as cis-acting but also as trans-acting translational regulatory elements.
As evidence of physiological relevance, we demonstrated that primary human skeletal muscle (hSkMc) cells utilize developmentally regulated variations in the ratio of STAU1-to-PKR during myogenesis to modulate the expression of the microRNA-binding protein LIN28 through the 3′ UTR IRAlus of LIN28 mRNA.7
Our study has opened up several new areas for future research. For example, STAU1 binding to those 3′ UTR IRAlus studied fails to detectably trigger SMD.7 It is unclear why 3′ UTR IRAlus fail to detectably trigger SMD, or if the apparent immunity to SMD is a feature of all 3′ UTR IRAlus mRNAs. Future studies that address the protein composition of 3′ UTR IRAlus mRNPs should provide insight into what mRNP configuration is required for SMD. As another area that is ripe for research, we as well as others4,5,7 have found that not all 3′ UTR IRAlus have the same effect on the nuclear-to-cytoplasmic distribution of their host mRNAs. Some cellular 3′ UTR IRAlus mRNAs are largely nuclear, while others are largely cytoplasmic.7 Furthermore, different 3′ UTR IRAlus mRNAs can manifest distinct affinities for ADAR1, STAU1, or PKR, resulting in variable post-transcriptional outcomes.7 Future studies that aim to define the sequences and structural motifs that are favored by different dsRBPs, as well as how the relative abundance of dsRBPs influences their dsRNA binding, will surely help our understanding of the complicated post-transcriptional regulatory web that controls 3′ UTR IRAlus-containing genes.
Acknowledgments
This work was supported by NIH GM074593.
Elbarbary RA, et al. Genes Dev. 2013;27:1495–510. doi: 10.1101/gad.220962.113.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27512
References
- 1.Berger A, et al. Prog Mol Subcell Biol. 2011;51:119–46. doi: 10.1007/978-3-642-16502-3_6. [DOI] [PubMed] [Google Scholar]
- 2.Gong C, et al. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen SK, et al. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen LL, et al. Cell Cycle. 2008;7:3294–301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 5.Capshew CR, et al. Nucleic Acids Res. 2012;40:8637–45. doi: 10.1093/nar/gks590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong C, Tang Y, Maquat LE. mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat Struct Mol Biol. 2013;20:1214–20. doi: 10.1038/nsmb.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbarbary RA, et al. Genes Dev. 2013;27:1495–510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonenberg N, et al. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]